Abstract
Background
Preterm birth is the second leading cause of death in children under the age of five years worldwide, but the etiology of many cases remains enigmatic. The dogma that the fetus resides in a sterile environment is being challenged by recent findings and the question has arisen whether microbes that colonize the fetus may be related to preterm birth. It has been posited that meconium reflects the in-utero microbial environment. In this study, correlations between fetal intestinal bacteria from meconium and gestational age were examined in order to suggest underlying mechanisms that may contribute to preterm birth.
Methods
Meconium from 52 infants ranging in gestational age from 23 to 41 weeks was collected, the DNA extracted, and 16S rRNA analysis performed. Resulting taxa of microbes were correlated to clinical variables and also compared to previous studies of amniotic fluid and other human microbiome niches.
Findings
Increased detection of bacterial 16S rRNA in meconium of infants of <33 weeks gestational age was observed. Approximately 61·1% of reads sequenced were classified to genera that have been reported in amniotic fluid. Gestational age had the largest influence on microbial community structure (R = 0·161; p = 0·029), while mode of delivery (C-section versus vaginal delivery) had an effect as well (R = 0·100; p = 0·044). Enterobacter, Enterococcus, Lactobacillus, Photorhabdus, and Tannerella, were negatively correlated with gestational age and have been reported to incite inflammatory responses, suggesting a causative role in premature birth.
Interpretation
This provides the first evidence to support the hypothesis that the fetal intestinal microbiome derived from swallowed amniotic fluid may be involved in the inflammatory response that leads to premature birth.
Introduction
Preterm birth is the major cause of perinatal morbidity and mortality and is a leading cause of death in children under the age of 5 years old worldwide [1]. The dogma for exclusive postnatal acquisition of microbes is shifting with increasing evidence that the infants' initial inoculum can be provided by maternal transmission before birth [2]. The mechanisms leading to preterm labor are not well understood; an integral role for microbiota in premature birth has been suggested [3], [4]. Microbiological evidence from placental tissue and amniotic fluid samples from preterm deliveries suggests that infection may contribute to approximately 25% of preterm births. Bacterial colonization rates are as high as 79% for birth at 23 weeks of gestation but considerably lower, at 11% at 31 to 34 weeks [4], [5].
Microbes often colonize amniotic fluid from mothers who deliver prematurely (regardless of ruptured or intact membrane), and the quantity of microbial DNA and markers of inflammation correlate inversely with gestational age [6], [7]. Various mechanisms of amniotic colonization have been described including the ascension and translocation of vaginal microbiota [8], [9], as well as via the bloodstream from non-reproductive tissues such as the oral gingiva [10]. The most widely considered paradigm is that once these microorganisms are inside the uterus, they result in the release of proinflammatory cytokines, prostaglandin, and matrix metalloproteases, which lead to cervical ripening, membrane rupture, uterine contractions and preterm birth [11]. It is unclear whether the resulting immune response derives maternally or from the fetus, but studies of blood spots obtained several days postnatally from infants born at different gestational ages suggest a fetal origin of the labor-triggering responses [12].
The site of origin of the fetal inflammatory response is unknown. However, given the higher sensitivity of fetal intestinal tissue to inflammatory stimuli than the sensitivity of mature intestine [13], inflammation-related induction of labor could very likely be derived from the fetal intestine. Fetuses swallow large quantities of amniotic fluid during the late second and third trimesters of pregnancy [14]. This suggests that in utero ingestion of microbes present in the amniotic fluid leads to the bacterial colonization of the fetal gut and incites an immune response resulting in the onset of labor. In order to investigate the proximal components of this mechanism, it is essential to evaluate the microbiome of the in utero fetal intestinal environment.
Several studies have shown that meconium is not sterile [15]–[17] and contact with microbes is associated with changes in the expression profile of innate immune genes of the fetal intestine [18]. If, indeed, microbes in amniotic fluid have contact with the intestine of the fetus and cause an inflammatory response, then detectable remnants, such as microbial DNA and markers of inflammation would be expected to be present in the meconium of these infants. By analyzing the meconium microbiome from infants of various gestational ages, microbial signatures that correspond with gestational age could indicate organisms that are involved in premature labor. Amniotic fluid is very difficult to obtain routinely at different gestational ages whereas meconium is readily accessible, and may be a reasonable alternative for evaluation of the in-utero microbial environment.
This study aims to determine if there are bacteria from the fetal intestine that correlate with prematurity and also to gain a better understanding of microbial establishment in the human intestine.
