Abstract
The nature and extent of mutational pleiotropy remain largely unknown, despite the central role that pleiotropy plays in many areas of biology, including human disease, agricultural production, and evolution. Here, we investigate the variation in 11,604 gene expression traits among 41 mutation accumulation (MA) lines of Drosophila serrata. We first confirmed that these expression phenotypes were heritable, detecting genetic variation in 96% of them in an outbred, natural population of D. serrata. Among the MA lines, 3385 (29%) of expression traits were variable, with a mean mutational heritability of 0.0005. In most traits, variation was generated by mutations of relatively small phenotypic effect; putative mutations with effects of greater than one phenotypic standard deviation were observed for only 8% of traits. With most (71%) traits unaffected by any mutation, our data provide no support for universal pleiotropy. We further characterized mutational pleiotropy in the 3385 variable traits, using sets of 5, randomly assigned, traits. Covariance among traits chosen at random with respect to their biological function is expected only if pleiotropy is extensive. Taking an analytical approach in which the variance unique to each trait in the random 5-trait sets was partitioned from variance shared among traits, we detected significant (at 5% false discovery rate) mutational covariance in 21% of sets. This frequency of statistically supported covariance implied that at least some mutations must pleiotropically affect a substantial number of traits (>70; 0.6% of all measured traits).
Keywords: evolution, pleiotropy, quantitative genetics, transcriptome, variational module
PLEIOTROPIC effects of mutations are a central component of theories that attempt to explain standing genetic variation in natural populations (Johnson and Barton 2005; Zhang and Hill 2005) and have widespread implications for human disease (Eyre-Walker 2010; Lawson et al. 2011; Sivakumaran et al. 2011) and agriculture (Hill 2010). Although the significance of pleiotropy has long been appreciated by biologists (Stearns 2010), only recently have data become available to characterize the general nature of pleiotropy (Wagner and Zhang 2011; Paaby and Rockman 2013). Evidence, predominantly from screens of gene knockouts, suggests most mutations affect few traits (Wang et al. 2010; Wagner and Zhang 2011). These null mutations are unlikely to be representative of naturally occurring mutations of smaller effect (Stern 2000). There are currently few data on pleiotropic covariance generated by naturally occurring mutations of small effect, such as those that are likely to underlie a substantial proportion of the genetic variation in quantitative traits (Johnson and Barton 2005; Yang et al. 2010).
If the molecular mechanism of pleiotropy is typically the involvement of a gene in multiple biological processes (type II pleiotropy), rather than multiple molecular functions of a gene (type I pleiotropy) (He and Zhang 2006; Wagner and Zhang 2011), then null mutations will represent an upper limit on the pleiotropy of naturally occurring, small effect mutations (Wagner and Zhang 2011). However, Hill and Zhang (2012a,b) have questioned the general conclusion that most alleles affect few traits, suggesting it might be an artifact of the statistical limitations of multiple-testing approaches. Furthermore, the distribution of pleiotropic effects of gain-of-function mutations has not been explicitly characterized. It therefore remains to be determined whether new, naturally occurring, allelic variants typically generate phenotypic covariance [i.e., exhibit developmental pleiotropy (Paaby and Rockman 2013)] among large or small sets of traits.
Gene expression traits lie at the interface between genotype and phenotype, and variation in gene expression levels is thought to contribute to intra- and interspecific diversity in other phenotypes (Britten and Davidson 1971; King and Wilson 1975; Carroll 2008; Wittkopp and Kalay 2012). These traits are ideal candidates for characterizing general patterns of pleiotropic effects because, as well as their evolutionary significance, they are associated with methods allowing simultaneous measurement of a large number of traits. The regulation of gene expression traits can be polygenic, with contributions from both cis- and trans-acting factors (West et al. 2007; Skelly et al. 2009; Grundberg et al. 2012). Trans-regulatory variants have been detected less often than cis variants, an observation suggested to reflect the stronger selection against trans-acting mutations (Wray 2007; Skelly et al. 2009). However, there is a known statistical bias against the detection of trans-regulatory expression QTL (eQTL) (Gilad et al. 2008; Nica and Dermitzakis 2013), which might be exacerbated by smaller phenotypic effects of trans variants (Grundberg et al. 2012). Few studies have addressed the natural mutational input to expression phenotypes (Halligan and Keightley 2009), with these suggesting either that few trans-acting mutations generate much of the variation (Denver et al. 2005) or, alternatively, that most mutations affect few expression traits, with very few mutations affecting up to a couple of hundred traits (Rifkin et al. 2005).
