Abstract
We present a two-layer hidden Markov model to detect the structure of haplotypes for unrelated individuals. This allows us to model two scales of linkage disequilibrium (one within a group of haplotypes and one between groups), thereby taking advantage of rich haplotype information to infer local ancestry of admixed individuals. Our method outperforms competing state-of-the-art methods, particularly for regions of small ancestral track lengths. Applying our method to Mexican samples in HapMap3, we found two regions on chromosomes 6 and 8 that show significant departure of local ancestry from the genome-wide average. A software package implementing the methods described in this article is freely available at http://bcm.edu/cnrc/mcmcmc.
Keywords: admixture, local ancestry, haplotype, linkage disequilibrium (LD), two-layer clustering
HAPLOTYPE variation is central to statistical and population genetics. Studies have revealed considerable sharing (Conrad et al. 2006) and significant variation (Liu et al. 2004) of haplotypes among populations. Since markers are linked on a haplotype, the makeup of haplotypes in a population produces unique patterns of linkage disequilibrium (LD): the dependence between markers’ marginal allele frequencies. Therefore, modeling LD is key to understanding haplotype variations. Many statistical models exist to model LD, but a model that can detect the structure of haplotypes is missing.
The most elegant model for LD is the coalescent with recombination (Kingman 1982; Hudson 1983) or the ancestral recombination graph (ARG). However, despite successful efforts on small-scale data sets (Wang and Rannala 2009), ARG remains notoriously hard to compute. Considerable efforts have been made to approximate ARG to allow computation on a large scale (Stephens and Donnelly 2000; Fearnhead and Donnelly 2002; Li and Stephens 2003; Scheet and Stephens 2006; Paul and Song 2010). Among them, the most successful is the PAC model of Li and Stephens (2003), which models a new haplotype as an imperfect mosaic of observed haplotypes to produce a conditional likelihood; the joint likelihood of all haplotypes is then approximated by the product of those conditionals. Using diffusion approximation, Paul and Song (2010) derived a similar likelihood that they called the conditional sampling distribution. A somewhat related approach is the clustering model (Scheet and Stephens 2006), which coalesces and condenses the observed haplotypes into a small number of (ancestral) haplotypes and models the observed haplotypes as imperfect mosaics of those condensed haplotypes.
These models assume haplotypes are sampled from a single source population and become ineffective when haplotypes are admixed. Admixed haplotypes have two scales of LD: the admixture LD between alleles in different source populations that typically spans a few to tens of centimorgans (Smith and O’Brien 2005) and the LD between alleles within each source population that typically spans a few tenths of a centimorgan. The HAPMIX model (Price et al. 2009) is among the first to model LD of admixed individuals, extending the PAC model to two source populations. This model is effective for inferring local ancestry of two-way admixtures (e.g., African-Americans), but it is not yet applicable to three-way admixtures such as Latinos. (In principle, however, HAPMIX should work with three-way admixtures.) Two recent examples of progress include LAMP-LD (Baran et al. 2012) and MULTIMIX (Churchhouse and Marchini 2013), both of which achieve similar performance to that of HAPMIX in inferring local ancestry of two-way admixtures and can handle three-way admixtures. However, HAPMIX and LAMP-LD both require phased haplotypes from source populations, and LAMP-LD and MULTIMIX both assume ancestries are fixed within a window of markers and switch only between windows. These methods often perform well for recent admixtures but underperform for distant admixtures, which implies limited ability to detect local ancestries of short track lengths. Distantly admixed individuals, such as Uyghurs whose admixture occurred >100 generations ago, are valuable for disease association (Xu and Jin 2008) and human genetic landscape studies (Li et al. 2009). Moreover, even for recent admixtures, there exists a nontrivial proportion of short ancestry segments. If we model the ancestral track length as exponentially distributed with mean 10 cM (equivalent to admixture that occurred 10 generations ago), then we expect to observe >9.5% of ancestry segments whose track lengths are <1 cM.
A different perspective of two scales of LD in admixture—one within a source population and one between different source populations—is structure on local haplotypes. Taking the two-way admixture as an example, haplotypes from two source populations may be condensed and structured into two groups, and a new haplotype is assigned probabilistically to a group based on its similarity with the (condensed) haplotypes in both groups. In fact, the local haplotype structure is a ubiquitous phenomenon in genetic data, and the admixture is just a more apparent example. Even among individuals sampled from a single source population, a set of local haplotypes might be enriched in one subset of individuals and a different set of local haplotypes enriched in another. For example, individuals of European descent may be separated according to whether they have different two-digit human leukocyte antigen (HLA)-A allele classes. Compared to the genetic difference between two alleles sampled from distinct ancestries, the genetic difference between two-digit HLA allele classes is more subtle. However, from the perspective of statistical modeling, these two scenarios are the same—both require detecting the structure of local haplotypes based on their similarities. None of the current methods is designed to handle this more delicate scenario.
In this study, we present a novel two-layer hidden Markov model (HMM) designed to learn the structure of local haplotypes. The new model uses two layers of latent clusters. In each layer, clusters are labeled to represent ancestry alleles, and multiple clusters of the same label over adjacent markers represent an ancestral haplotype. In a nonrecombined region, the upper layer aims to capture structure near the root of a coalescent tree, whereas the lower layer aims to capture haplotype variation near the tip. Recombination is approximated by cluster switching within each layer. The lower-layer clusters are fuzzy mosaics of the upper-layer clusters, and haplotypes in the observed data are fuzzy mosaics of the lower-layer clusters. The fuzziness results from mutations and uncertainty of inheritance; the mosaics are results of historic recombinations. Existing cluster-based models use single-layer clusters. For example, fastPHASE (Scheet and Stephens 2006) and Beagle (Browning and Browning 2007) use, equivalently, the lower-layer clusters to model ancestral haplotypes; and STRUCTURE (Pritchard et al. 2000) equivalently uses the upper-layer clusters to model ancestry populations. Although seemingly incremental, the two-layer model has an attractive feature that is not available in a single-layer model—detecting structure of haplotypes. The upper-layer clusters represent different groups (populations) and the lower-layer clusters represent group-specific haplotypes (Figure 1). Thus we may infer local ancestries by condensing and grouping local haplotypes into different groups and assigning a local haplotype probabilistically into groups.
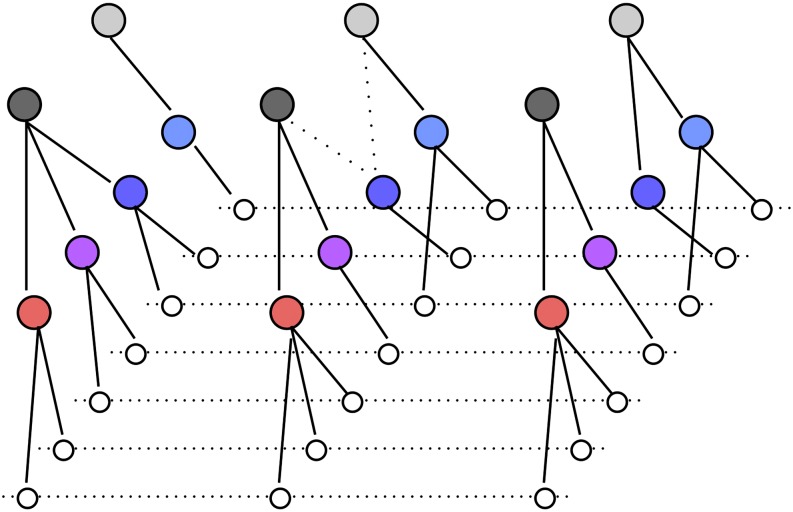
Figure 1.