Methods
Study Patients
Written informed consent was obtained from the infants' parents and investigations were conducted according to the principles expressed in the Declaration of Helsinki. The study including consent procedure was approved by the UF/Shands Institutional Review Boards. IRB# is 386–2008. Meconium was collected from 52 infants at three University of Florida hospitals. The gestational age of subjects ranged between 23 and 41 weeks. Samples were collected from diapers with sterile spatulas, placed in sterile tubes, and frozen at −80°C within 12 hours. ‘Hands on care’ of infants was scheduled every 3 hours, so meconium samples were collected up to 3 hours from the time stool was passed. Additionally, all samples were collected within 48 hours of birth. Metavariables were recorded and analyzed for these samples (Table S1 in File S1).
DNA Extraction & Amplification
DNA was extracted from approximately 200 mg of meconium sample using FastSpin DNA extraction kit for soil (MP Biomedicals, Santa Ana, CA). DNA was quantified using Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE). Amplification of the V4 region of 16S rRNA gene was performed using 515F and 806R primers20 flanked with Ion Torrent sequencing barcodes and adaptors (Ion Torrent Systems Inc., Guilford, CT). PCR reactions were prepared with 25 µl of GoTaq colorless Master Mix (Promega, Madison, WI), 10 µM of primers, 20 ng of sample DNA adjusted to 50 µl total volume with sterile, nuclease-free water. Samples were then purified using PCR Purification Kit (Qiagen, Hilden, Germany), quantified fluorometrically with QuBit dsDNA High Sensitivity (Invitrogen, Life technologies Inc., Carlsbad, CA), and fragment size was verified on an agarose gel. Neither DNA nor amplification was detected in blank control samples, and any meconium samples in which amplification was not detected were determined to be sterile.
Sequencing and Data Processing
Multiplexed sequencing reactions were performed on the Ion Torrent PGM platform using four Ion 318™ Chips and the Sequencing 300 bp kit protocol (Ion Torrent Systems Inc., Guilford, CT). An average of 3.9 million reads per chip were generated (average read length of 214 bp). Reads longer than 300 bases were separated by barcode and trimmed using sickle (https://github.com/najoshi/sickle.git); 72.3% of reads were retained after quality filtering. An average of 162,994 (±76,173) reads per sample remained with an average read length of 250 bp (±66 bp). Sequenced reads for non-sterile samples (N = 35) have been deposited on MG-RAST (ID 5358).
Reads were assigned to Operational Taxonomic Units (OTUs) by aligning to the RDP-TaxCollector database with a minimum identity of 95% (USEARCH); 82.6% (±6.6%) of reads were classified in any given sample. OTU contingency tables were created using the taxonomic description at the Domain, Phylum, Class, Order, Family, Genus, and Species levels as denoted by the taxonomic description in the TaxCollector database. Tables were created from USEARCH output and filtered such that each OTU had a minimum of 100 reads in any meconium sample.
Perhaps the largest potential confounder in this work is the possibility for contamination. Sterility of sample collection and processing is supported by the result that 32·7% of all samples had no detectable 16S rRNA and, hence, were assumed to be sterile. The microbes found in meconium samples related closely to those detected by others in amniotic fluid (see below). Furthermore, previous microbial studies of meconium showed that surface community structure of meconium was not distinguishable from the interior suggesting contact with skin is not a primary contributor to the meconium microbiome and hence not an appreciable confounder [19].
Statistical Analysis
Identification of associations between the presence of 16S rRNA or preterm labor (PTL) and categorical metavariables were assessed using Fisher's exact test (Table S2 in File S1). Confounding variables associated with gestational age were identified (Table S3 in File S1) and further evaluated. Non-metric multidimensional scaling (NMDS) using Bray-Curtis distance and diversity analyses (Shannon-Weaver index and Chao 1 richness) were performed on rarefied count data (sample size of 47,000 reads) using the phyloseq [20] package in R. Analysis of similarity (ANOSIM) was used to assess which variables best accounted for microbiome variability. When necessary, gestational age was treated as a categorical variable and grouped by <33 weeks or >33 weeks based on findings that the most serious illnesses associated with preterm birth usually occur in infants born <33 weeks. Spearman correlation analyses and two-sided Mann-Whitney tests were calculated from proportion of total reads using R; all correlations and tests with p<0·05 were considered significant. All analyses and graphics were generated in R version 2.15.0 (R project, Statistical Software): http;//www.R-project. org, and ggplot2 package, respectively.