Here, we used a microarray platform in Drosophila serrata to measure 11,604 gene expression phenotypes. We assayed expression traits across 41 inbred lines, established from an inbred common ancestor and diverged through the accumulation of new mutations for 27 generations. We first characterized the distribution of mutational heritability and determined the underlying distribution of allelic effects generating that heritability. Then, using a mixed-modeling approach, we determined the mutational covariance among multiple expression traits, in randomly allocated sets, to estimate the frequency of mutational pleiotropy.
Materials and Methods
Populations and data collection
Mutation accumulation lines:
As described in McGuigan et al. (2011), we established 100 mutation accumulation (MA) lines from a laboratory-adapted population of D. serrata, which had previously been subjected to 13 generations of full-sib inbreeding. The MA lines were subsequently maintained through a further 27 generations of brother–sister mating. Based on mutation rate data from D. melanogaster, scaled to the slightly larger D. serrata genome size [0.215 pg (Gregory and Johnston 2008)], we predicted an average per MA line accumulation of 34 mutations, composed of 19.6 [confidence interval: 16.8; 22.8] single-nucleotide mutations (Keightley et al. 2009) and 14.8 [C.I.: 3.4; 52.2] indels and complex mutations (Haag-Liautard et al. 2007).
For each of 45 extant MA lines, on a single day, four replicate brother–sister pairs were initiated in separate vials, and 40 virgin male offspring were collected from these replicate vials (∼10 males per vial). Use of replicate rearing vials, assigned at random across MA lines, ensured that gene expression variation due to microenvironmental variation among vials was not confounded with the variation of interest in this study, the mutational (genetic) variance among inbred lines. Males were held in groups of 5 until 4 days posteclosion, when two replicate RNA extractions per MA line were conducted, each on a random subsample of 20 males. All MA lines were reared simultaneously, and all RNA extractions were performed on the same day. Following snap-freezing on liquid nitrogen, total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA) and purified using RNeasy kits (QIAGEN, Valencia, CA), all according to manufacturer’s instructions.
Outbred population:
To place the mutational variation in context, we compared the inherited (among-line) variance in the MA experiment to that observed among a set of inbred lines established to capture the standing genetic variation in a natural population. Forty-two iso-female lines were founded by wild-caught D. serrata from Brisbane, Australia. Lines were inbred via full-sib mating for 15 generations. As with the MA lines, we used replicate rearing vials that ensured among-line variance was not confounded with within-line common environment variance. In the generation before RNA sampling, each line was propagated in three replicate vials by five virgin female and three virgin male parents. Virgin male offspring were collected from all of these vials on the same day and held in the same fashion as in the MA lines. Two replicate RNA extractions per line were conducted on random subsamples of thirty 3-day-old males per replicate, following the same protocol as in the MA lines. The outbred population lines and RNA collection are described in further detail in Allen et al. (2013).
Microarray hybridization
Microarrays were designed from a D. serrata EST library (Frentiu et al. 2009); sequences (length ≥200 bp, n = 11,383) are available in the Transcriptome Shotgun Archive (GAHN00000000.1) and (length <200 bp, n= 283) directly from the authors upon request. The microarray was manufactured by NimbleGen (Roche) and images were analyzed with NimbleScan (Roche). Microarrays contained 20K random probes plus 11,631 features from ESTs, targeted by five 60-mer oligonucleotide probes; each probe appeared twice on each microarray. A single sample was hybridized to each array, with a single color (Cy-3). Twelve arrays appeared on each slide, and samples were randomly assigned to a slide, with the stipulation that replicate extractions from a single line did not appear on the same slide. cDNA synthesis, labeling, hybridization, and microarray scanning were performed by the Centre for Genomics and Bioinformatics, Bloomington, Indiana.
