Abstract
DNA methylation is a chromatin modification that contributes to epigenetic regulation of gene expression. The inheritance patterns and trans-generational stability of 962 differentially methylated regions (DMRs) were assessed in a panel of 71 near-isogenic lines (NILs) derived from maize (Zea mays) inbred lines B73 and Mo17. The majority of DMRs exhibit inheritance patterns that would be expected for local (cis) inheritance of DNA methylation variation such that DNA methylation level was coupled to local genotype. There are few examples of DNA methylation that exhibit trans-acting control or paramutation-like patterns. The cis-inherited DMRs provide an opportunity to study the stability of inheritance for DNA methylation variation. There was very little evidence for alterations of DNA methylation levels at these DMRs during the generations of the NIL population development. DNA methylation level was associated with local genotypes in nearly all of the >30,000 potential cases of inheritance. The majority of the DMRs were not associated with small RNAs. Together, our results suggest that a significant portion of DNA methylation variation in maize exhibits locally (cis) inherited patterns, is highly stable, and does not require active programming by small RNAs for maintenance.
DNA methylation may contribute to heritable epigenetic information in many eukaryotic genomes. In this study, we have documented the inheritance patterns and trans-generational stability for nearly 1000 DNA methylation variants in a segregating maize population. At most loci studied, the DNA methylation differences are locally inherited and are not influenced by the other allele or other genomic regions. The inheritance of DNA methylation levels across generations is quite robust with almost no examples of unstable inheritance, suggesting that DNA methylation differences can be quite stably inherited, even in segregating populations.
Keywords: DMR, inheritance pattern, transgenerational stability, near-isogenic lines, maize
MANY organisms exhibit abundant phenotypic diversity within a species. This observed diversity is often attributed to genetic variation, but there is a large part of this diversity that cannot be explained by genetic polymorphisms alone (Manolio et al. 2009). It has been proposed that epigenetic variation could be one component of this missing heritability (Petronis 2010). DNA methylation is a well-studied contributor to epigenetic information in many eukaryotes. In plants, DNA methylation occurs in three sequence contexts, CG, CHG, and CHH (where H = A, T, or C). It is widely accepted that de novo establishment of DNA methylation is often guided by small RNA (RNA-directed DNA methylation, RdDM), while maintenance of DNA methylation is performed by different methyltransferases, namely MET1 (METHYLTRANSFERASE 1) for CG methylation, CMT3 (CHROMOMETHYLASE 3), and DRM1/2 (DOMAINS REARRANGED METHYLTRANSFERASE 1 and 2) for non-CG methylation (CHG and CHH) (Chan et al. 2005). Variation in DNA methylation profiles has been observed in many species (Vaughn et al. 2007; Eichten et al. 2011b, 2013; Regulski et al. 2013; Schmitz et al. 2013a,b), and there are examples where epigenetic variation can lead to phenotypic change (Cubas et al. 1999; Manning et al. 2006).
There are multiple mechanisms that can give rise to variation in DNA methylation (Richards 2006). DNA methylation variation may be the result of pure epigenetic variation (Richards 2006) that is not dependent upon any sequence change. There is evidence for pure epigenetic changes in several plants (Becker et al. 2011; Eichten et al. 2011b; Schmitz et al. 2011; Havecker et al. 2012). It is unclear how pure epigenetic variation could occur, but failure to faithfully maintain methylation patterns by methyltransferase has been proposed as one possible mechanism (Schmitz et al. 2011). Alternatively, DNA methylation variation could be dependent on genetic polymorphisms such as structural rearrangements (Melquist et al. 1999) or polymorphic transposon insertions (Martin et al. 2009; Eichten et al. 2012; Yang et al. 2013) that occur near the affected region. DNA methylation variation could also be triggered by signals from trans-acting loci (Cao and Jacobsen 2002; Riddle and Richards 2002; Bender 2004). About 85% of the maize genome is composed of transposable elements (TEs) (Schnable et al. 2009), and composition and organization of transposable elements often vary across maize inbred lines (Fu and Dooner 2002; Brunner et al. 2005). Maize also has a relatively high abundance of 22-nt small RNAs (Nobuta et al. 2008) and exhibits substantial variation in small RNA composition among different maize lines (Barber et al. 2012). This variation in transposable elements and small RNA provided ample opportunity for variation in DNA methylation of nearby sequences.
As we seek to gain a better understanding of the role of DNA methylation variation in natural phenotypic variation it is important to understand the inheritance patterns and trans-generational stability for DNA methylation. While genetic variation is highly heritable and exhibits well-known inheritance patterns, it is possible that DNA methylation may exhibit metastable patterns (Regulski et al. 2013). Studies in Arabidopsis comparing unselected descendants 30 generations later with their ancestors found that DNA methylation patterns are largely heritable, but examples where methylation changes at single cytosines or over larger regions were also found (Becker et al. 2011; Schmitz et al. 2011). The rate of change for DNA methylation status at a single cytosine is substantially higher than the SNP rates but the rate of DNA methylation changes for larger (∼100 bp) regions is relatively low. Studies using segregating recombinant inbred lines (RILs) population in maize and soybean also suggested the occurrence of unstable methylation over generations, although with variable rates (Eichten et al. 2013; Regulski et al. 2013; Schmitz et al. 2013a). The goal of this study was to characterize the inheritance patterns of differentially methylated regions (DMRs) and estimate their stability in a near-isogenic population of maize plants.
Materials and Methods
Materials
B73-Mo17 near-isogenic lines (NILs) used in this study were grown from germplasm described in Eichten et al. (2011a). The NILs were developed from two lines, B73 and Mo17. Specifically, the B73-Mo17 hybrids were backcrossed to either recurrent parent for three generations. The two sets of backcrossing populations were then self-pollinated for three to six generations to generate the NIL population. Seventy-one NILs together with the two parental inbreds, B73 and Mo17 were grown in a controlled environment with a cycle of 15 hr of light and 9 hr of dark. The samples were placed using a randomized design. Each NIL was planted in one pot, while B73 and Mo17 were each planted in three pots. Three seeds were planted in each pot. At the 19th day after planting, the third leaf from one healthy plant was harvested and used for DNA extraction.