Results
Gestational age and bacterial colonization of meconium
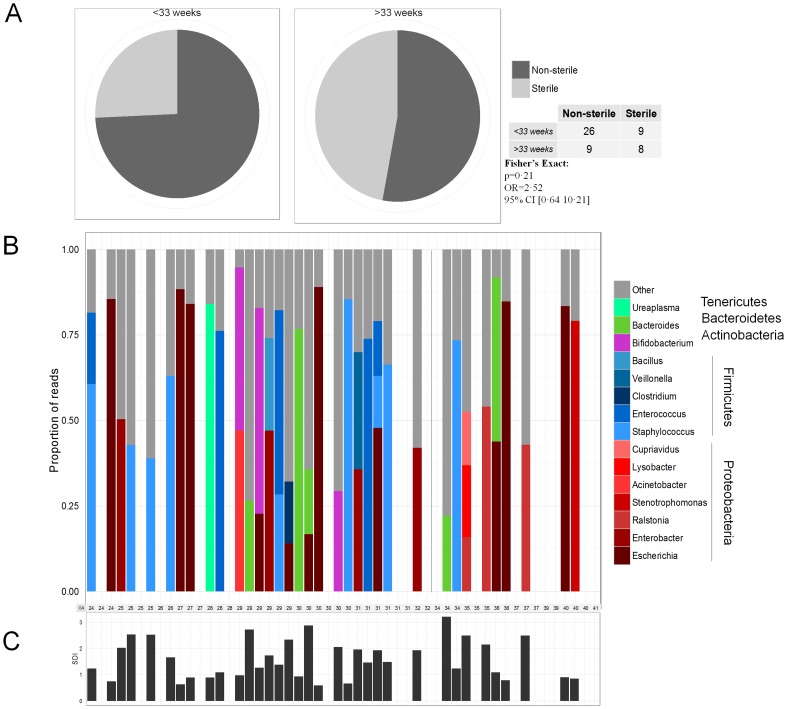
Of 52 infants sampled, 16S rRNA was amplified from meconium DNA of 35 subjects (67·3%). A greater percentage of meconium samples were non-sterile in subjects of <33 weeks gestational age (74·3%; N = 35) compared to subjects >33 weeks gestational age (52·9%; N = 17) (Figure 1A), but this was not significant (p = 0·21; Fisher's Exact test). No significant associations were detected between the presence of 16S rRNA or PTL with administration of maternal antibiotics, infant antibiotics, gestational age, mode of delivery, anticipated mode of feeding (breast versus formula), or gender (Table S2 in File S1).
Figure 1. Gestational age and bacterial colonization of meconium.
(A) A larger percentage of meconium samples from infants <33 weeks gestational age tend to be colonized (74·3%; N = 35) compared to infants of >33 weeks gestational age (52·9%; N = 17). (B) The bacterial composition of meconium is dominated by few genera; on average the most abundant genera in any given sample comprised 57·3±22·5% of reads; <33 and >33 weeks is displayed by the black, dashed line. (C)Dominant genera contribute to low diversity, measured by Shannon index; this is indicative of a founding population. Furthermore, gestational age was not correlated with Shannon diversity index (Spearman: rho = 0·03, p = 0·85).
Dominant genera characterize meconium microbial community – low diversity regardless of gestational age
The microbiome of all non-sterile samples in this study were dominated by a particular genus (Figure 1B), regardless of gestational age. Any one dominant genus accounted for 18%–89% of the microbiome of a given subject. Spikes of Firmicutes-classified genera (shades of blue in Figure 1B) occurred more frequently prior to 33 weeks gestational age (9 of 26 in non-sterile subjects <33 weeks and 1 of 9 in >33 weeks gestational age; Figure 3C, Table 1). No association between several variables and the dominance of particular genera or diversity was detected with the exception of mode of delivery (Table S4 in File S1). The microbiome of infants delivered by cesarean section was more diverse by both Shannon and Chao measurements (p = 0·047 and p = 0·032, respectively; Table S4 in File S1)
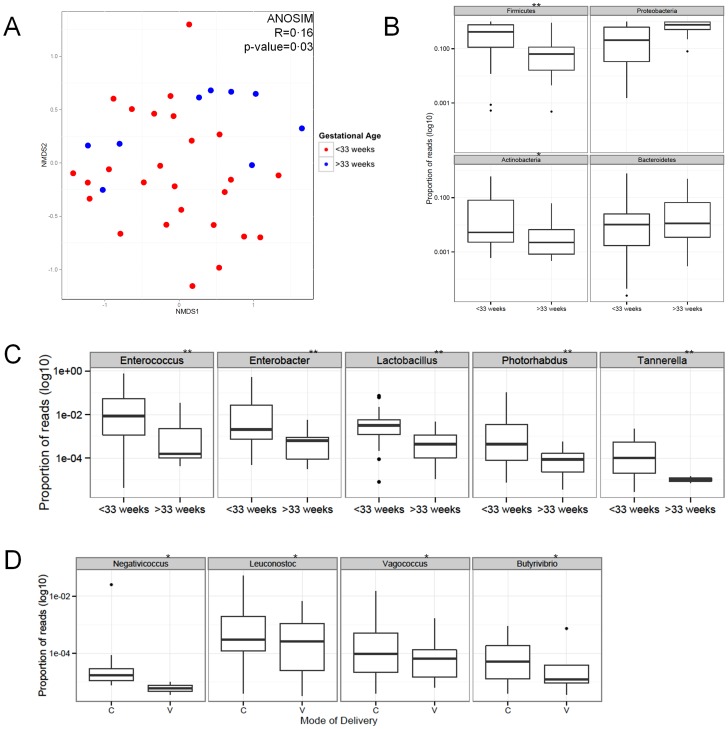
Figure 3. Inflammatory marker S100A12 was correlated with gestational age.