Quality control of the array data was performed using the BioConductor “oligo package” probe level models (Gentleman et al. 2004; Carvalho and Irizarry 2010; Draghici 2012) and using the experimental metrics report provided by NimbleGen (Roche Nimblegen 2011). Due to poor quality data (e.g., high mean empty signal or strong local stains) in at least one replicate of a line, we discarded several lines, resulting in data from 41 MA lines and 30 outbred population lines retained for analysis. Due to lack of probe replication, 27 features on the microarrays were not included in analyses, resulting in 11,604 traits for further analysis. The expression data are available through the NCBI Gene Expression Omnibus (Edgar et al. 2002; Barrett et al. 2011) (outbred lines, GSE45801; MA lines, GSE49815).
Statistical analyses
Our analytical strategy for this large data set centered on using a series of mixed models of increasing complexity to elucidate the extent of pleiotropy among gene expression traits. Analyses of microarray data often begin with preprocessing to remove technical variation, such as signal intensity variation among different arrays or binding efficiency variation among different probes targeting the same gene. Here, we determined the average log10 signal intensity of the two replicates of each probe on each array, but rather than any further preprocessing, we used the mixed models to partition the variation of interest (among MA or outbred population lines) from other experimental sources of variation (Ayroles and Gibson 2006). As a consequence of our experimental design, overall differences in signal intensity among arrays will contribute to within-line (among-replicate) variation, but not to among-line variance. We chose to analyze the five probe means for each gene, rather than take an overall mean (or median), because our mixed-model approach enabled us to consider all this information in the model rather than discard the variation that may be present among different probes targeting the same gene. Prior to analysis, we standardized (mean = 0; SD = 1) the log10 mean expression data for each trait. The standardization to unit variance facilitated both comparison between data sets (putting the data on the common scale of heritability) and the multivariate analyses, where large differences in the scale of each trait within a single model can inhibit convergence.
We observed a segregating factor in the MA lines that must have been present in the ancestor and was therefore not a product of mutation during the experiment (Supporting Information, File S1). To prevent this factor from contributing to estimates of mutational (among-line) variance, a fixed effect was fitted in all analyses to remove the mean difference in trait expression between the two groups of lines with the two alternative forms of this segregating factor.
Univariate statistical analyses:
Mixed models were implemented within a restricted maximum-likelihood framework in SAS (v. 9.3), using the basic univariate model
| (1) |
where, for each gene, the standardized log10 mean expression, Y, of the kth replicate probe in the jth replicate extraction and hybridization of the ith line is partitioned into variance due to the random factors of (MA or outbred population) Line, representing the among-line variance (VB); Rep(licate), within-line (VW), variance between replicate extractions and hybridizations of the same line; and the residual (error), ε, variance among the five replicate probes per gene. Statistical support for among-line variance of greater than zero was determined through a log-likelihood ratio test (LRT) (1 d.f.) for each gene, comparing two nested models in which the random effect of Line was included or not included. The P-values from this LRT were then used to establish significance at a false discovery rate (FDR) (Benjamini and Hochberg 2000; Storey and Tibshirani 2003) of 0.05, using the MULTTEST procedure in SAS.