Graphic representation of the two-layer model. White circles connected by dotted lines are seven haplotypes over three markers (for simplicity, haplotypes are assumed to be observed instead of diplotypes). Colored circles are lower-layer latent clusters representing ancestral haplotypes; gray circles are upper-layer latent clusters that enforce structure on haplotypes. Circles that share the same color (or gray level) have the same labels over three markers. Two dotted lines between latent clusters indicate that the lower cluster is shared between two upper clusters. Refer to Figure 2 for a numerical example.
Local ancestries of admixed individuals provide important information for disease association mapping (Smith and O’Brien 2005) and demographic history (Johnson et al. 2011). It is an important subject that has attracted much recent attention (Patterson et al. 2004; Tang et al. 2006; Sundquist et al. 2008; Price et al. 2009; Baran et al. 2012; Churchhouse and Marchini 2013). One way to infer local ancestry is to use ancestry informative markers (AIMs)—markers whose allele frequencies have large differences among populations (Smith et al. 2004). Local ancestry inference using AIMs has a low resolution because AIMs are relatively scarce. On the other hand, haplotypes provide richer information that is complementary to the AIMs. Taking an extreme example, if one population has 50% A-T and 50% T-A haplotypes whereas another population has 50% A-A and 50% T-T haplotypes, there would be no difference in the marginal allele frequencies between the two populations, while the two-marker haplotypes are very informative. The two-layer model uses local haplotypes in source populations to define population features for each small genomic region, based on which admixed haplotypes are assigned probabilistically to different populations. These genomic regions are not prespecified; instead, they are learned from data. Compared to methods that group markers in windows and allow only ancestral switches between windows (Baran et al. 2012; Churchhouse and Marchini 2013), our method performs better because prespecified windows may conflict with actual ancestral switches.
Methods and Models
For ease of presentation, we assume haplotypes are observed. By integrating out phase, our model applies directly to diploid individuals (Appendix). We assume readers have some basic knowledge of the HMM or are familiar with classic LD models such as PAC (Li and Stephens 2003) and fastPHASE (Scheet and Stephens 2006).
The two-layer HMM
We assume the numbers of upper- and lower-layer clusters are S and K, respectively, and denote N the number of haplotypes and M the number of markers. For each individual i, let be the latent state of the upper and lower clusters at marker m. Here takes values in 1 , … , S and and takes values in 1 , … , K; a lower cluster k associates with a parameter θmk to represent ancestral allele frequency. We may drop the superscript when referring to an arbitrary individual.
The main HMM:
The emission of an observed haplotype marker of individual i at marker m from a lower-layer cluster is modeled as
| (1) |
where ξ is the collection of parameters associated with the HMM (details will follow), and θmk is the allele frequency associated with lower-cluster k at marker m. The complete data likelihood has the form
| (2) |
The Markov transition of the latent states tries to capture the following intuitions: a haplotype copies mosaically from (ancestral) haplotypes in one source population and then may switch to another source population and copy mosaically from its haplotypes. The upper-layer switch probabilities j determine how frequently switches occur between different source populations and the lower-layer switch probabilities r determine how frequently switches occur between ancestral haplotypes within each source population. Thus, the model accommodates two scales of LD observed in admixed individuals. We have at the first marker
| (3) |
and the Markov transitions
| (4) |
where is the probability that individual i jumps to upper-cluster s given the jump occurs, and βmsk is the probability an individual jumps to lower-cluster k given the jump occurs and the upper-cluster being s. Note α(i) is an individual specific S vector to denote the admixture proportion, and β is an M × S × K tensor shared by all individuals. The I(a = b) is an indicator function.
We made three assumptions on the transition matrix of the hidden states. First, given the switch occurs between marker m − 1 and m, is independent of and is independent of This assumption, used by previous models (Li and Stephens 2003; Scheet and Stephens 2006), reduces the number of parameters and simplifies computation. Second, given the switch occurs, takes values according to α(i) and only according to α(i); on the other hand, given the switch occurs and takes values according to β, which is a function of m, but not i. This accommodates the fact that LD patterns are heterogeneous across markers. Third, we assume that if the upper layer switches, then the lower layer must switch; however, the lower layer can switch even if the upper layer does not switch. This encourages the upper-layer-specific LD patterns.
In the main HMM, the upper-layer latent state Xm contributes only to transitions of latent states (through β) and does not contribute to emitting an observed genotype or θ estimates (likelihood does not involve allele frequencies associated with Xm). This works well when K, the number of lower-layer clusters, is not too large, but less well for a large K. To stabilize θ estimates for a large K, we use an ancillary HMM to model the upper-layer clusters emitting θ.
The ancillary HMM:
Given estimates of θ, we assume the ancillary HMM is independent of the main HMM. The ancillary HMM is a single-layer HMM where the K ancestral haplotypes (the θ matrix) are assumed as observed (recall K is the number of lower clusters). The latent state of the kth ancestral haplotype at marker m, denoted by represents which population (upper cluster) the ancestral marker descends from; takes values in 1 , … , S (recall S is the number of upper clusters), and it associates with an allele frequency parameter η. Here we use W instead of X to denote the upper-layer cluster because X belongs to the observed genotypes and W belongs to the inferred ancestral haplotypes. We model emission of θmk from as
| (5) |
where Beta(x, a, b) denotes a Beta density with parameters a, b. This emission is adapted from the Balding–Nichols model (Balding and Nichols 1995). The original model is designed to model population divergence, and hence F is specified through Fst values (a measurement of allele frequency divergence) between different populations. In our context, we use it as a “random effect model” to stabilize θ estimates. For computational convenience, we set F = 1 (Appendix). Treating θmk as observed, the complete data likelihood has the form
| (6) |
The transition of the latent states is modeled as
| (7) |
where the jump probabilities ρ are unrelated to the jump probabilities of the main HMM.
Model fitting
In the main HMM, the collection of parameters ξ contains allele frequencies θ (an M × K matrix) and β (an M × S × K matrix), α (an N × S matrix), and j and r (both M vectors). In the ancillary HMM, the set of parameters contains η (an M × S matrix), a (a K × S matrix), and ρ (an M vector). We briefly discuss how to estimate these parameters using expectation maximization (EM), focusing on the main HMM. For details, please refer to the Appendix.
For an arbitrary individual i, we write the forward probability φ(m, s, k) = p(h1:m, Xm = s, Ym = k|ξ) and the backward probability ψ(m, s, k) = p(hm+1:M|Xm = s, Ym = k|ξ); both probabilities can be computed analytically. The posterior probabilities of the latent states at each marker are p(Xm = s, Ym = k|h, ξ) ∝ φ(m, s, k)ψ(m, s, k). We then compute quantities to update the model parameters, which are ancestral allele frequencies θ and the Markov transition parameters α, β, j, and r. For θ, we follow the classical approach to derive updates by optimizing the expected complete data (observed and latent) log-likelihood, conditioning on the previous estimates of ξ. For Markov transition parameters, we identify and compute sufficient statistics (the expected number of switches to each cluster pair); the updates are functions of those sufficient statistics. All updates can be computed analytically or numerically and require no sampling. Upon convergence of EM, we have ξ*.