(A) Non-metric multidimensional scaling ordination plot depicting the relatedness of the bacterial communities from all meconium samples; communities from >33 week infants (blue) clustered more closely than those from <33 week infants (red). Analysis of similarity (ANOSIM) revealed that gestational age (<33 and >33 weeks) had the largest effect on meconium microbial structure (R = 0·16; p-value = 0·03). (B) Of the four predominant phyla, the relative abundance of Firmicutes and Actinobacteria was correlated with low gestational age (**p<0·01 & *p<0·05, respectively). (C) Genera negatively correlated with gestational age (**p<0·01) are presented. (D) Genera associated with mode of delivery (*p<0·05) were observed, though these differences are not as pronounced as the genera associated with gestational age.
Table 1. Phyla, family, and genera taxonomy significantly correlated with gestational age (*p<0·05, **p<0·001; Spearman correlation) are reported.
| Phylum | Family | Genus | |||||||||
| OTU | <33 weeks | ≥33 weeks | p-value | OTU | <33 weeks | ≥33 weeks | p-value | OTU | <33 weeks | ≥33 weeks | p-value |
| Firmicutes** | 44·5 (±17·6) | 17·1 (±14·5) | 0·006 | Bacillaceae* | 2·50 (±2·5) | 0·76 (±0·95) | 0·031 | Bacillus* | 2·48 (±2·78) | 0·70 (±1·87) | 0·032 |
| Staphylococcaceae* | 13·30 (±11·9) | 4·71 (±8·89) | 0·036 | Staphylococcus* | 13·28 (±13·21) | 4·71 (±17·8) | 0·036 | ||||
| Enterococcaceae** | 8·77 (±9·8) | 0·43 (±0·53) | 0·007 | Enterococcus** | 8·67 (±11·04) | 0·41 (±1·04) | 0·007 | ||||
| Vagococcus* | 0·08 (±0·15) | 0·01 (±0·04) | 0·018 | ||||||||
| Lactobacillaceae** | 0·83 (±0·81) | 0·07 (±0·08) | 0·003 | Lactobacillus** | 0·82 (±0·91) | 0·07 (±0·15) | 0·003 | ||||
| Leuconostocaceae* | 0·42 (±0·52) | 0·02 (±0·02) | 0·018 | Leuconostoc* | 0·42 (±0·60) | 0·02 (±0·04) | 0·016 | ||||
| Clostridiaceae* | 1·56 (±1·8) | 0·35 (±0·50) | 0·030 | Clostridium* | 1·54 (±1·98) | 0·33 (±0·96) | 0·033 | ||||
| Peptostreptococcaceae* | 1·15 (±2·0) | 0·03 (±0·05) | 0·033 | ||||||||
| Veillonellaceae* | 2·46 (±3·1) | 0·39 (±0·59) | 0·030 | Veillonella* | 2·27 (±3·44) | 0·30 (±0·99) | 0·020 | ||||
| Negativicoccus* | 0·001 (±0·25) | 0·0001 (±0·00) | 0·011 | ||||||||
| Erysipelotrichaceae* | 1·26 (±2·5) | 0·05 (±0·05) | 0·049 | ||||||||
| Actinobacteria* | 7·6 (±7·6) | 1·1 (±1·0) | 0·012 | Bifidobacteriaceae* | 5·52 (±6·7) | 0·35 (±05·6) | 0·013 | Bifidobacterium* | 5·47 (±7·65) | 0·35 (±1·11) | 0·016 |
| Proteobacteria | 35·4 (±17·9) | 65·6 (±16·5) | 0·523 | Enterobacteriaceae* | 26·32 (±16·9) | 15·20 (±15·9) | 0·024 | Enterobacter** | 6·35 (±7·99) | 0·06 (±0·14) | 0·002 |
| Citrobacter* | 0·19 (±0·23) | 0·03 (±0·06) | 0·014 | ||||||||