We emphasize three points concerning the interpretation of these variance components. First, the among-line variance for each expression trait from model (1) differs from that commonly estimated for standard quantitative traits because it is based on samples of pooled RNA from 20–30 individuals, an approach commonly used for individually small organisms, such as vinegar flies (Rifkin et al. 2005; Ayroles et al. 2009; Yang et al. 2011). Individuals within an inbred line are assumed to be genetically nearly identical after >15 generations of full-sib inbreeding (Lynch and Walsh 1998), and there is therefore little biological difference in how VB would be interpreted here vs. the more common situation where phenotypes are taken from single individuals. Second, the VW variance component contains both a biological and a technical source of error, which are completely confounded. Three or four different vials (common environments) were sampled from each line, then two independent RNA samples were taken, and each sample was hybridized to a different array. The confounding of different sources of biological and technical error into the VW variance component does not adversely affect the estimation of VB. VW is not considered further in our analyses, with the exception that is it used to enable heritability to be calculated (see below).
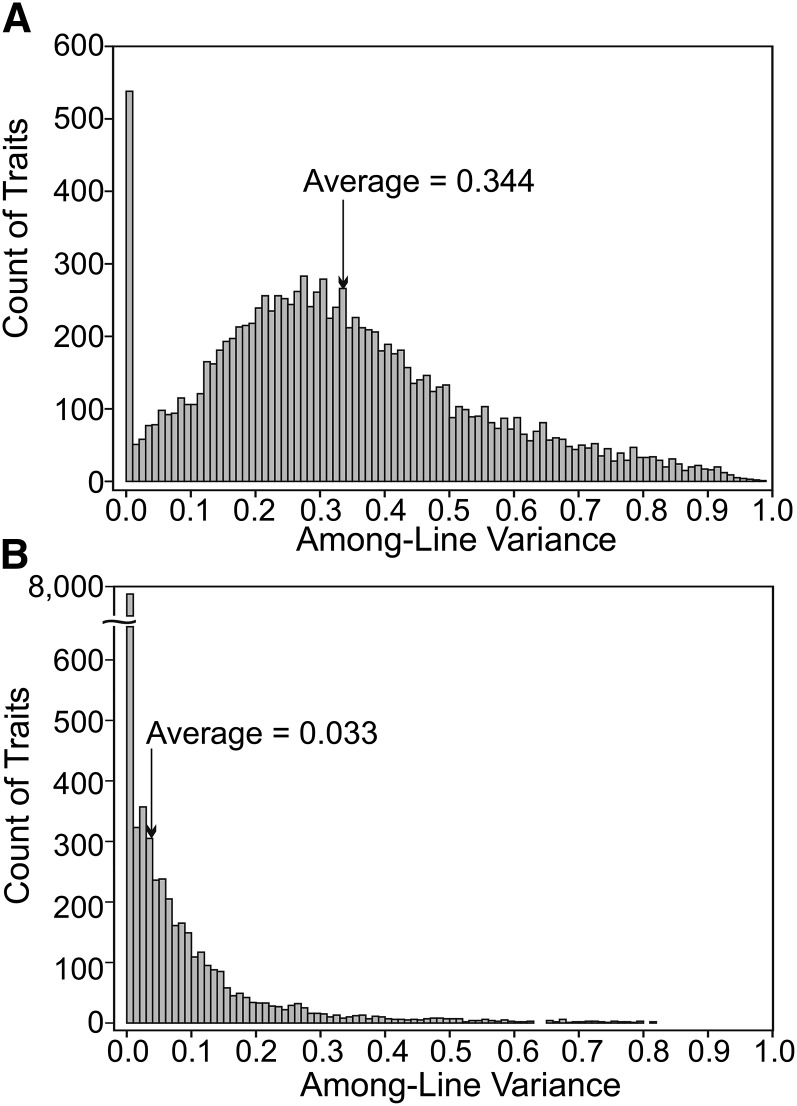
Finally, the residual error was estimated among the means of each of the five replicate probes, an approach that treats the expression of different probes of the same gene as estimates of the same biological trait. We detected consistent differences in mean signal intensity among the five replicate probes of each gene in the outbred population data, but not in the MA line data. We therefore included a fixed effect for probe in model (1) when comparing the two data sets (Figure 1), but not in subsequent analyses of the MA data. Inclusion of this fixed effect resulted in slightly fewer expression traits associated with a zero among-line variance component (67.5% with the fixed effect vs. 70.8% without: compare Figure 1 and Figure 2).