Constraint on cluster switches:
Estimating switch probabilities j and r is more difficult for two reasons. First, a large jm (or rm) estimate in a previous iteration often results in a large estimate in the current iteration, and, as a consequence, the choice of initial values of j and r influences heavily the point at which they converge. Second, j and r are not completely identifiable. If both αs and βmsk are close to 1, then a large probability in either jm or rm results in a similar likelihood. We overcome these difficulties by putting constraints on j and r; the constraints are derived from the coalescent theory.
Define where is the lower-layer cluster switch rate, where Ne is the effective population size and cm is the genetic distance between markers (m − 1) and m. We approximate cm by assuming 1 cM spans 1 Mb. Recall from the coalescent theory (cf. Ewens 2004) that is the mean coalescent time for k lineages; we then have Assuming N ≫ K, then δl ≈ 1/K. This leads to a natural choice for constraint on (and hence r). For example, if Ne = 10,000, , and K = 10, then we may apply the constraint In practice, we directly estimate rm and compute rescale to match the constraint, and reestimate rm.
Define where One might be tempted to follow a similar coalescent argument to specify δu, but unlike δl, which is robust to recent demographic history because it pertains to an “ideal” ancestral population, δu is heavily influenced by demographic histories (for example, admixture generations) and the coalescent argument becomes ineffective. As a workaround, we constrain j through the admixture generation γ. In practice, we first estimate jm to compute and then use to rescale to reestimate jm. Defining ancestry track length as then λ and γ follow a simple relationship γλ = 100.
Inference and computation:
We are interested in the upper cluster dosage for each individual i, defined as which is the posterior estimate of local ancestry at marker m; its genome-wide average is the admixture proportion. To investigate structure of haplotypes, we are also interested in computing conditional dosage for lower clusters, defined as
After trial and error, we arrived at the following ways to improve model-fitting performance:
Because the dimension of ξ is high and standard EM procedures tend to converge to a local mode instead of the global mode, it is useful to average inferences over multiple EM runs.
It is helpful to initialize parameters with values that preserve symmetry; e.g., θmk ≈ 0.5, and βmsk ≈ 1/K for all values of m, s, and k, respectively. The initial values can be simulated from symmetric Beta or Dirichlet distributions with large rates.
The training data from source populations can be either phased or unphased. The difference is small when phasing is accurate and the computation with phased data is faster (linear vs. quadratic in numbers of upper-layer clusters S and lower-layer clusters K). However, when phasing is less accurate, for example, pure statistical phasing without help of transmission, using unphased data is preferred. By default, we assume phased training data sets are used, except in the analysis of Mexican samples or where noted.
The common practice used in imputation (cf. Guan and Stephens 2008), for which one first fits the model to the training data from source populations and then runs the forward–backward algorithm once on the admixed individuals, tends to produce spurious ancestry switches in spikes; performing additional EM steps using both source samples and admixed samples (joint model fitting) reduces spurious ancestry switches. We recommend joint model fitting.
Metrics for performance:
We used two metrics to measure performance of local ancestry inference: the mean deviation and Pearson’s correlation. An individual’s local ancestry can be expressed by an M × S matrix, where M is the number of markers and S is the number of upper-layer clusters (or source populations). The column stacking of the matrix produces a vector x. The mean deviation is defined as where xm is the actual value, is an inferred value, |⋅| denotes absolute value, and L = MS. Pearson’s correlation is computed using x and
Choice of parameters:
The companion software is easy to use—users need to specify only three parameters: the number of upper clusters S, the number of lower clusters K, and the admixture generations γ. For local ancestry inference, S is clear a priori. For example, S = 2 for African-Americans and S = 3 for Latinos. We used K = 5S in our study, but the method is robust to a wide range of K values. We demonstrate this through examples. For a set of simulated two-way admixed individuals, we used S = 2 and K = 5, 10, or 20 to fit the model. K = 10 and K = 20 outperform K = 5 for both deviation and correlation, especially for correlation; the difference between K = 10 and K = 20 is small (Supporting Information, Table S1).
As a rule, we recommend averaging results over multiple choices of γ. In general, γ = 10 for African-American samples, γ = 20 for Latinos, and γ = 100 for Uyghurs appear to be good choices. In our simulation studies, the local ancestry inference is robust to γ up to a multiple of 2; however, γ affects the smoothness of the local ancestry inference. We simulated two-way admixed individuals with admixture generation γ = 100 and fitted the model using γ = 50, 100, and 200, respectively. For all individuals, small values of γ produce smoother local ancestry estimates than those obtained from large values of γ. But, for all three choices of γ, the main ancestry blocks were inferred well. Taking one individual as an example, the deviation estimates for three choices of γ were 0.067, 0.062, and 0.092, with γ = 100 performing the best and γ = 200 performing the worst, presumably because the metric is sensitive to smoothness. Three Pearson’s correlations are 0.934, 0.947, and 0.932 for three choices of γ. As a comparison, the deviation estimate for HAPMIX is 0.067 and the correlation is 0.939. Although quantitatively similar to our method, HAPMIX does miss a major ancestry block in the middle (Figure S1).
Simulating admixed individuals:
The procedure we used to simulate three-way admixed individuals is similar to what is used in HAPMIX (Price et al. 2009) for two-way admixtures. For a given admixture generation γ, we compute the average ancestral track length λ = 100/γ and then t = 1000λ (a region of 1 Mb contains ∼1000 HapMap SNPs). We randomly choose three haplotypes, hc, hy, and ha, from Utah residents with Northern and Western European ancestry (CEU), Yoruba in Ibadan, Nigeria (YRI), and Han Chinese in Beijing, China, CHB and Japanese in Tokyo, Japan (CHB+JPT) populations, respectively, and copy from the three haplotypes to form a new admixed haplotype by repeating the following three steps: (1) let s be the current position on a genome and generate a number w according to an exponential distribution with mean t; (2) copy SNPs (s, s + w] from ha with probability δ1, from hy with probability δ2, and from ha with probability δ3 = 1 − δ1 − δ2; and (3) increase s by w; finish if s exceeds the total number of SNPs. Two admixed haplotypes are paired randomly to form a diploid individual. The markers are then thinned to match the Illumina 650K SNP chip. The two-way admixture can be simulated accordingly. We chose (0.8, 0.2) as the target two-way admixture proportions and (0.6, 0.2, 0.2) for three-way admixture proportions. Note that the simulated admixture proportions vary due to a finite number of SNPs.
Summary of symbols and notations:
For the convenience of the reader, we summarize the symbols used in the Methods and Models and Appendix sections in Table 1.