| Erwinia* | 0·33 (±0·36) | 0·03 (±0·07) | 0·013 | ||||||||
| Klebsiella* | 0·60 (±0·64) | 0·05 (±0·12) | 0·022 | ||||||||
| Raoultella* | 0·30 (±0·30) | 0·21 (±0·44) | 0·035 | ||||||||
| Pantoea* | 0·36 (±0·38) | 0·03 (±0·05) | 0·011 | ||||||||
| Photorhabdus** | 0·98 (±1·34) | 0·01 (±0·01) | 0·002 | ||||||||
| Oxalibacteraceae | 0·01 (±0·01) | 1·08 (±1·23) | 0·436 | Oxalicibacterium* | 0·001 (±0·00) | 0·56 (±1·38) | 0·030 | ||||
| Bacteroidetes | 6·0 (±8·0) | 9·6 (±8·4) | 0·329 | Porphyromonadaceae | 0·20 (±0·17) | 0·09 (±0·12) | 0·087 | Tannerella** | 0·02 (±0·02) | 0·00 (±0·0) | 0·004 |
The mean percent (%) relative abundance and standard deviation for each taxonomic name for meconium samples from infants <33 weeks and >33 weeks gestational age are indicated.
Meconium microbiome is most suggestive of amniotic fluid origin
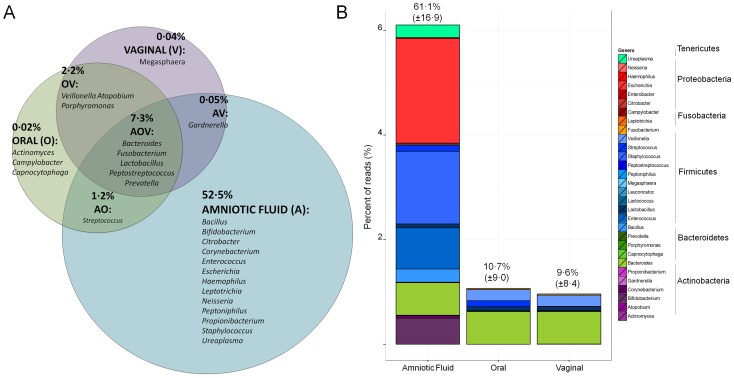
To consider the potential sources contributing to the microbiota found in infant meconium, the meconium microbiome data here were compared to data from various published microbiome studies (Figure 2). Thirty genera found present in studies of amniotic fluid [6], [7], colostrums [21], vaginal canal [22], oral cavity [22], and meconium of full term infants [15] were also identified in meconium samples of this study (Figure S1 in File S1). Genera common between amniotic fluid and the meconium samples of this study accounted for a greater relative abundance (61·1%±16·9) than other sources included in the comparison. Genera identified in the oral and vaginal cavities of pregnant women accounted for 10·7% (±9·0) and 9·6% (±8·4), respectively.
Figure 2. Meconium microbiome is most suggestive of amniotic fluid origin.
(A) The average percent relative abundance in meconium samples of this study for genera reported in amniotic fluid, and the oral and vaginal cavities of pregnant women6,7,27 are displayed by the Venn diagram which distinguishes unique and shared maternal environments of genera. (B) The potential total mean contribution and standard deviation of any particular maternal locale (amniotic fluid5,6, oral21, or vaginal21), and the phyletic distribution of contributing genera is shown in the stacked bar plot. The color assignment is as follows: Actinobacteria = purple; Bacteroidetes = green; Firmicutes = blue; Fusobacteria = orange; Proteobacteria = red; Tenericutes = aquamarine.
Confounding variables associated with gestational age
Birth weight, mode of delivery, and infant antibiotic exposures were associated with low gestational age (p<0·05; Table S3 in File S1). No other recorded metadata collected were significantly associated with gestational age.