Figure 1.
The distribution of genetic variance for 11,604 expression traits in two experimental populations of Drosophila serrata. (A) Standing genetic variance among 30 inbred lines derived from an outbred population. (B) Genetic variance among 41 MA lines, founded from a single inbred ancestral line, after 27 generations of mutation accumulation. Both graphs are on the scale of broad-sense heritability, in the case of the MA lines, cumulative across the 27 generations.
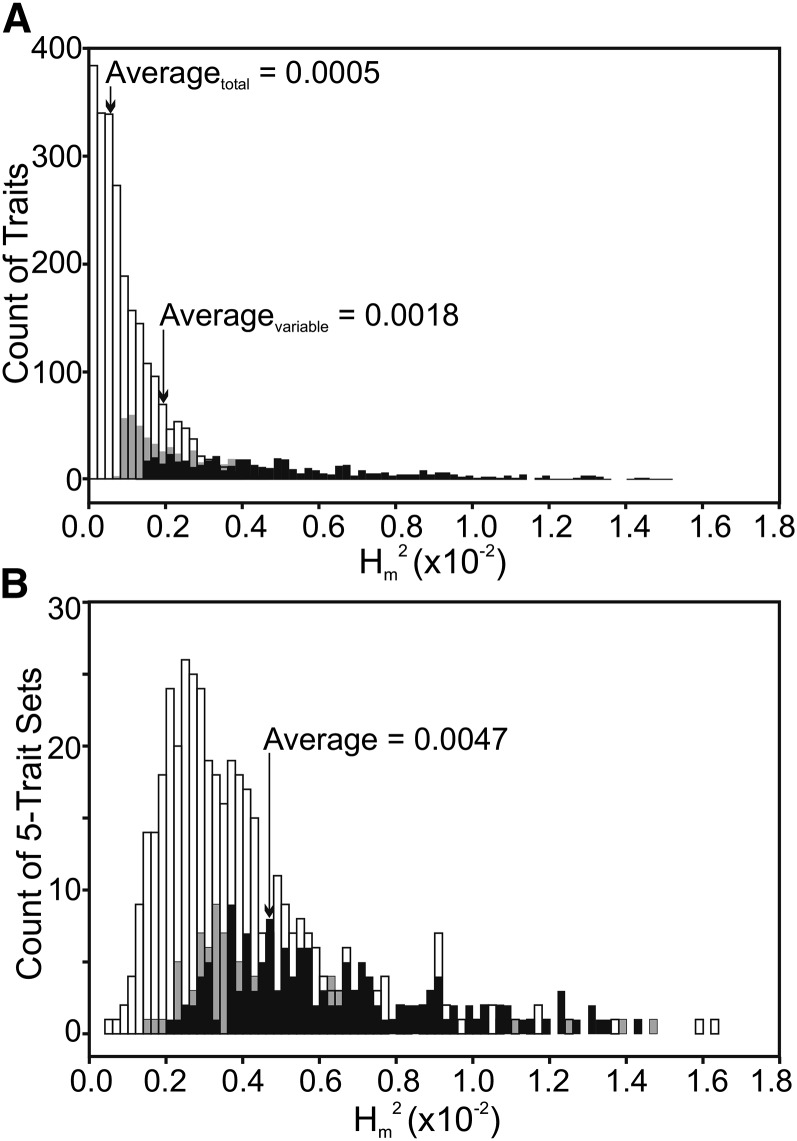
Figure 2.
The distribution of per-generation broad-sense mutational heritability in the 3385 variable traits. Open bars represent nonsignificant estimates, while shaded and solid bars, respectively, represent estimates significant in log-likelihood ratio tests (LRT) at P < 0.05 but not at 5% FDR and estimates that are significant at FDR < 0.05. (A) Mutational heritability of individual expression traits. The average heritabilities of all 11,604 traits (averagetotal) and of the 3385 variable traits (averagevariable) are indicated. (B) The heritability of the common eigenvector, mmax, of each of the 677 five-trait sets created from the 3385 variable traits presented in A.