Array design and data collection
A custom 12 × 270,000 array was designed to assess methylation levels at 4763 DMRs identified using a 2.1 M array platform in a previous study (Eichten et al. 2013). The 12 × 270,000 array was designed to provide a higher resolution of the 4763 DMRs by including probes from the 2.1 M array as well as a series of newly designed probes to increase the density of assessed regions. The 12 × 270,000 array contains 12 subarrays, each of which has a total of 271,144 experimental probes. These included 106,800 probes from the 2.1 M array platform that are within the DMRs; 110,743 probes that are newly designed to fit in between the original probes and 5-kb surrounding regions; 32,024 probes that are selected from the 2.1-M platform and can be used as control; 13,504 probes that show differential hybridization signal between B73 and Mo17 inbreds, which can be used to validate NIL genotypes; and 8073 replicated probes found on chromosome 7 to validate array quality. Array data were collected using the method described in Eichten et al. (2013).
Analysis of array data
The raw array image was aligned and quantified using NimbleScan software (Roche). Pair files with quantification data for each probe were then exported from NimbleScan and imported into an R environment. Two CGH ratios were then calculated to check genotypes of each NIL using the genotyping probes on the array that showed big hybridization difference between B73 and Mo17: the ratio of the Cy3 signal from each NIL to that from B73, and the ratio of the Cy5 signal from each NIL to that from B73. The NILs with confirmed genotypes were used for following analysis. The signals for those NILs were normalized using a two-step array normalization method. Signal was first normalized within each array to minimize space effect and then normalized across arrays to minimize batch effect. Array raw data as well as normalized data were deposited in National Center for Biotechnology Information GEO under accession no. GSE51567.
Classification of DMR inheritance
The methylation level for each DMR in each NIL, B73, and Mo17, was determined by averaging the signal of all probes within the DMR. A Student’s t-test was then performed between B73 and Mo17, each with three biological replicates. DMRs that showed significant difference at the P < 0.05 level were considered to be differentially methylated between B73 and Mo17. Those significant DMRs were then tested between the B73-like NILs and Mo17-like NILs using a t-test, with each NIL type having >30 samples for each DMR. The DMRs showing significant difference at P < 0.05 level (Benjamini and Hochberg corrected P-value) was used to classify cis, trans, and complex inheritance patterns, while the DMRs that did not show significant difference were used to differentiate paramutation from unclassified patterns. The DNA methylation levels for each DMR were also assessed using whole-genome bisulfite sequencing data from B73 and Mo17 as previously reported (Eichten et al. 2013). To be classified as a paramutation-like DMR, we require the methylation level of the NIL population to be similar to one parent (P ≥ 0.05) but significantly different from the other parent (P < 0.05).
To differentiate DMR with cis, trans, or complex inheritance pattern, we used the informative NILs, which were defined as NILs that had an introgression at the genomic location of the DMR. Only DMRs that had at least three informative NILs were used for inheritance classification. For those DMRs, the genotype of each NIL was predicted based on the methylation level using the linear discriminant analysis (LDA) function from R and were compared to the inferred genotypes based on another prior study (Eichten et al. 2011a). A probability parameter was then calculated for each DMR based on the observed pattern of prediction using the function pk(1 − p)n−k, where n is the number of informative NILs, k is the number of NILs with methylation matching local genotypes, p is the probability of methylation matching local genotype and was considered to be 0.5. DMRs with a probability >0.13 were classified as complex DMRs. DMRs with a probability <0.13 were classified into cis DMR if all/most of NILs were matching local genotype, and trans DMRs if all/most of NILs were matching background genotype.
Methylation level and genotype in the intermated B73-Mo17 (IBM) RIL population (Lee et al. 2002) was from previous studies (Eichten et al. 2013; Li et al. 2013). The study of Eichten et al. (2013) measured methylation level using the original probes on the 2.1-M array platform, while both original and newly synthesized probes were used to calculate methylation level in this study. The classification criteria for inheritance patterns in the RILs were as described in Eichten et al. (2013).
Identification of cis DMR with instability of DNA methylation levels
To identify cis DMRs with instability for DNA methylation levels, all the NILs including the NILs with or without introgression at the interested DMR were used. The genotypes of those NILs were predicted based on the methylation level of the DMR using the LDA function in R. A probability that the predicted genotype is different from the observed genotype based on other genotyping technology was obtained. If this probability was <0.8, no instability occurred; if >0.8, a putative instability was defined. This analysis was then replicated for each individual probe within the DMR. If the percentage of probes showing instability is >60%, instability was defined for that NIL at the DMR.
To identify unstable DMRs with high confidence, we applied the following criteria: (1) the probability of instability was raised to 0.95 instead of 0.8; (2) parental value was required to span 0 for better separation of B73- or Mo17-like genotypes; (3) the number of NILs with instability at an interested DMR was required to be no more than three since methylation levels for DMRs with more than three unstable cases generally show a large overlap between B73-like and Mo17-like NILs; (4) after removing NILs with putative instability, Fisher’s linear discriminant (FLD) was calculated for the remaining NILs, and the best FLD was used to separate them into two groups. This value was required to be at least 12 for better separation of the two groups and the genotype within one group is B73 and the other group is Mo17. Application of these criteria resulted in 22 cases of instability from 19 DMRs.
Analysis of small RNA data
Small RNA data from a previous study (Regulski et al. 2013, GEO accession no. GSE39232) was processed using the FASTX toolkit to remove 3′ adapters, control sequencing quality and select small RNA size between 21 nt and 24 nt. Small RNA mapped to tRNA, rRNA, and miRNA was removed from the analysis. The remaining small RNA was mapped to maize genome, allowing up to 1000 mapping locations. For small RNA that was mapped to multiple locations, the abundance of that small RNA was normalized to the number of mapping locations. Small RNA was then clustered based on the DMR unit. The small RNA levels for 21/22-nt and 23/24-nt class were summarized separately for each DMR. DMRs with at least five reads were regarded to be covered by small RNA. Small RNA read counts were then normalized using DESeq (Anders and Huber 2010), which was also used to identify DMRs with differentially expressed small RNA between B73 and Mo17.