Microbiome differences correlated with gestational age
Non-metric multidimensional scaling and analysis of similarity (ANOSIM) using Bray-Curtis distance revealed that microbial communities of <33 weeks gestational age group were different from that of >33 weeks gestational age group (R = 0·16; p = 0·03) (Figure 3A). Additionally, ANOSIM was used to evaluate the effects of collected metavariables on the dispersion of samples in the ordination plot and to further assess the influence of potential confounding factors on the microbiome (Table S5 in File S1). Gestational age had the largest influence on microbial community structure (R = 0·161; p = 0·029), while mode of delivery (MOD) had a lesser but significant effect as well (R = 0·100; p = 0·044).
Proteobacteria and Firmicutes comprised the majority of average reads classified at the phylum level, 35·4% (±17·9%) and 44·5% (±17·6%), respectively in <33 weeks and 65·6% (±16·5%) and 17·1% (±14·5%), respectively in >33 weeks (Table 1). The relative abundances of Firmicutes and Actinobacteria were negatively correlated with gestational age (p-values of 0·006 and 0·02, respectively; Figure 3B, Table 1).
All significant correlations detected were negatively correlated with gestational age with the exception of Oxalicibacterium (Table 1). Taxonomic families within the phylum Firmicutes correlated with gestational age included: Bacillaceae, Staphylococcaceae, Enterococcaceae, Lactobacillaceae, Leuconostocaceae, Clostridiaceae, Peptostreptococcaceae, Veillonellaceae, and Erysipelotrichaceae. At the genus level (Fig. 3C), the strongest correlations were attributed to Enterococcus (p-value 0·007) and Lactobacillus (p-value 0·003) and comprised an average of 8·67% (±11·04%) versus 0·41% (±1·04%) for Enterococcus and 0·82% (±0·91%) versus 0·07% (±0·15%) for Lactobacillus in <33 weeks and >33 weeks, respectively. Among Actinobacteria, Bifidobacterium was significantly correlated (p-value 0·016), comprising 5·47% (±7·65%) compared to 0·35% (±1·11%) of total reads for <33 and >33 weeks, respectively. Proteobacteria genera that were significantly correlated with gestational age were primarily Enterobacteriaceae and those with the strongest correlations included Enterobacter (p-value 0·002) and Photorhabdus (p-value 0·002) with an average relative abundance of 6·35% (±7·99%) versus 0·06% (±1·14%) for Enterobacter and 0·98% (±1·34%) versus 0·01% (±0·01%) for Photorhabdus in <33 and >33 weeks, respectively. Found in even lower abundance (mean 0.02% in <33 weeks, and not detected at all in >33 weeks samples), Tannerella was identified in these samples and correlated with low gestational age (p-value 0.004).
Microbiome differences associated with mode of delivery
Based on ANOSIM results (reported above and Figure 3A) and diversity analysis showing that the meconium microbiome is related to with mode of delivery and accounts for differences in diversity (Table S4 in File S1), genera associated with mode of delivery were identified using Mann-Whitney test (Table S6 in File S1). These genera include Negativococcus (p = 0·029), Leuconostoc (p = 0·036), Vagococcus (p = 0·036), and Butyrivibrio (p = 0·044) and were found to have a greater relative abundance in cesarean-delivered infants (Figure 3D). All of these genera are Firmicutes and had a low relative abundance.
Discussion
Recent evidence supporting a paradigm shift away from the dogma that the womb is sterile and that the human infant is thus born sterile is likely to have major applications in human health and disease [2]. A major implication of these findings relates to preterm birth. Accordingly, we hypothesized that swallowed infected amniotic fluid may incite an intestine-derived inflammatory response that induces preterm labor and that the microbes that relate to this response can be non-invasively evaluated in meconium. Here several findings are presented that support this hypothesis.
Differences in microbial colonization are associated with gestational age. The data in this study show that regardless of gestational age, the meconium microbiome is usually characterized by a high relative abundance of a specific genus (Figure 1B), and the microbial communities differ between <33 and >33 weeks. The former supports the notion that initial intestinal colonization is characterized by a founding species which becomes increasingly diverse with age as exposure to several factors affecting the microbiome increases [23]. The difference in community structure can be explained by specific genera that are negatively correlated with gestational age (Table 1). These observations suggest that composition, rather than colonization alone, is a determinant in gestational age.