The mutational variance was estimated as VM = VB/2t, where t was the number of generations (here, 27 generations of mutation accumulation) (Lynch and Walsh 1998). Because VB was estimated on the scale of phenotypic standard deviations (data were variance standardized before analysis), here VM = H2M, which is the mutational heritability. This estimate of H2M is likely to be downwardly biased (underestimated) by the inclusion of technical (among-hybridization and among-probe) as well as environmental (within-line) variance in the denominator.
The shape of the distribution of mutational effects on phenotypes has been characterized relatively few times (Keightley and Halligan 2009), and our experiment provided the opportunity to investigate general patterns across many traits. For the 3385 traits with mixed-model estimates of nonzero among-line variance (see Results) we obtained the best linear unbiased predictors (BLUPs) of each MA line from model (1); BLUPs from the mixed models are estimates of the mutational effects in each line, once other sources of variance have been partitioned appropriately. BLUPs are centered on the population mean expression for each gene, with positive and negative deviations of individual lines representing increased and decreased expression, respectively. To investigate the magnitude of mutational effects, we took the absolute value of each BLUP. Using maximum likelihood, implemented in the UNIVARIATE procedure of SAS, we fitted gamma distributions, with a threshold of 0 (Eyre-Walker and Keightley 2007; Keightley and Halligan 2009), to these absolute BLUP values. This analysis assumes that there are not multiple mutations in the same gene within each line. If mutations are occurring at random throughout the genome, then over the relatively short duration of this MA experiment (27 generations), it is reasonable to assume that mutations accumulating within a given MA line occurred in different genes. It is important to note, however, that our analysis cannot distinguish a single mutation from multiple mutations affecting a polygenic trait. Again, given the relatively few mutations expected in each line (∼34; see above), unless an expression trait is highly polygenic it is likely that single mutations will contribute to line BLUP deviations in most cases.
Measuring mutational pleiotropy:
To ensure that we did not downwardly bias the detection of pleiotropy (Hill and Zhang 2012a,b), we included all traits with nonzero estimates of variance (3385) in our analyses of mutational covariance, irrespective of statistical confidence in the estimate of mutational variance for these individual traits. We build up a picture of the general patterns of covariance among traits (and thus prevalence and extent of pleiotropy), beginning by considering the simplest form of pleiotropy, that affecting just 2 traits, and building to consider sets of 5 traits. Multivariate mixed models enabled us to estimate the genetic covariance between expression traits in a variance-component form that has a standard interpretation for any pair in a multivariate set of quantitative traits and has well-known statistical properties (Lynch and Walsh 1998). In both the bivariate and the multivariate analyses (outlined below), traits are assigned to their bi- or multivariate trait set completely randomly, without reference to their biological function. There is no a priori expectation that these traits, chosen at random from across the transcriptome, should covary, unless pleiotropy is extensive.
We did not exhaustively sample the 5,725,727 bivariate combinations of variable expression traits; rather, we looked for general patterns through a subsample of 6768 random pairs of traits, a large, arbitrary number of random pairs representing 0.1% of all possible pairs. Each bivariate pair was subjected to the multivariate form of model (1),
| (2) |
where Zl and Zr are design matrices for the line and replicate within-line random effects. We modeled the covariance structure among traits at the line (δl) and replicate (δr) levels, using unstructured 2 × 2 covariance matrices, and ε was a diagonal matrix containing the residual (among-probe) variances for each trait. Standing variation was partitioned out via a fixed effect as in model (1). We again used LRTs to determine statistical support for mutational covariance between two traits, comparing the log-likelihood of the unconstrained model (in which covariance was estimated) with that of a model where the covariances were constrained to zero, and only the two individual trait variances were estimated (a diagonal matrix model). The P-values from these LRT were used to determine the 5% FDR level.