Table 1. Symbols and their brief definitions.
| Catagory | Symbols | Values | Definition |
|---|---|---|---|
| Constant | S | Integer | No. upper clusters |
| K | Integer | No. lower clusters | |
| N | Integer | No. individuals | |
| M | Integer | No. markers | |
| Main HMM | (1, … , S) | Individual i’s upper cluster at marker m | |
| (1, … , K) | Individual i’s lower cluster at marker m | ||
| θmk | [0, 1] | Lower cluster allele frequency | |
| [0, 1] | |||
| βmsk | [0, 1] | ||
| jm | [0, 1] | Probability that Xm switches labels | |
| rm | [0, 1] | Probability that Ym switches labels | |
| Ancillary HMM | (1, … , S) | Haplotype k’s upper cluster at marker m | |
| ηmk | [0, 1] | Upper cluster allele frequency | |
| [0, 1] | |||
| ρm | [0, 1] | Probability that Wm switches labels | |
| Derived parameter | γ | Integer | Admixture generations |
| λ | Real | Ancestry track length | |
| ξ, ξ* | (θ, α, β, j, r, η, a, ρ) | Collection of all parameters | |
| φ(i)(m, s, k) | Real | Forward probability of individual i | |
| ψ(i)(m, s, k) | Real | Backward probability of individual i | |
| Data | 0, 1 or missing | Haplotype individual i at marker m | |
| 0, 1, 2 or missing | Diplotype individual i at marker m |
When a superscript is omitted, it stands for an arbitrary individual; when a subscript is omitted or substituted with a dot, it stands for a collection of parameters of that coordinate. For example, denotes genotype at marker m of individual i; g(i) denotes genotypes at all markers of individual i; and g denotes genotypes at all markers of an arbitrary individual. In addition, we may use to denote a subset of genotypes from marker m to marker n of individual i.
Results
Structure of haplotypes
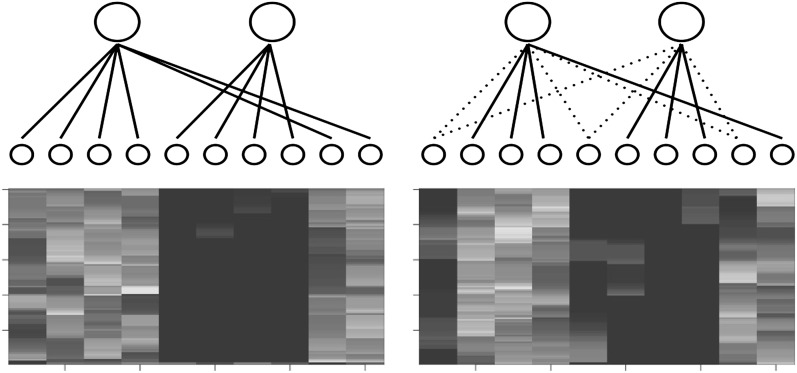
The two-layer model can detect the structure of (ancestral) haplotypes. To illustrate this, we took chromosome 2 of unrelated CEU and YRI individuals (120 haplotypes each) from HapMap2 (International Hapmap Consortium 2007) and fitted the two-layer model with S = 2, K = 10, and γ = 100, ignoring their population labels. Then, we computed the conditional dosage (conditioning on Xs = 1), which, we recall, is defined as The conditional dosages for two typical regions (100 SNPs each) are plotted in Figure 2. In one region, the lower clusters are split clearly (but not evenly) between two upper-layer clusters; in the other, the lower-layer clusters are split but less clearly with some lower clusters shared between two upper clusters. This example demonstrates that the two-layer model can indeed detect the structure of (ancestral) haplotypes. Moreover, Figure 2 illustrates that some local haplotypes are population specific whereas others are shared between populations. This local haplotype sharing is an intrinsic feature of genetic data (Conrad et al. 2006), and the two-layer model can learn this feature, which is of particular importance in local ancestry inference.
Figure 2.
Structure of haplotypes. Each row denotes a SNP, and each column denotes a lower-layer haplotype in our model. We chose two typical regions, each containing 100 SNPs. The plot shows the lower-cluster dosage conditional on the left upper cluster (conditional dosage). Brighter pixels indicate larger dosages. A solid edge connecting to the left upper cluster indicates the average (over 100 SNPs in the region) conditional dosage is >80% of total dosages; a solid edge connecting to the right upper cluster indicates the conditional dosage is <20% of total dosages. A dotted line indicates edge uncertainty.
Figure 2 underpins the most important difference between our model and the HAPMIX model. The HAPMIX model assumes to have contemporary—not ancestral—haplotypes as training data from each source population; this is equivalent to having fixed and exclusive edges between an upper-layer cluster and lower-layer clusters in our model. In our two-layer model, however, the edges are learned from data and are not predetermined; an edge can emerge and disappear along a chromosome and a lower-layer cluster can have multiple edges connecting to upper-layer clusters, which naturally captures local haplotype sharing. As a comparison, local haplotype sharing is not a natural part of the HAPMIX (Price et al. 2009) model, and a miscopy parameter is introduced and (somewhat) arbitrarily specified to adapt to the local haplotype sharing feature of the data.
Local ancestry inference
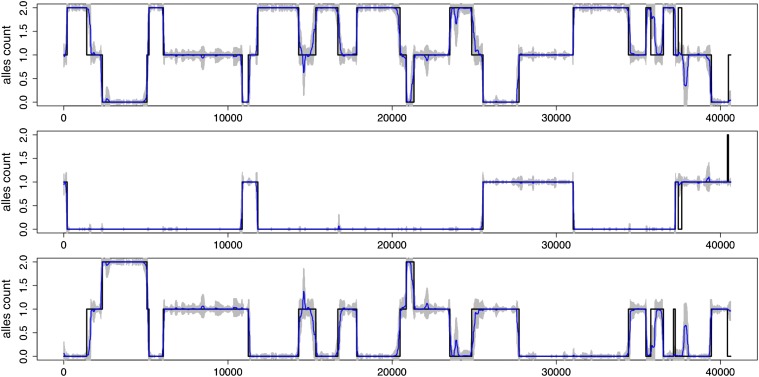
We first demonstrate that our method achieves exceptional accuracy in local ancestry inference. We simulated a three-way admixed individual, using the procedure described in the Methods and Models section with γ = 20 (equivalently, λ = 5 cM), and then fitted the two-layer model (S = 3, K = 15) using this individual and individuals from source populations, excluding haplotypes used to simulate the admixed individual. (We used 100 haplotypes from CEU, 100 from YRI, and 160 from East Asian of HapMap2 as source haplotypes.) Figure 3 compares the actual and inferred local ancestries: the local ancestry of a three-way admixed individual was inferred with exceptional accuracy. The estimated ancestral allele dosages often have large uncertainties at markers where the estimates differ from the true values. This suggests that, when combining results over multiple EM runs, the estimates may be weighted by their uncertainty, e.g., inverse of variance. Note that, for a diploid individual, our method can compute the probabilistic assignment to all possible pairs of ancestries at each marker, allowing us to quantify the mean and variance of the estimated ancestry dosages. The admixture proportions were also accurately inferred (Figure S2).
Figure 3.
Inference of local ancestry. The plot shows the results of a typical EM run for local ancestry inference of a three-way admixed individual. Each panel shows ancestry allele dosages (y-axis), one for each source population, along the chromosome (x-axis). Black lines in each panel are the true values, and blue lines are estimated mean dosages. Gray bars on top of blue lines reflect ±2 SD of the estimated mean dosages. At each marker, y-values on lines of the same color sum to 2.
Comparison with HAPMIX and LAMP-LD:
Next, we compared our method with two state-of-the-art methods used in local ancestry inference: HAPMIX (for two-way admixture) and LAMP-LD (for three-way admixture). We used two metrics in our comparison—mean deviation and Pearson’s correlation between the inferred and actual local ancestries for each simulated admixed individual (see Methods and Models section for their definitions).