By comparing the combined relative abundances of genera found in environments possibly contributing to an in utero environment (i.e. vaginal, oral) as well as the amniotic fluid, it was found that a larger relative abundance of genera from preterm baby meconium were shared with amniotic fluid samples than from other reported microbial niches (Figure 2). This similarity suggests that meconium is more indicative of amniotic fluid bacterial communities than other niches. Also of interest is that genera attributed to oral and vaginal environments accounted for approximately the same relative abundance in meconium samples (∼10%), which indicates that both potential origins of intrauterine infection and subsequent fetal gastrointestinal colonization can occur and are potentially doing so simultaneously. In addition previous studies have suggested that an altered vaginal flora is implicated in preterm birth, but targeted antibiotic therapy against vaginosis related bacteria has not been beneficial in preventing preterm labor [24]. Another finding that supports the in-utero origin of the meconium microbiome is that the composition of microbes in feces changes markedly from meconium after the first postnatal week [23].
Genera negatively correlated with gestational age (p-value <0·01) and representing >5% in infants <33 weeks of the total bacterial population are Enterococcus and Enterobacter. Both of these genera have been associated with inciting inflammatory responses in prematurely born infants [25]. Two other genera, Lactobacillus and Photorhabdus, represented a lower proportion of the bacterial community (∼0·8–1·0%in <33 weeks) but had a greater relative abundance in <33 week samples. It has been shown that lactic acid bacteria induce interleukin-6 (IL-6) production and non-specific immunity [26] In addition, the cytokine IL-6 is higher in amniotic fluid of preterm versus full term births [27]. Although the ability of the increased Lactobacillus population observed in this particular study on inciting IL-6 production has not been investigated, taken together, these findings provide a reasonable candidate mechanism. The increased presence of Lactobacillus in the meconium of preterm infants suggests that it was also present in amniotic fluid and thus may be involved in triggering events leading to preterm labor. Additionally, Tannerella has been implicated as a marker for periodontal infection and preterm birth [28] and the detection of Tannerella in samples <33 weeks but not in samples >33 weeks further suggests a role of fetal colonization by oral microbes in preterm birth.
Mode of delivery did have a minor but observable effect on the microbiome (indicated by the ANOSIM results and an increased diversity in cesarean delivered infants); however, this effect is not as great as gestational age. Although, early gestational age was associated with cesarean delivery (p = 0·009; Table S3 in File S1), when mode of delivery was compared to the relative abundance of different genera, the resulting significant genera were different from those correlated with gestational age. This indicates that mode of delivery provides a unique community that is not influenced by gestational age. Others have reported on microbiome changes due to mode of delivery [29], though the differences observed due to mode of delivery are reported from infants that are several days old, not necessarily meconium. While the gut microbiome is undoubtedly, heavily influenced by mode of delivery, we argue that such differences are observable but are not the primary source in meconium and suggest that meconium provide an easily accessible indicator of the in utero microbial environment which is likely to provide insight to the etiology of preterm birth.
In summary, this study generates a novel hypothesis in that it associates microbes found in meconium with prematurity. The known immunoreactivity of the fetal intestine along with the presence of these microbes suggests a fetal intestinal origin for some cases of spontaneous preterm labor. Further identification and characterization of these organisms and their interaction with the host should lead to novel therapies for prevention of many cases of prematurity.
Supporting Information
Supporting tables and figures.