The next step in our characterization of pleiotropy was a set of multivariate analyses. Each of the 3385 variable traits was assigned to one of 677 five-trait sets. Traits were assigned at random and each trait was assigned to only one of the 677 matrices. A trait set size of five was chosen as a compromise between model convergence (models with more traits sometimes did not converge, and convergence times were prohibitively long) and the desirability of searching for covariance among as many traits as are often used in evolutionary studies of standard metric traits.
The five-trait sets were subjected to a multivariate linear mixed model of the same form as model (2), but where the covariance (δl and δr) among traits was modeled using a reduced rank factor-analytic structure:
| (3) |
Here, ψ was a diagonal matrix containing specific variances for each trait (i.e., the mutational variance unique to a trait and not shared with other traits in the model), and Λ was a lower triangular matrix of factor loadings for a single factor capturing the common variance among the five traits (Meyer 2009; McGuigan and Blows 2010). For the among-line model term, multiplying Λ by its transpose (as in Equation 3) gives the mutational covariance matrix, M, which can be diagonalized. In these models, this matrix has a single dimension, represented by the eigenvector mmax, with the amount of mutational variance in the trait set that is due to the pleiotropic covariance described by the eigenvalue of mmax. The presence of a common factor (nonzero eigenvalue of mmax) reveals pleiotropic effects on at least two of the five traits.
We used two approaches to test for significant mutational covariance among traits. First, we used a LRT to contrast a model that estimated the common mutational variance with a model in which only trait-specific mutational variances were estimated. This LRT has 5 d.f. and can be considered a conservative test for the presence of mutational covariance because the precise mixture of chi-square distributions is unknown (Self and Liang 1987; Littell et al. 2006). Second, we used a randomization test in which the data for each trait were shuffled among MA lines, while keeping replicate measures within each line together. This shuffling approach ensured that estimates of variance of each trait remained the same at the among-line, within-line, and residual levels, but covariances among traits were disrupted. This shuffled data set, subdivided into the same five-trait sets, was then subjected to analysis through the multivariate (factor analytic) mixed model in exactly the same way as described above for the observed data. We used paired t-tests to determine whether the observed and shuffled data differed in their estimates of mutational variance, represented by the eigenvalue of mmax.
We also used a paired t-test to determine whether the observed and shuffled data differed in the extent of pleiotropy. We normalized the vector mmax to unit length (mmaxT mmax = 1) and took the square of the largest contribution of an expression trait to this normalized vector as a metric of the extent of pleiotropy of each five-trait matrix. Trait contributions were squared to allow us to consider both positive and negative covariance among traits. This metric scales from 1.0, where only one trait contributed to mmax and there was no mutational covariance with the other traits, down to 0.2, where all five traits have equal variances and contribute equally to the eigenvector.
To estimate the mutational heritability of mmax, we first determined the phenotypic variance in mmax: VP = VB + VW + VE, where VB was the eigenvalue of mmax, and the variance at the within line (VW) and residual (VE) levels was determined by projecting the normalized vector through the within-line and residual covariance matrices obtained from model (2). We then applied the equation
| (4) |
where t is the number of generations of mutation accumulation (here, 27 generations).
Results
Characteristics of mutational variation in individual expression traits
The distributions of standing genetic and mutational variance were markedly different from one another (Figure 1). As observed in other taxa (Ayroles et al. 2009; Skelly et al. 2009; Nica and Dermitzakis 2013), most of the 11,604 expression traits were heritable (8782 had significant among-line variance at 5% FDR) in the outbred population of D. serrata (Figure 1A). A small proportion (489, 4.2%) of traits were invariant (i.e., had an among-line variance component of zero in the mixed-model analysis). In contrast, the mutational variance had a distinct, L-shaped distribution, where the majority of traits were invariant among MA lines (Figure 1B).