Quantitative PCR validation of unstable DMRs
The exact same DNA used for microarray analysis was used in methylation-sensitive quantitative PCR (qPCR) assays to validate the instability of DMRs observed in the microarray analysis. Genomic DNA (1 µg) was digested using the methylation-dependent restriction enzyme FspEI (New England Biolabs) following the manuals. A mock digestion using 50% glycerol substituting the enzyme was performed in parallel as the control. The digests were performed at 37° for 8 hr followed by enzyme inactivation at 65° for 20 min. After digestion, 240 µl water was added to the 30-µl digest, and 8.8 µl was used for qPCR in a 20-µl reaction system using the SsoFast EvaGreen Supermix (Bio-Rad, no. 172-5201). qPCR was performed on a CFX384 Real-Time PCR system using the following condition: 98° for 4 min to activate enzyme, followed by 40 cycles of 98° for 10 sec and 58° for 10 sec. All primers used in qPCR are listed in supporting information, Table S1.
Results
DNA methylation analyses in a B73-Mo17 near-isogenic line population using a custom array
Previous research has utilized meDIP-array profiling of DNA methylation to identify DMRs among different maize inbred lines (Eichten et al. 2011b, 2013). We were interested in documenting the inheritance patterns and stability of these DMRs in biparental populations of maize lines. A custom multiplex microarray format that included the same probes originally used to discover the DMRs as well as additional probes within these regions and various control probes was designed and synthesized (Materials and Methods, Figure 1A, and Figure S1). The custom array was used to measure DNA methylation level in B73 and Mo17 in three biological replicates, along with a panel of 71 B73-Mo17 NILs (Eichten et al. 2011a), each of which was measured in a single sample (Figure 1B). The NILs included 34 lines using B73 as recurrent parent and 37 lines using Mo17 as recurrent parent. The NILs were developed by three rounds of backcrosses to the recurrent parent followed by three to six generations of self-pollination (Eichten et al. 2011a). This is expected to result in NILs that have ∼93% of the genome inherited from the recurrent parent and ∼7% from the donor parent with relatively low levels of residual heterozygosity. The 4763 DMRs surveyed by this array platform were discovered through comparisons of DNA methylation levels in ∼20 different maize inbred lines (Eichten et al. 2013). As the NILs are derived from B73 and Mo17, we filtered the DMRs to those displaying differences between the three biological replicates of those two genotypes. There were 962 DMRs that exhibit significant differences (P < 0.05) between B73 and Mo17 and subsequent analyses focus on this set of 962 DMRs (Table S2).
Figure 1.
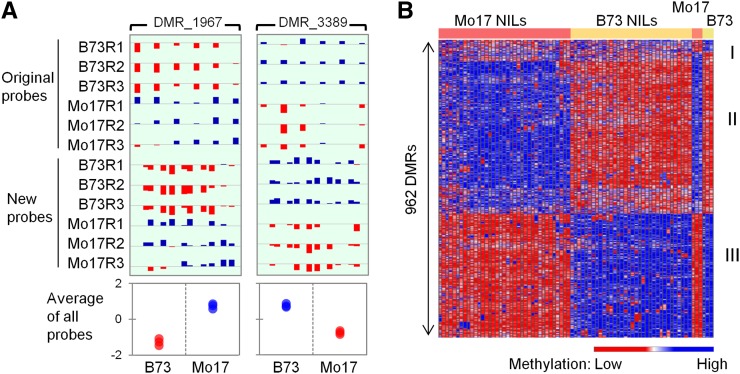
Methylation assays in B73, Mo17, and their near-isogenic lines (NILs). (A) Comparison of original and new probes for two DMRs in inbred lines B73 and Mo17. Each inbred had three biological replicates. Each bar in the top panel represents a probe within the DMR. The height of the bar represents methylation level, with hyper- and hypomethylation level colored as blue and red, respectively. Note the new probes were in between the original probes. In the bottom panel, the average methylation level of all probes was shown, with each dot representing one biological replicate of either B73 or Mo17. (B) Methylation level of 962 DMRs in B73, Mo17, and NILs. Each row represents a DMR, and each column represents a line. High and low methylation level was colored by blue and red, respectively. The color-coded bar at the top shows line types. B73 NILs are using B73 as recurrent parent, Mo17 NILs are using Mo17 as recurrent parent. The DMRs were grouped into three classes, I–III. Class I did not show difference between NILs with B73 genotype and NILs with Mo17 genotype, although they were different between B73 inbred and Mo17 inbred. Classes II and III were different in both comparisons, with class II having high methylation in Mo17 genotype and class III having high methylation in B73 genotype.
The majority of the 962 DMRs are 200–1000 bp in length and contain at least three probes (Figure S2, A and B). These DMRs are evenly distributed across the maize genome. In total, the DMRs represent ∼797 kb (0.034%) of the 2.3-Gb maize genome, and 25% of them overlap an annotated gene (Table S2). Hierarchical clustering of DNA methylation levels for the 962 DMRs in all samples suggested that the DMRs could be roughly divided into three groups (Figure 1B). Group I (77 DMRs) showed significant difference between inbred lines B73 and Mo17, but no difference between B73-NILs and Mo17-NILs. Groups II and III (885 DMRs) showed significant difference between B73 and Mo17, and also between B73-NILs and Mo17-NILs. They differed in which genotype had a higher DNA methylation level. The high level of consistency between B73 vs. Mo17 and B73-NILs vs. Mo17-NILs (885/962, ∼92%) suggested that most of the DMRs are heritable in the NIL population. We thus proceed to classify the inheritance patterns of those DMRs.
Inheritance patterns for DMRs in NIL population
A series of criteria were employed to classify the inheritance patterns in the NIL population for the 962 significant DMRs between B73 and Mo17 (Figure 2 and Table S2). Although the classification of inheritance patterns was based solely upon the analysis of DNA methylation levels in NILs we also used information from whole-genome bisulfite sequencing of B73 and Mo17 (Eichten et al. 2013; Figure 2A and Table S2) and microarray-based DNA methylation analysis in a set of 17 intermated B73-Mo17 RILs (Eichten et al. 2013; Figure 2B and Table S3) to further characterize and validate the DMRs and their inheritance patterns.