For comparison with HAPMIX, we simulated three sets (10 individuals in each set) of two-way admixed individuals with γ = 10, 20, and 100 (corresponding to the ancestry track lengths of λ = 10, 5, and 1 cM, respectively). The difficulty in inferring local ancestry increases as the admixture generation increases. The results of our method were obtained with S = 2 and K = 10 and averaged over 10 independent EM runs. The results of HAPMIX were obtained using its default parameters. Both methods used 100 haplotypes from CEU and 100 haplotypes from YRI as source haplotypes; the haplotypes used to simulate admixed individuals are excluded from the source haplotypes. Table 2 summarizes the results. For easier problems (λ = 10 and 5 cM or, equivalently, γ = 10 and 20), when both methods perform well, HAPMIX performs slightly but not significantly better (two-sample t-test, P = 0.52 and 0.63 for deviation, and P = 0.20 and 0.09 for correlation), whereas for harder problems (λ = 1 cM or, equivalently, γ = 100), our method outperforms HAPMIX (P = 5 × 10−4 for deviation and P = 2 × 10−5 for correlation). Our method has some practical advantages over HAPMIX:
Table 2. Comparison with HAPMIX for two-way admixture.
| Metrics | 1 cM | 5 cM | 10 cM | Methods |
|---|---|---|---|---|
| Deviation | 0.104 ± 0.013 | 0.034 ± 0.015 | 0.019 ± 0.001 | Two-layer |
| 0.126 ± 0.011 | 0.030 ± 0.013 | 0.017 ± 0.001 | HAPMIX | |
| Correlation | 0.891 ± 0.018 | 0.963 ± 0.020 | 0.971 ± 0.012 | Two-layer |
| 0.844 ± 0.019 | 0.973 ± 0.016 | 0.980 ± 0.010 | HAPMIX |
We used two metrics: deviation (the smaller the better) and correlation (the larger the better). We simulated 10 admixed individuals under three different average ancestral track lengths (in centimorgans). Each cell includes the mean ± SD. See main text for more details.
It cleanly handles missing data, whereas HAPMIX does not allow missing data.
It does not require a recombination map as an input, whereas HAPMIX requires a highly accurate recombination map. In fact, our method can be used to infer the recombination rate, a potential application we might document elsewhere.
It can directly work with diploid data, whereas HAPMIX requires haplotypes from source populations. When the phasing of individuals from source populations is imperfect (e.g., statistical phasing without the help of transmission), our method has an advantage.
We compared our method with LAMP-LD for three-way admixed individuals. Similar to the comparison with HAPMIX, we simulated three sets (10 individuals in each set) of three-way admixed individuals with γ = 10, 20, and 100, which produced the mean ancestral track lengths of 10, 5, and 1 cM, respectively. The results of our method were obtained with S = 3 and K =15 and averaged over 10 independent EM runs. The results of LAMP-LD were obtained with default parameters. Both methods used 100 haplotypes from CEU, 100 haplotypes from YRI, and 160 haplotypes from East Asian (CHB+JPT) as source haplotypes; the haplotypes used to simulated admixed individuals are excluded from the source haplotypes. Table 3 summarizes our results.
Table 3. Comparison with LAPM-LD for three-way admixture.
| Metrics | 1 cM | 5 cM | 10 cM | Methods |
|---|---|---|---|---|
| Deviation | 0.155 ± 0.010 | 0.043 ± 0.009 | 0.020 ± 0.006 | Two-layer |
| 0.192 ± 0.024 | 0.046 ± 0.014 | 0.022 ± 0.012 | LAMP-LD | |
| Correlation | 0.859 ± 0.020 | 0.961 ± 0.013 | 0.981 ± 0.005 | Two-layer |
| 0.721 ± 0.035 | 0.934 ± 0.021 | 0.966 ± 0.016 | LAMP-LD |
We used two metrics: deviation (the smaller the better) and correlation (the larger the better). We simulated 10 admixed individuals under three different average ancestral track lengths (in centimorgans). Each cell includes the mean ± SD. See main text for more details.
Similar to the comparison with HAPMIX, for more difficult problems (λ = 1 cM or, equivalently, γ = 100), our method outperforms LAMP-LD (deviation P = 6 × 10−4 and correlation P = 2 × 10−8). For easier problems (λ = 10 and 5 cM or, equivalently, γ = 10 and 20), both methods perform similarly if measured by deviation (P = 0.69 or 0.67). There is a marked difference in performance if measured by Pearson’s correlation—our method outperforms LAMP-LD (P = 0.01 for γ = 10 and P = 3 × 10−3 for γ = 20). A closer look revealed that LAMP-LD tends to make more mistakes on small regions of a few hundred SNPs (Figure S3). We suspect that this has to do with grouping markers into windows, even though the recommended window size [50−100 SNPs (Baran et al. 2012)] is smaller than the size of often misidentified regions. In addition, LAMP-LD appears to be very certain everywhere, which can be misleading.
Computation speed:
We compared the speed of our method with that of HAPMIX and LAMP-LD. For each method, we used the same parameters as those that produced the results presented in this article. The run time was obtained from a desktop computer with an Intel Xeon CPU X5690 of 3.47 GHz; all programs used a single core. For two-way admixture we compared with HAPMIX. With two sets of source haplotypes of 100 each and 10 simulated diploid individuals of 7,616 SNPs, HAPMIX took 201 sec with its default parameters, while our method took 118 sec with S = 2 and K = 10 for a single EM run of 30 steps. For three-way admixture we compared with LAMP-LD. With three sets of source haplotypes of 100, 100, and 160 and 10 simulated diploid individuals of 6,983 SNPs, LAMP-LD took 218 sec with its default parameters, while our method took 538 sec with S = 3 and K = 15 for a single EM run of 30 steps.
Local ancestry of Mexican samples
We applied our method to infer the local ancestries of Mexican samples in both HapMap3 (International Hapmap Consortium 2010) and the 1000 genomes (1000G) projects (1000 Genomes Project Consortium 2010). In these analyses, we used only markers that are present in all source populations, and those that are absent in one source population were removed from the study.