(PDF)
Funding Statement
The study was funded by United States National Institutes of Health grant # HD059153. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Goldenberg RL, Culhane JF, Iams JD, Romero R (2008) Epidemiology and causes of preterm birth. Lancet 371: 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Funkhouser LJ, Bordenstein SR (2013) Mom knows best: the universality of maternal microbial transmission. PLoS Biol 11: e1001631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Muglia LJ, Katz M (2010) The enigma of spontaneous preterm birth. N Engl J Med 362: 529–535. [DOI] [PubMed] [Google Scholar]
- 4.Onderdonk AB, Hecht JL, McElrath TF, Delaney ML, Allred EN, et al. (2008) Colonization of second-trimester placenta parenchyma. Am J Obstet Gynecol 199: : 52 e51–52 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Watts DH, Krohn MA, Hillier SL, Eschenbach DA (1992) The association of occult amniotic fluid infection with gestational age and neonatal outcome among women in preterm labor. Obstet Gynecol 79: 351–357. [DOI] [PubMed] [Google Scholar]
- 6. DiGiulio DB (2012) Diversity of microbes in amniotic fluid. Semin Fetal Neonatal Med 17: 2–11. [DOI] [PubMed] [Google Scholar]
- 7. DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, et al. (2008) Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS ONE 3: 33056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goldenberg RL, Culhane JF, Iams JD, Romero R (2008) Epidemiology and causes of preterm birth. Lancet 371: 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salafia CM, Vogel CA, Vintzileos AM, Bantham KF, Pezzullo J, et al. (1991) Placental pathologic findings in preterm birth. Am J Obstet Gynecol 165: 934–938. [DOI] [PubMed] [Google Scholar]
- 10. Goepfert AR, Jeffcoat MK, Andrews WW, Faye-Petersen O, Cliver SP, et al. (2004) Periodontal disease and upper genital tract inflammation in early spontaneous preterm birth. Obstet Gynecol 104: 777–783. [DOI] [PubMed] [Google Scholar]
- 11. Romero R, Espinoza J, Gonçalves LF, Kusanovic JP, Friel L, et al. (2007) The role of inflammation and infection in preterm birth. Semin Reprod Med 25: 21–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Skogstrand K, Hougaard DM, Schendel DE, Bent NP, Svaerke C, et al. (2008) Association of preterm birth with sustained postnatal inflammatory response. Obstet Gynecol 111: 1118–1128. [DOI] [PubMed] [Google Scholar]
- 13. Nanthakumar NN, Fusunyan RD, Sanderson I, Walker WA (2000) Inflammation in the developing human intestine: A possible pathophysiologic contribution to necrotizing enterocolitis. Proc Natl Acad Sci U S A 97: 6043–6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gilbert WM, Brace RA (1993) Amniotic fluid volume and normal flows to and from the amniotic cavity. Semin Perinatol 17: 150–157. [PubMed] [Google Scholar]
- 15. Jiménez E, Marín ML, Martín R, Odriozola JM, Olivares M, et al. (2008) Is meconium from healthy newborns actually sterile? Res Microbiol 159: 187–193. [DOI] [PubMed] [Google Scholar]
- 16. Madan JC, Salari RC, Saxena D, Davidson L, O'Toole GA, et al. (2012) Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch Dis Child Fetal Neonatal Ed 97: F456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mshvildadze M, Neu J, Schuster J, Theriaque D, Li N, et al. (2010) Intestinal microbial ecology in premature infants assessed with non-culture-based techniques. J Pediatr 156: 20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rautava S, Luoto R, Salminen S, Isolauri E (2012) Microbial contact during pregnancy, intestinal colonization and human disease. Nat Rev Gastroenterol Hepatol 9: 565–576. [DOI] [PubMed] [Google Scholar]
- 19. Gosalbes MJ, Llop S, Valles Y, Moya A, Ballester F, et al. (2013) Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin Exp Allergy 43: 198–211. [DOI] [PubMed] [Google Scholar]
- 20. McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8: e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, et al. (2012) The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr 96: 544–551. [DOI] [PubMed] [Google Scholar]
- 22. Srinivasan U, Misra D, Marazita ML, Foxman B (2009) Vaginal and oral microbes, host genotype and preterm birth. Med Hypotheses 73: 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moles L, Gomez M, Heilig H, Bustos G, Fuentes S, et al. (2013) Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One 8: e66986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brocklehurst P, Gordon A, Heatley E, Milan SJ (2013) Antibiotics for treating bacterial vaginosis in pregnancy. Cochrane Database Syst Rev 1: CD000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stewart CJ, Marrs EC, Magorrian S, Nelson A, Lanyon C, et al. (2012) The preterm gut microbiota: changes associated with necrotizing enterocolitis and infection. Acta Paediatr 101: 1121–1127. [DOI] [PubMed] [Google Scholar]
- 26. Miettinen M, Vuopio-Varkila J, Varkila K (1996) Production of human tumor necrosis factor alpha, interleukin-6, and interleukin-10 is induced by lactic acid bacteria. Infect Immun 64: 5403–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Menon R, Camargo MC, Thorsen P, Lombardi SJ, Fortunato SJ (2008) Amniotic fluid interleukin-6 increase is an indicator of spontaneous preterm birth in white but not black Americans. Am J Obstet Gynecol 198: e1–7. [DOI] [PubMed] [Google Scholar]
- 28. Jarjoura K, Devine PC, Perez-Delboy A, Herrera-Abreu M, D'Alton M, et al. (2005) Markers of periodontal infection and preterm birth. Am J Obstet Gynecol 192: 513–519. [DOI] [PubMed] [Google Scholar]
- 29. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, et al. (2010) Delivery mode shaoes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. PNAS 107(26): 11971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting tables and figures.
(PDF)