Figure 2.
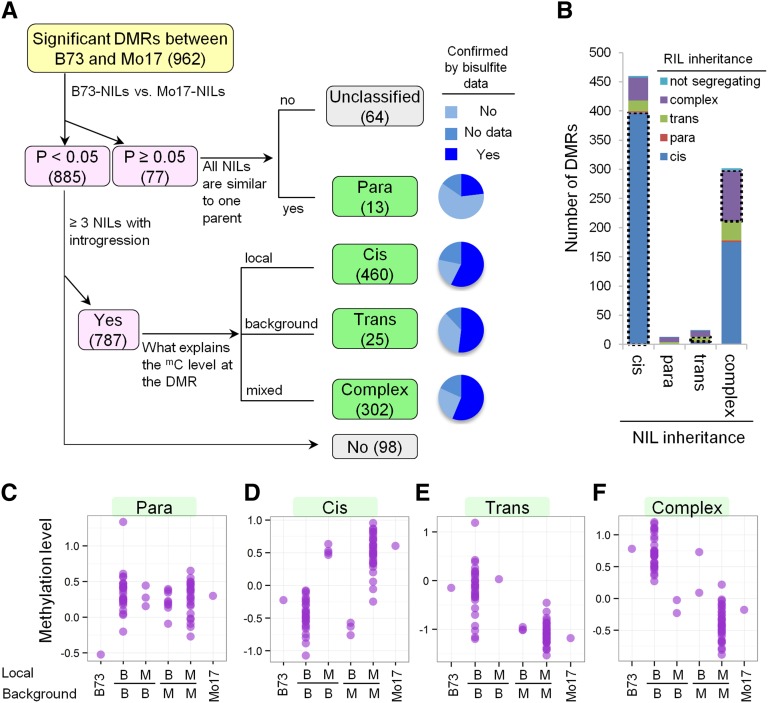
DMR inheritance patterns in NIL population. (A) Flow chart to show the classification methods of DMR inheritance. Four major patterns were classified: cis, trans, paramutation-like, and complex patterns. The pie charts on the right side show the results by comparing parental difference from the array platform and that from whole genome bisulfite sequencing data. (B) Number of DMRs with each inheritance pattern in NIL and RIL populations. DMRs that show the same inheritance patterns in both NIL and RIL population are in dotted boxes. (C–F) Examples of DMR with different inheritance patterns. Each purple dot represents one line: B73, Mo17, or a NIL. B, B73; M, Mo17.
There are 77 DMRs that do not exhibit significant differences between B73-like NILs and Mo17-like NILs. These 77 DMRs were further classified based on whether the NILs exhibit consistent DNA methylation levels similar to one parent (n = 13, paramutation like) or whether they exhibit “unclassified” patterns (n = 64). Paramutation-like DMRs were those where the methylation of the entire NIL population was similar to the methylation state of one parent, but significantly different from that of the other parent (Figure 2, A and C). There were only 13 DMRs that exhibited evidence for paramutation-like behavior in the NIL population. The evidence to support these DMRs from bisulfite sequencing is relatively weak as only 3/13 exhibit similar parental differences in bisulfite analyses (Table S2). The analysis of DNA methylation levels for these 13 DMRs in the RIL population does not support the classification of paramutation-like behavior (Figure 2B, Table S2 and Table S3). Instead, many of these DMRs exhibit complex inheritance patterns in the RILs. The combined results of the NIL and RIL analyses find little evidence for significant numbers of DMRs that exhibit strong paramutation-like inheritance. It is possible that some of these loci exhibit paramutation that is transient or has incomplete penetrance. The other 64 DMRs that do not exhibit significant differences between B73-like and Mo17-like NILs were described as unclassified. These may be the result of highly stochastic behavior or differences in the DNA methylation level between the B73 and Mo17 used for this study and the exact B73 and Mo17 used as parents for the NIL population.
The remaining 885 DMRs exhibited significant differences between the B73-like NILs and the Mo17-like NILs. The number of informative NILs, in which a line contains an introgression covering the DMR, was identified for each DMR. Based on the number of NILs (71) and the genome structure of the NIL population (∼7% introgression) it is expected that 4–5 informative NILs will be included for each of the DMRs (Figure S3). The regulation of the methylation level at those introgression loci are expected to include examples of simple Mendelian control located near the DMR itself (cis or local inheritance) or located at unlinked genomic locations (trans or remote inheritance), as well as examples of complex, polygenic control of DNA methylation. To classify these types of inheritance patterns, DMRs with at least 3 informative NILs (787 of the 885) were further studied (Figure 2A). The analysis of DNA methylation for the informative NILs with an introgression allowed a test of whether the local genotype (cis) or the genetic background (trans) of the majority of the informative NILs was correlated with DNA methylation level at a particular DMR. In some cases the DNA methylation levels were not significantly associated with either the local genotype or the genetic background and these were classified as “complex” inheritance patterns that could reflect control from both cis and trans or hypervariable patterns (Figure 2A).
The most prevalent class of mapped DMRs (460 of 787; 58%) exhibit DNA methylation states that agree with the local genotype suggesting cis-acting control or inheritance of DNA methylation (Figure 2D). Methylation difference in 73% of these DMRs was confirmed by bisulfite sequencing (Figure 2A and Table S2). We also found that ∼86% of the DMRs that were classified as having cis inheritance in the NIL population were also classified as having cis inheritance in the RIL population (Figure 2B, Table S2 and Table S3). A relatively small portion of the mapped DMRs (25 of 787; 3.2%) exhibit DNA methylation states that appear to be controlled by the genetic background rather than the local genotype and are classified as trans-regulated DMRs (Figure 2, A, B, and E). Bisulfite sequencing data confirmed parental differences for 59% of those trans DMRs (Table S2). Nearly one-third (8 of 25) of the trans DMRs were also classified as having trans-acting control in the RIL population (Figure 2B, Table S2, and Table S3). There were a substantial number of complex DMRs that exhibit significant differences in DNA methylation, which was further confirmed by bisulfite data (69%), but they could not be unambiguously classified as cis or trans inheritance patterns (Figure 2F). These complex DMRs generally showed less parental difference than cis DMRs (Figure S4), which makes it more difficult to correlate methylation with local/background genotype, leading to some misassignment of cis or trans DMRs into this category. Indeed we found that the majority of DMRs classified as complex based on the NIL population are actually classified as cis or trans in the RIL population (Figure 2B, Table S2, and Table S3). There was little evidence for major differences in the context of DNA methylation differences or in the location of the DMRs relative to genes and transposons for DMRs that exhibited cis, trans, or complex inheritance patterns (Figure S5).