HapMap3 samples:
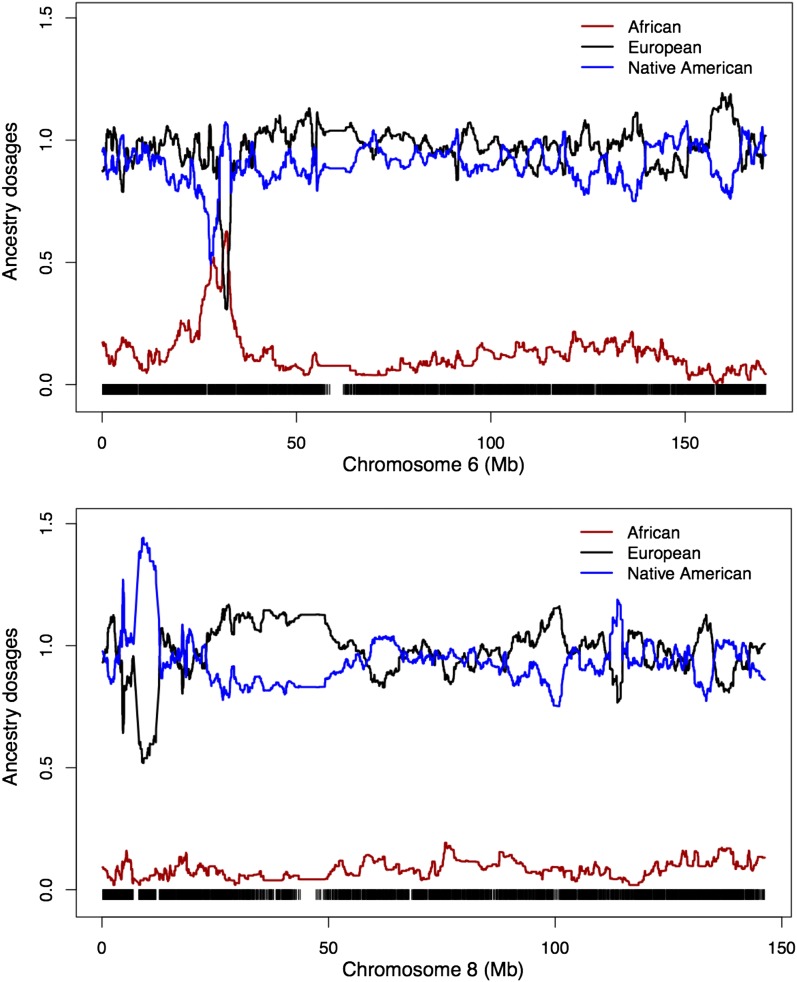
We used 112 diplotypes from CEU and 147 diplotypes from YRI in HapMap3 and 35 diplotypes from Maya and Pima in the Human Genetic Diversity Panel (HGDP) (Li et al. 2008) as three source populations (denoted as SP1) to infer the local ancestry of 58 Mexican samples from HapMap3 (all diplotypes). We fitted the model with S = 3, K = 15, and γ = 10, 20, or 50 on each chromosome separately. The mean ancestry proportions for CEU, YRI, and Native Americans are 0.495, 0.048, and 0.457, respectively, consistent with those reported by others (Johnson et al. 2011; Churchhouse and Marchini 2013). In examining local ancestral allele dosages, we found two regions that had significant departures from the genome-wide averages (Figure 4). Perhaps not very surprisingly, one is within the MHC region on chromosome 6, and the other is located on chromosome 8p23.1, a region known to harbor a large inversion. The region with elevated African ancestry on chromosome 6 contains two peaks that are located at 27.99−28.78 Mb and 30.93−32.44 Mb, respectively, both of which have African allele dosages >0.5. Assuming binomial sampling and approximating sample mean with normal distribution, we obtained a P-value <10−30 for African ancestry to reach above 0.50 allele dosages. Similarly for the region on chromosome 8 we computed a P-value <10−8 for Native American ancestry to reach above 1.44 (a P-value ≈ 2 × 10−7 for European ancestry to reach below 0.52).
Figure 4.
Regions whose local ancestries depart from the genome-wide averages. The y-axis is the average ancestral allele dosages over 58 Mexican samples. Black segments at the bottom indicate SNPs, whose coordinates are from NCBI Build 36.
1000G samples:
We also analyzed Mexican samples in the 1000G. Using identity by state, we identified 29 (of 66 total) samples that overlap with HapMap3 Mexican samples. For SNPs that are typed in both projects, there is a high genotype concordance for all 29 samples (average Hamming distance <0.002). We inferred the local ancestries of these 66 samples, using 234 CEU and 230 YRI haplotypes in 1000G and 35 diplotypes of Maya and Pima in HGDP as three source populations (denoted as SP2). We found the following:
Not surprisingly, the two regions on chromosomes 6 and 8 also show significant departure from the genome-wide averages in these samples.
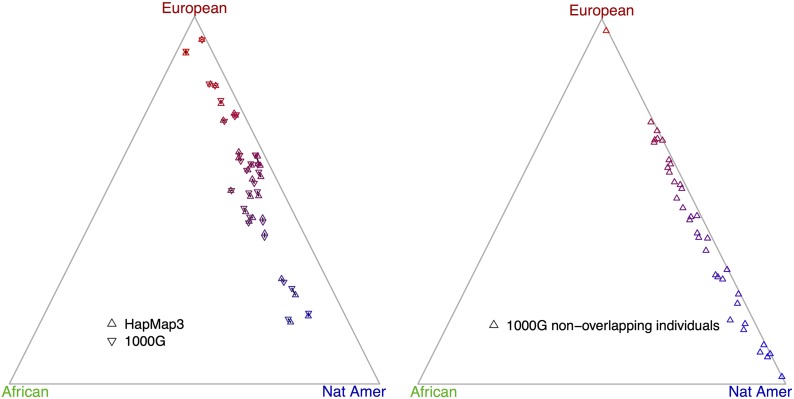
Among 29 overlapping individuals, the inferred admixture proportions have a high concordance between two choices of source populations SP1 and SP2 (Figure 5). Because we used unphased CEU and YRI in HapMap3 as source populations (SP1) for HapMap3 Mexican samples and used phased CEU and YRI in 1000G as source populations (SP2) for 1000G Mexican samples, this high concordance suggests, indirectly, that the phasing of CEU and YRI in 1000G is reliable.
The 37 nonoverlapping individuals in 1000G have an average smaller European ancestry proportion of 41.9% compared to 56.6% of those 29 overlapping individuals (Figure 5), and this difference is not likely caused by random sampling (permutation test P < 0.004).
Figure 5.
Admixture proportions of 1000G Mexican samples (chromosome 2). The left plot shows the concordance of 29 overlapping individuals genotyped in HapMap3 and 1000G. Two points that belong to the same individual are connected by a short segment. The right plot shows the remaining 37 Mexican samples in 1000G. Each individual has inferred admixture proportions, a triplet (x, y, z) with x + y + z = 1. A unique point can be determined when each component represents distance to an edge of an equilateral triangle.
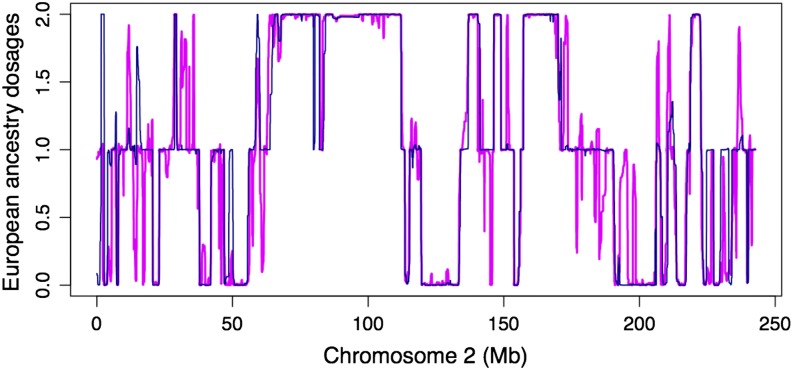
Since 1000G provides phased haplotypes for Mexicans, we therefore inferred the local ancestries of these haplotypes, using three source populations, SP2. The inferred local ancestries have excessive ancestry switches compared to those using unphased diplotype data (Figure 6). These excessive switches are likely caused by imperfect phasing—when using diplotypes our method integrates out phase uncertainties. Phasing admixed individuals is a difficult problem. Our results suggest, indirectly, that there is room for improvement in this area and we anticipate the two-layer model will make meaningful contributions.
Figure 6.
Comparison between phased and unphased 1000G data. The plot shows the inferred European ancestry allele dosages (y-axis) of a typical Mexican individual. The x-axis denotes SNPs. The blue (pink) line denotes inferred values using unphased (phased) 1000G data. Excessive ancestry switches of the pink line indicate imperfect phasing.