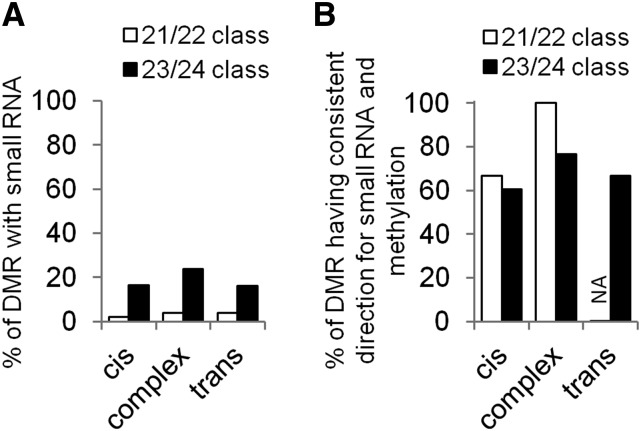
The relatively small number of trans-controlled DMRs suggests that most DMRs are inherited through maintenance of DNA methylation patterns as opposed to active targeting through RNA-directed DNA methylation pathway or other means. Recent studies have described small RNAs in shoot tissue of B73 and Mo17 (Regulski et al. 2013). The overlap of these small RNAs with the DMRs monitored in this study was analyzed (Figure 3). A relatively small portion of the DMRs are covered by small RNAs: ∼5% by 21/22-nt and ∼20% by 23/24-nt small RNA (Figure 3A and Table S2). The DMRs with complex inheritance patterns exhibit slightly higher small RNA coverage than DMRs with cis or trans inheritance patterns. It was somewhat surprising that only a small proportion (1/25 by 21/22 nt; 4/25 by 23/24 nt) of the trans DMRs had small RNA coverage. The lack of small RNA coverage in trans DMRs could be due to the existence of other trans-acting factors or potential that the important small RNAs are not expressed in the tissues surveyed for small RNA abundance. The concordance of DNA methylation and small RNA coverage was assessed to determine if the genotype with higher DNA methylation also had higher small RNA coverage. An agreement in direction between DNA methylation levels and small RNA coverage was observed for both 21/22-nt and 23/24-nt small RNA classes and for all DMR inheritance patterns (Figure 3B). Together, these results suggested a role of small RNA in establishing/maintaining the methylation difference at a subset of the DMRs, but reveals that the majority of differential DNA methylation is not associated with small RNA presence.
Figure 3.
Small RNA coverage at DMR with different inheritance patterns. (A) Percentage of DMRs with small RNA. (B) Percentage of DMRs with concordance between small RNA level and methylation level in the same parental line. The small RNA data were generated in a study by Regulski et al. (2013). Small RNA was isolated from 5 days after planting coleoptile tissue from two genotypes, B73 and Mo17, which are the same lines used for NIL development in this study. Small RNAs were grouped into two classes based on their size: 21/22-nt class and 23/24-nt class. NA, not available, which is because the trans DMRs covered by small RNA do not show significant difference on small RNA levels between the two parental lines B73 and Mo17.
Stability of DMRs across generations
A major focus of this study was to better understand the stability of inheritance for DNA methylation variation. Here, we were able to study the stability of DNA methylation variation in a segregating population that has gone through four to seven generations of self-crossing (the exact number of generations for a particular DMR cannot be determined since any particular allele may have been present in the F1 or introduced in any of the three backcrosses) (Eichten et al. 2011a). For this purpose, only the cis DMRs were included since it is difficult to differentiate between instability and presence of trans-acting factors for the other DMRs. We hypothesized that if the transmission process is stable, the methylation level at cis DMR would be coupled to the local genotype for all of the NILs studied; otherwise, rare examples of NILs where methylation levels do not match local genotype would be observed. Thus, a comparison of the DNA methylation level and the genotype for all NILs would allow the discovery of NILs with potential instability.
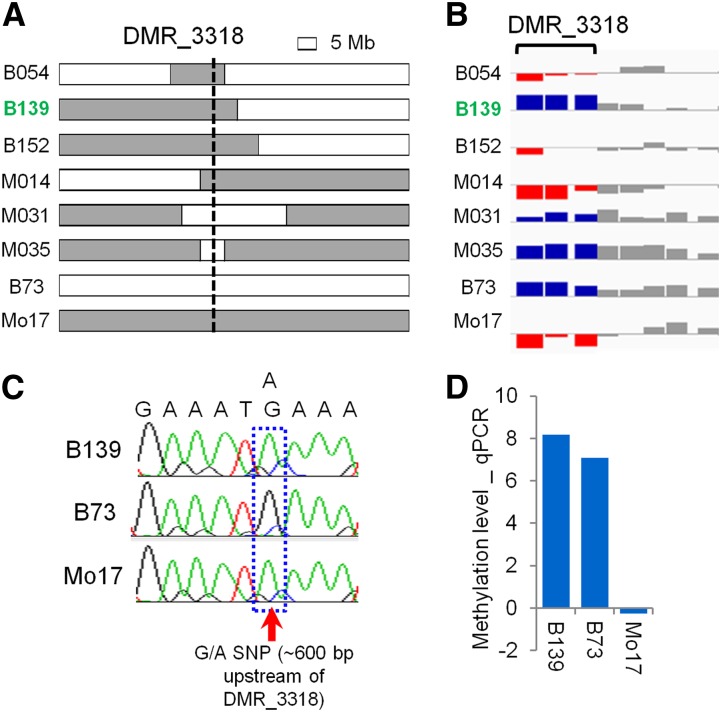
It is possible that false-positive examples of unstable DNA methylation could result from a miscalled genotype due to residual heterozygosity in the NILs or technical error in genotyping measurement. We sought to exclude unstable DMRs resulting from genotyping error by checking the genotypes of the NILs showing instability using the genotyping probe on this array platform. In addition, we assessed DNA methylation levels for every single probe within the DMR and required >60% of probes within a DMR to support an instability to minimize possible technical artifacts in array measurement. This resulted in a total of 368 unstable cases out of a total of 30,671 tests (∼1.2%). Seventeen examples of unstable inheritance (from 12 DMRs) were independently tested using qPCR-based analysis of DNA methylation (Figure S6; Materials and Methods). While the methyl-sensitive qPCR was able to validate the parental difference and cis inheritance pattern of these DMRs, it did not support the unstable DNA methylation for a particular NIL genotype that was suggested by the microarray profiling. In each case, the DNA methylation level found using methyl-sensitive qPCR matches the local genotype (Figure S6). To determine whether there are any examples of instability for cis DMRs a more stringent set of criteria (Materials and Methods) were implemented to identify the most robust examples of unstable DNA methylation inheritance. This led to an identification of 19 DMRs with 22 cases of instability (some DMRs showed instability in more than one NIL). Methylation levels at 15 DMRs involving 18 unstable cases were assessed using methyl-sensitive qPCR (Figure S7). Only 1 of these putative 18 cases was confirmed (Figure 4 and Figure S7). It occurred at DMR_3318 in a B73-NIL with Mo17 introgression at the DMR, B139 (Figure 4A). B139 showed similar methylation level as B73 based on both the array and qPCR measurement (Figure 4, B and D), but it had a Mo17 genotype based on both the genotyping probes on the array and Sanger sequencing (Figure 4, A and C). The mismatch between methylation level and genotype indicated that a switch of methylation level happened after the hybridization of B73 and Mo17.
Figure 4.
Validation of instability at DMR_3318. (A) Genotypes of eight representative lines. The vertical dashed line indicates the position of DMR_3318. B73 genotype is shown as white bar and Mo17 as gray bar. (B) Methylation level for the same lines as in A, based on MeDIP-array. Each bar represents a probe. Blue and red bars represent high and low methylation level, respectively. Probes outside DMR_3318 are shown in gray. (C) Genotype of B139, B73 inbred, and Mo17 inbred at DMR_3318 based on Sanger sequencing. (D) Methyl-sensitive qPCR-based measurement of methylation level at DMR_3318.
Discussion
There is a substantial interest in understanding the contribution of DNA methylation variation toward heritable natural variation, response to environmental conditions, as well as development. This study is focusing on studying the inheritance patterns and stability for DNA methylation variation that occurs between two parental inbreds. Nearly 1000 DMRs between the B73 and Mo17 inbreds were assessed in 71 NILs. This provided an opportunity to study the frequency of different types of inheritance patterns as well as the stability of inheritance of locally (cis) controlled DMRs. Understanding these two issues would provide insights into frequency changes of (epi)alleles over time within a population, as well as the contribution of DNA methylation to less stable inheritance of information.
Inheritance patterns for DNA methylation variation
One of the particularly intriguing aspects of DNA methylation is the potential for unexpected segregation behaviors. For example, there are instances in which there can be communication of epigenetic state between alleles at a single locus as in paramutation (Chandler 2007) or communication between homologous loci located in distinct genomic positions as in PAI silencing (Melquist and Bender 2003). We were interested in determining the frequency at which DMRs would exhibit inheritance patterns that resemble paramutation or trans-acting control as opposed to cis (local) inheritance. NILs provide advantages as the successive rounds of backcrossing result in maintained heterozygosity within introgression regions, providing ample opportunity for allelic interactions to occur. Second, the ability to test the effect of the genetic background (essentially all potential trans-factors) is relatively easy in a NIL population.
In our analysis of 962 DMRs, only a very small subset (13) exhibit strong evidence for paramutation-like patterns. While we have referred to these as “paramutation-like” it is also possible that these patterns could be observed due to a rare DNA methylation loss/gain in the parental plants and this would not represent a paramutation-like event. It is worth noting that our definition of paramutation-like inheritance was relatively strict. We required that the paramutation would occur at high frequency and then be highly stable. There are some examples of paramutation, such as the B-I locus that exhibit full penetrant and stable behaviors (Chandler and Stam 2004). However, there are other examples of paramutation, such as the p1 and R-r:std locus, for which less stable behavior is observed (Chandler and Stam 2004; Chandler and Alleman 2008; Hollick 2010). It is quite possible that the relatively strict criteria that we used have failed to identify some examples of paramutation-like patterns of inheritance for DMRs. Regulski et al. (2013) noted a slightly higher rate of paramutation-like behavior. They did not require the same level of penetrance or stability to be classified as paramutation-like and found that up to 10% of DMRs exhibit potential paramutation-like inheritance (Regulski et al. 2013). The analysis of inheritance of DNA methylation patterns in epiRILs suggests that allelic communication of DNA methylation states may occur for some loci but not others (Johannes et al. 2009; Reinders et al. 2009; Schmitz and Ecker 2012). While some loci that have reduced DNA methylation in ddm1 mutant plants can regain DNA methylation, when functional DDM1 is reintroduced there are many other loci that fail to become remethylated (Johannes et al. 2009). The behavior of the DMRs in these epiRILs indicated that many DMRs are not influenced by paramutation-like behavior, reinforcing the view that not all DNA methylation variation can be communicated between alleles.
Among the 787 total DMRs that could be assessed for cis/trans inheritance patterns for DNA methylation, we found that 58% exhibit cis inheritance and 3.2% exhibit trans inheritance patterns. The remaining DMRs could not be conclusively classified as cis or trans and are classified as having complex patterns of inheritance. These complex DMRs may reflect a number of distinct biological processes. In some cases these may be examples of cis or trans inheritance that did not meet our statistical criteria for these classifications. Another potential explanation for the complex inheritance patterns of some of these DMRs could be multilocus control of DNA methylation levels. It is possible that a trans-acting factor and the local genotype interact to influence DNA methylation. A recent study (Schmitz et al. 2013b) using genome-wide association scan mapping of DNA methylation in Arabidopsis documented a number of examples in which multiple loci influenced a particular DMR. Finally, it is worth noting that some of the complex patterns of inheritance could be the result of unstable inheritance of DNA methylation patterns.