Discussion
We have presented a two-layer HMM to detect structure of local haplotypes and demonstrated its usefulness in local ancestry inference. The prevailing model for admixture is the one-pulse model [or “immediate admixture” model (Ewens and Spielman 1995)], where haplotypes from two source populations mixed once some generations ago and continued to admix afterward without influx of additional haplotypes from source populations. In reality, however, this assumption is overly simplified. Treating the mixing generation γ as a parameter, the two-layer model can average results over multiple choices of mixing generations. This makes our method applicable to the scenario of continuously mixing, which is perhaps a more realistic model for admixture.
Our method can directly work with diploid data and thus eliminates phase uncertainty that often plagues other methods. This is particularly useful for local ancestry inference of Latinos, as high-quality Native American haplotypes are unavailable. Our method performs significantly better than other methods for ancestry segments of ≤1 cM, as demonstrated in both simulated and real data analysis. Because of the high resolution, our method discovered an interesting phenomenon—departure of local ancestry from the genome-wide averages. Although it makes biological sense for the two regions—the MHC region and a large inversion on chromosome 8—to show significant departure from genome-wide averages, we nonetheless caution readers not to generalize the conclusions to Mexican populations or Latinos in general, unless these are confirmed after analyzing much larger data sets.
The two-layer model extends the fastPHASE (Scheet and Stephens 2006) model from a single source population to multiple source populations; indeed, if the number of upper-layer clusters is set to 1, then the two-layer model reduces to the fastPHASE model. On the other hand, the two-layer model extends the STRUCTURE (Pritchard et al. 2000) model from independent markers to densely linked markers; if markers are assumed independent and the numbers of upper and lower clusters are equal and each lower cluster is assumed to descend deterministically from an upper cluster, then the two-layer model reduces to the STRUCTURE model. As an integration of STRUCTURE and fastPHASE models, the two-layer model enforces and learns the structure of local haplotypes. Because the structure of haplotypes is a ubiquitous phenomenon in genetic data, the two-layer model has many other potential applications:
Using lower-cluster dosages, we can compute pairwise local haplotype sharing, defined as the probability of two haplotypes descending from the same lower clusters, which reflects genetic relatedness between haplotypes. Preliminary studies suggest that local haplotype sharing can be used to impute HLA alleles and detect genetic associations.
As the two-layer model can infer the local ancestry with high accuracy, it is reasonable to speculate that it will also be effective in genotype imputation and phasing for admixed individuals.
Our method can directly estimate cluster-switch rates between adjacent markers, and this permits the inference of recombination rates and hotspots, which will be particularly useful for admixed individuals.
Aggregating is an effective method for detecting rare variant associations (Li and Leal 2008). For admixed individuals, it would be helpful to aggregate rare variants of the same local ancestries.
Because a diploid individual has two sets of latent states (one for each haplotype), our EM algorithm is quadratic in both numbers of upper clusters S and numbers of lower clusters K and linear in numbers of individuals and markers. This potentially limits the two-layer model’s applicability. With phased data in source populations, the computation is fast because our EM algorithm is linear in S and K for a haploid individual. It is a challenge to find a linear algorithm that is as accurate as the quadratic algorithm when fitting our model to diploid individuals; nevertheless, we are actively investigating this possibility. The recent progress concerning linear algorithms to fit the PAC model (Delaneau et al. 2012) is extremely encouraging. Note that this quadratic computational challenge might disappear in the near future due to the recent development of methods such as phase-seq (Yang et al. 2011), which produces genomic sequences completely phased across an entire chromosome.
Supplementary Material
Acknowledgments
The author thanks Paul Scheet for helpful discussions regarding the θ update used in Scheet and Stephens (2006) and Alex Renwick and Hanli Xu for the results of HAPMIX. Robert Waterland and John Belmont read and commented on an early version of this article. Mark Meyer, Jennifer Coon, and Joanne Salman’s suggestions improved the grammar and spelling of the manuscript. Nancy Cox suggested that the author double check the consistency of strands between different data sets; her suggestion led to the discovery of a bug that flipped alleles of A/T, C/G SNPs when their minor allele frequencies are close to 0.5, which subsequently produced some spurious results reported in an early draft. The author thanks two anonymous reviewers for their constructive comments that greatly improved clarity of the presentation. The author was supported in part by U.S. Department of Agriculture/Agricultural Research Service award 6250-51000-052.
Appendix: Expectation Maximization
We first outline the EM algorithm, assuming the haplotypes are observed. Given an initial guess of parameters ξ*, the complete data likelihood, denoting is
| (A1) |
The new estimate of ξ is
| (A2) |
Update ξ* = ξ and iterate the procedure until ξ* converges.
To elaborate on the EM algorithm: conditioning on ξ*, the posterior distribution of can be computed for each i. To estimate ξ, one can either sample many paths from (the hard EM) or integrate out analytically (the soft EM). Intuitively, the soft EM will perform better because it does not introduce sampling variation. However, with the hard EM only forward probabilities need to be computed to sample from p(Z(i)|h(i), ξ*). More importantly, computational tricks may be applied on the sampled paths to avoid possible traps of local optimum. In this article we use the soft EM for model fitting and report possible computational improvement elsewhere.
A diploid individual has two sets of latent states at each marker, which indicate the upper- and lower-layer cluster membership (we drop the superscript for the individual and this should cause no confusion). The conditional likelihood for the ith individual is with “emission”
| (A3) |
where
| (A4) |
and μ = 4Nν is the scaled mutation rate. In the implementation we used μ = 0.001. Note the one-to-one correspondence between t. and θm. and that we implicitly assumed Hardy–Weinberg equilibrium in the emission.
Forward and Backward Recursion
In what follows, every probability statement is conditioned on ξ*. The forward recursion follows,
| (A5) |
where and
| (A6) |
| (A7) |
| (A8) |
| (A9) |
All summation with dummy variables needs to be done only once. This is the benefit of the parameterization for Markov transition described in this article. The overall complexity of the forward and backward recursion is O(MS2K2) for diploid individuals and O(MSK) for haploid individuals.
Note The backward recursion follows,
| (A10) |
where and
| (A11) |
| (A12) |
| (A13) |
| (A14) |
The posterior of latent states at each marker for each individual can be computed via
| (A15) |
and renormalize to have
Update θ
To update parameters in each EM step, we solve for each component x of ξ,
| (A16) |
Assume we have both diploid g and haploid h individuals in our data. For diploid individuals, at marker m, write Let for k = 0, 1, 2. Similarly, for haploid individuals, at marker m, write Let for k = 0, 1. Let
| (A17) |
Take the derivative with respect to θmj and sum over k for diploid individuals to get
| (A18) |
for each j = 1, … , K (recall K is the number of lower-layer clusters). We have K equations with K unknowns and we can solve numerically for tj and hence θmj. To do so, we need the Jacobian J(t.) = (djk), where
| (A19) |
and
| (A20) |
We can solve J(t(n))(t(n + 1) − t(n)) = −F(t(n)) for the unknown t(n + 1) − t(n).
Compared to the update used in Scheet and Stephens (2006), this update for θ does not directly involve its value in the previous iteration. Perhaps unwilling to solve a linear system repetitively, Scheet and Stephens (2006) used an approximation to the last terms of Equation A18,
| (A21) |
where
| (A22) |
which can be computed by approximating tj and tk with values in the previous iteration. Denote
| (A23) |
and we have
| (A24) |
and solve to get
| (A25) |
With (A25) as a starting point only a few iterations are needed to estimate θ using the numerical method described earlier. Note, however, that solving the linear system has complexity O(K3), which makes the complexity of model fitting to be O(max(MS2K2, MK3)).