The relative paucity of DMRs that exhibit inheritance patterns reflecting trans-acting control has been observed in other analyses of DMR inheritance. Previous studies of maize DMRs in a RIL population (Eichten et al. 2013) found relatively few trans inheritance patterns. The mapping of methyl-QTL in a soybean RIL population found that only 2.5% of the methyl-QTL mapped to trans-acting genomic regions while the other 97.5% mapped near the DMR (Schmitz et al. 2013a). However, association mapping of DNA methylation variation in Arabidopsis found more examples of trans control than cis control. It is worth noting that the expected nature of many examples of trans inheritance patterns may inhibit the ability to accurately document them using many molecular approaches. One mechanism for trans-acting control of DNA methylation is that a homologous sequence located in another genomic position can generate a siRNA that can trigger RNA-directed DNA methylation as observed at the PAI locus (Melquist and Bender 2003). Note that in this case, there are multiple closely related sequences in a genome. In our analyses, the DMRs were discovered through meDIP-array analysis and the microarray probes were required to be single copy (no other sequence with >90% identity found in the maize B73 reference genome). This may have removed many of the examples of potential trans-acting control from the survey. The trans-acting control that we are most likely to identify in our dataset are cases in which Mo17 contains a homologous sequence that is lacking from the B73 reference genome or examples in which the extra copy of a sequence is missing from the reference genome assembly. We did attempt to map the genetic position of the trans-acting factor in the NILs and were able to find a putative genomic region for 17 of the 25 trans DMRs. Six of these 17 DMRs also exhibit trans inheritance patterns in RILs and map to the same genomic locations. However, our attempts to validate these examples of trans-acting control were confounded by related sequences. Methyl-sensitive qPCR assays for many of the trans DMRs revealed amplification of multiple related sequences in at least one of the genotypes. The difficulties in identifying and validating trans-acting control of DNA methylation will not only be limited to the approaches used in this work, but also apply to other approaches using sequencing technology. Whole-genome bisulfite sequencing studies often require unique alignments of reads to a single genomic position. If the locus that generates the trans-acting signal is highly similar it may prevent the ability to quantify DNA methylation and detect variation using whole genome bisulfite sequencing.
Many studies of inheritance patterns for natural variation in DNA methylation have found a large number of examples of DMRs with cis or local inheritance (Vaughn et al. 2007; Eichten et al. 2013; Regulski et al. 2013; Schmitz et al. 2013a,b). In addition, the analysis of inheritance of altered DNA methylation patterns in epiRILs has found relatively stable inheritance of allelic DNA methylation patterns that are induced by ddm1 or met1 loss of function (Johannes et al. 2009; Reinders et al. 2009). The cis inheritance of DNA methylation can reflect two different potential processes. In some cases the cis inheritance pattern could actually reflect cis-acting genetic variation that directs the DNA methylation. For example, the insertion of a transposon in one allele could result in DNA methylation of neighboring sequences. Schmitz et al. (2013b) noted that a number of the methyl-QTL that map in cis contain structural variation or TE insertions in Arabidopsis. This type of cis inheritance reflects the genetic influences on DNA methylation and has been described as obligatory epialleles (Richards 2006). Alternatively, cis inheritance could reflect highly stable inheritance of DNA methylation information in the absence of any causative genetic changes, i.e., pure epiallele (Richards 2006; Eichten et al. 2011b).
Stability of inheritance for cis-controlled DNA methylation variation
Our dataset provided an opportunity to study the stability for the inheritance of DNA methylation over four to seven generations in a biparental population. The DMRs that generally exhibit cis inheritance pattern were analyzed to determine if there were any NILs that exhibit an unexpected association between genotype and methylation status for a particular DMR. A small number of unstable DNA methylation states relative to genotype were identified (1.2% of all potential cases). However, attempts to validate these examples of unstable behavior were not successful (Figure S6). A further attempt to validate only the most robust examples only found evidence to support 1 of the 18 examples (Figure S7). This suggests that DMRs that show local inheritance are quite stably inherited. Given that we could only validate one example of instability the estimated “mutation” rate for DNA methylation level of a DMR would be approximated as 1/(30,671 tests × 7 generations) [∼4.6 × 10−6]. Due to the small numerator, this rate might be a quite unreliable estimate of the true rate. The analysis of DNA methylation in Arabidopsis accessions that have been grown for 30 generations without selection revealed that the estimated mutation rate per mCG per generation is 4.5 × 10−4 (Schmitz et al. 2011). However, in this study the rate of DMR changes (regional changes in DNA methylation level rather than single base changes) was much lower, consistent with the idea that methylation mutation rate over a large region is smaller than single sites (Becker et al. 2011; Schmitz et al. 2011).
Although we do observe quite stable inheritance for some DMRs, we cannot conclude that all DMRs will exhibit stable behavior. DMRs that exhibit unstable inheritance would not be classified as having cis inheritance patterns. Instead, these examples would be classified as complex patterns. Alternatively, highly unstable DMRs may not even be discovered in many experiment designs. The 962 DMRs used for this study were selected because they exhibit significant differences in three biological replicates of B73 compared to three biological replicates of Mo17. If a DMR is highly unstable, then it may not exhibit consistent behavior within the inbred parents and may not even be selected for analysis within the population. Many of the other studies on inheritance of DMRs find that at least a subset of the DMRs exhibit stable behavior that can be mapped to cis or trans genomic regions (Schmitz et al. 2013a,b) but in each case there are also a number of the DMRs that cannot be attributed to particular causes. These may be analogous to the DMRs that we classified as complex and may reflect a subset of DMRs with high levels of instability. Future work is important to more clearly document the range of stabilities exhibited by natural variants for DNA methylation.
Supplementary Material
Acknowledgments
Evan Starr assisted with the DNA isolations and microarray hybridizations for this study. Computational support was provided by the Minnesota Supercomputing Institute and the Texas Advanced Computing Center. This work was supported by a grant from the National Science Foundation to N.M.S. (DBI-1237931).
Footnotes
Communicating editor: J. Birchler
Literature Cited
- Anders S., Huber W., 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber W. T., Zhang W., Win H., Varala K. K., Dorweiler J. E., et al. , 2012. Repeat associated small RNAs vary among parents and following hybridization in maize. Proc. Natl. Acad. Sci. USA 109(26): 10444–10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker C., Hagmann J., Müller J., Koenig D., Stegle O., et al. , 2011. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480: 245–249. [DOI] [PubMed] [Google Scholar]
- Bender J., 2004. DNA methylation of the endogenous PAI genes in Arabidopsis. Cold Spring Harb. Symp. Quant. Biol. 69: 145–153. [DOI] [PubMed] [Google Scholar]
- Brunner S., Fengler K., Morgante M., Tingey S., Rafalski A., 2005. Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17(2): 343–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Jacobsen S. E., 2002. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12(13): 1138–1144. [DOI] [PubMed] [Google Scholar]
- Chan S. W.-L., Henderson I. R., Jacobsen S. E., 2005. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6: 351–360. [DOI] [PubMed] [Google Scholar]
- Chandler V. L., Stam M., 2004. Chromatin conversations: mechanisms and implications of paramutation. Nat. Rev. Genet. 5: 532–544. [DOI] [PubMed] [Google Scholar]
- Chandler V., 2007. Paramutation: from maize to mice. Cell 128(4): 641–645. [DOI] [PubMed] [Google Scholar]
- Chandler V., Alleman M., 2008. Paramutation: epigenetic instructions passed across generations. Genetics 178(4): 1839–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P., Vincent C., Coen E., 1999. An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401: 157–161. [DOI] [PubMed] [Google Scholar]
- Eichten S. R., Foerster J. M., de Leon N., Ying K., Yeh C.-T., et al. , 2011a B73-Mo17 near-isogenic lines demonstrate dispersed structural variation in maize. Plant Physiol. 156(4): 1679–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten S. R., Swanson-Wagner R. A., Schnable J. C., Waters A. J., Hermanson P. J., et al. , 2011b Heritable epigenetic variation among maize inbreds. PLoS Genet. 7(11): e1002372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten S. R., Ellis N. A., Makarevitch I., Yeh C.-T., Gent J. I., et al. , 2012. Spreading of heterochromatin is limited to specific families of maize retrotransposons. PLoS Genet. 8(12): e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten S. R., Briskine R., Song J., Li Q., Swanson-Wagner R., et al. , 2013. Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell 25(8): 2783–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Dooner H. K., 2002. Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99(14): 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker E. R., Wallbridge L. M., Fedito P., Hardcastle T. J., Baulcombe D. C., 2012. Metastable differentially methylated regions within Arabidopsis inbred populations are associated with modified expression of non-coding transcripts. PLoS ONE 7(9): e45242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick J. B., 2010. Paramutation and development. Annu. Rev. Cell Dev. Biol. 26: 557–579. [DOI] [PubMed] [Google Scholar]
- Johannes F., Porcher E., Teixeira F. K., Saliba-Colombani V., Simon M., et al. , 2009. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 5(6): e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Petsch K., Shimizu R., Liu S., Xu W. W., et al. , 2013. Mendelian and non-Mendelian regulation of gene expression in maize. PLoS Genet. 9(1): e1003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Sharopova N., Beavis W. D., Grant D., Katt M., et al. , 2002. Expanding the genetic map of maize with the intermated B73 x Mo17 (IBM) population. Plant Mol. Biol. 48(5–6): 453–461. [DOI] [PubMed] [Google Scholar]
- Manning K., Tör M., Poole M., Hong Y., Thompson A. J., et al. , 2006. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38: 948–952. [DOI] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Troadec C., Boualem A., Rajab M., Fernandez R., et al. , 2009. A transposon-induced epigenetic change leads to sex determination in melon. Nature 461: 1135–1138. [DOI] [PubMed] [Google Scholar]
- Melquist S., Luff B., Bender J., 1999. Arabidopsis PAI gene arrangements, cytosine methylation and expression. Genetics 153(1): 401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melquist S., Bender J., 2003. Transcription from an upstream promoter controls methylation signaling from an inverted repeat of endogenous genes in Arabidopsis. Genes Dev. 17: 2036–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K., Lu C., Shrivastava R., Pillay M., De Paoli E., et al. , 2008. Distinct size distribution of endogenous siRNAs in maize: evidence from deep sequencing in the mop1–1 mutant. Proc. Natl. Acad. Sci. USA 105(39): 14958–14963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronis A., 2010. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature 465: 721–727. [DOI] [PubMed] [Google Scholar]
- Reinders J., Wulff B. B. H., Mirouze M., Marí-Ordóñez A., Dapp M., et al. , 2009. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 23: 939–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski M., Lu Z., Kendall J., Donoghue M. T. A., Reinders J., et al. , 2013. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 23: 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards E. J., 2006. Inherited epigenetic variation—revisiting soft inheritance. Nat. Rev. Genet. 7: 395–401. [DOI] [PubMed] [Google Scholar]
- Riddle N. C., Richards E. J., 2002. The control of natural variation in cytosine methylation in Arabidopsis. Genetics 162(1): 355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R. J., Schultz M. D., Lewsey M. G., O’Malley R. C., Urich M. A., et al. , 2011. Transgenerational epigenetic instability is a source of novel methylation variants. Science 334(6054): 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R. J., Ecker J. R., 2012. Epigenetic and epigenomic variation in Arabidopsis thaliana. Trends Plant Sci. 17(3): 149–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R. J., He Y., Valdés-López O., Khan S. M., Joshi T., et al. , 2013a Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 23: 1663–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R. J., Schultz M. D., Urich M. A., Nery J. R., Pelizzola M., et al. , 2013b Patterns of population epigenomic diversity. Nature 495: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., et al. , 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956): 1112–1115. [DOI] [PubMed] [Google Scholar]
- Vaughn M. W., Tanurdžić M., Lippman Z., Jiang H., Carrasquillo R., et al. , 2007. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 5(7): e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Li Z., Li W., Ku L., Wang C., et al. , 2013. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 110(42): 16969–16974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.