Update Markov Transition Parameters
To estimate Markov transition parameters, following Scheet and Stephens (2006), we introduce latent state transitions (jumps) Jim and Rim that occurred between marker m − 1 and m at upper and lower layers for individual i. Denote Jims the number of upper-layer jumps to and Rimsk the number of lower-layer jumps to and Recognizing that Jims and Rimsk are sufficient for α, β, j, and r, we have
| (A26) |
where one may recall that S is the number of upper-layer clusters.
In what follows, when a latent state in forward or backward probabilities was substituted by a dot, then that component was summed over. Note that p(g(i)|ξ*) = φ(M, ⋅, ⋅, ⋅, ⋅) and
| (A27) |
and
| (A28) |
Second,
| (A29) |
with each component being
| (A30) |
| (A31) |
| (A32) |
Finally, special treatment is needed at marker m = 1. For each s, k set
| (A33) |
and renormalize such that where d = 2, 1 for diploid and haploid individuals, respectively. Set
Ancillary HMM
The expected complete data log-likelihood is given as
| (A34) |
where pmjs is the sth upper-cluster dosage of the jth haplotype at marker m. From the Balding–Nichols model (Balding and Nichols 1995), we have
| (A35) |
Combining the above two equations and dropping the m in notation, we have for an arbitrary marker
| (A36) |
This suggests that we add (Fηs − 1)pjs to the top and (F − 2)pjs to the bottom of (A25) to estimate θj,
| (A37) |
where Γ is a digamma function. When F > 1, we use recurrence relation Γ(x + 1) = 1/x + Γ(x) twice to get
| (A38) |
Because η ∈ [0, 1], we may use at x = Fηs + 2 and x = F(1 − ηs) + 2 to solve for ηs numerically. When F = 1, however, we may use the reflection formula Γ(1 − ηs) − Γ(ηs) = π cot(πηs) to solve for ηs analytically.
The forward and backward probabilities of the ancillary HMM and other parameter estimates are simply special cases of the main HMM.
Footnotes
Communicating editor: C. Sabatti
Literature Cited
- Balding D. J., Nichols R. A., 1995. A method for quantifying differentiation between populations at multi-allelic loci and its implications for investigating identity and paternity. Genetica 96: 3–12. [DOI] [PubMed] [Google Scholar]
- Baran Y., Pasaniuc B., Sankararaman S., Torgerson D. G., Gignoux C., et al. , 2012. Fast and accurate inference of local ancestry in Latino populations. Bioinformatics 28(10): 1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S. R., Browning B. L., 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81(5): 1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchhouse C., Marchini J., 2013. Multiway admixture deconvolution using phased or unphased ancestral panels. Genet. Epidemiol. 37(1): 1–12. [DOI] [PubMed] [Google Scholar]
- Conrad D. F., Jakobsson M., Coop G., Wen X., Wall J. D., et al. , 2006. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat. Genet. 38(11): 1251–1260. [DOI] [PubMed] [Google Scholar]
- Delaneau O., Marchini J., Zagury J., 2012. A linear complexity phasing method for thousands of genomes. Nat. Methods 9(1): 179–181. [DOI] [PubMed] [Google Scholar]
- Ewens, W. J., 2004 Mathematical Population Genetics 1: Theoretical Introduction (Interdisciplinary Applied Mathematics), Ed. 2. Springer-Verlag, Berlin/Heidelberg, Germany/New York. [Google Scholar]
- Ewens W. J., Spielman R. S., 1995. The transmission/disequilibrium test: history, subdivision, and admixture. Am. J. Hum. Genet. 57(2): 455–464. [PMC free article] [PubMed] [Google Scholar]
- Fearnhead P., Donnelly P., 2002. Approximate likelihood methods for estimating local recombination rates. J. R. Stat. Soc. B 64: 657–680. [Google Scholar]
- Guan Y., Stephens M., 2008. Practical issues in imputation-based association mapping. PLoS Genet. 4(12): e1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 1983. Properties of a neutral allele model with intragenic recombination. Theor. Popul. Biol. 23: 183–201. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium , 2007. A second generation human haplotype map of over 3.1 million snps. Nature 449(7164): 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium , 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467(7311): 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N. A., Coram M. A., Shriver M. D., Romieu I., Barsh G. S., et al. , 2011. Ancestral components of admixed genomes in a mexican cohort. PLoS Genet. 7(12): e1002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman J. F. C., 1982. On the genealogy of large populations. J. Appl. Probab.19(A): 27–43. [Google Scholar]
- Li B., Leal S. M., 2008. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 83(3): 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Cho K., Kidd J. R., Kidd K., 2009. Genetic landscape of Eurasia and admixture in Uyghurs. Am. J. Hum. Genet. 85: 934–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Z., Absher D. M., Tang H., Southwick A. M., Casto A. M., et al. , 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319(5866): 1100–1104. [DOI] [PubMed] [Google Scholar]
- Li N., Stephens M., 2003. Modeling linkage disequilibrium, and identifying recombination hotspots using SNP data. Genetics 165: 2213–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Sawyer S. L., Mukherjee N., Pakstis A. J., Kidd J. R., et al. , 2004. Haplotype block structures show significant variation among populations. Genet. Epidemiol. 27(4): 385–400. [DOI] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium, 2010. A map of human genome variation from population-scale sequencing. Nature 467(7319): 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Hattangadi N., Lane B., Lohmueller K. E., Hafler D. A., et al. , 2004 Methods for high-density admixture mapping of disease genes. Am. J. Hum. Genet. 74(5): 979–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. S., Song Y. S., 2010. A principled approach to deriving approximate conditional sampling distributions in population genetics models with recombination. Genetics 186: 321–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Tandon A., Patterson N., Barnes K. C., Rafaels N., et al. , 2009. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 5(6): e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P., Stephens M., 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78: 629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. W., O’Brien S. J., 2005. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nat. Rev. Genet. 6(8): 623–632. [DOI] [PubMed] [Google Scholar]
- Smith M. W., Patterson N., Lautenberger J. A., Truelove A. L., McDonald G. J., et al. , 2004. A high-density admixture map for disease gene discovery in African Americans. Am. J. Hum. Genet. 74: 1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., Donnelly P., 2000. Inference in molecular population genetics. J. R. Stat. Soc. Ser. B Stat. Methodol. 62(4): 605–635. [Google Scholar]
- Sundquist A., Fratkin E., Do C. B., Batzoglou S., 2008. Effect of genetic divergence in identifying ancestral origin using HAPAA. Genome Res. 18(4): 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Coram M., Wang P., Zhu X., Risch N., 2006. Reconstructing genetic ancestry blocks in admixed individuals. Am. J. Hum. Genet. 79(1): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Rannala B., 2009. Population genomic inference of recombination rates and hotspots. Proc. Natl. Acad. Sci. USA 106(15): 6215–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Jin L., 2008. A genome-wide analysis of admixture in Uyghurs and a high-density admixture map for disease-gene discovery. Am. J. Hum. Genet. 83(3): 322–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Chen X., Wong W. H., 2011. Completely phased genome sequencing through chromosome sorting. Proc. Natl. Acad. Sci. USA 108(1): 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.