The lack of genetic variation among MA lines did not reflect nonexpression of these genes. Almost all genes (90.2%) were expressed in all 41 MA lines (Figure S1), consistent with other data on whole-body expression of adult male Drosophila (Ayroles et al. 2009; Wyman et al. 2010). Rather, the invariance of transcript abundance among MA lines was consistent with low segregating variation in the inbred ancestral line and with the estimated 34 mutations per MA line being insufficient to generate detectable mutational variance in most genes. It is important to note that our results do not imply that traits invariant among MA lines were not subject to mutation: the heritability of these traits in the lines derived from the outbred population (Figure 1A) indicates that they are, but, by chance, mutations were not observed for them in the current study.
Overall, the average heritability of all expression traits was 0.0005; considering only the 3385 traits with nonzero estimates of among-line variance in model (1) (i.e., the traits for which there is some evidence that a mutation might have occurred), the average mutational heritability was 0.0018 (Figure 2A). Log-likelihood ratio tests supported significant heritability for 8.9% (1035) of all traits, 533 (4.6%) of which remained significant at a 5% FDR.
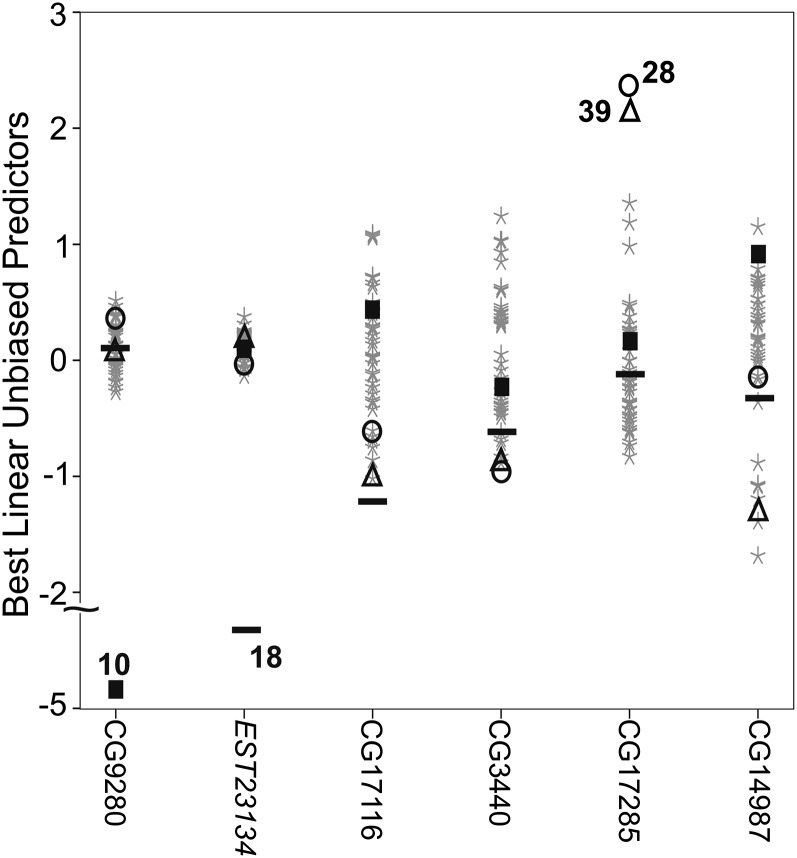
The distribution of phenotypic effects of individual MA lines is illustrated for six expression traits in Figure 3. For some, relatively few, traits (e.g., CG9280 and EST23134), the among-line variance (H2M = 0.014 and 0.011, respectively) was generated by a large phenotypic deviation (> 2 phenotypic standard deviations) of one MA line (lines 10 and 18, respectively, in Figure 3), reflected in gamma shape parameters of <1 (k = 0.81 and 0.66, respectively). Other traits (e.g., Figure 3; CG17285, k = 1.01, H2M = 0.012; CG14987, k = 1.26, H2M = 0.011) were characterized by multiple distinct clusters of lines, suggesting putative mutations of moderate effect (1–2 phenotypic SD) on trait variance. Finally, for many traits (e.g., Figure 3; CG3440, k =2.54, H2M = 0.009; CG17116, k = 1.55, H2M = 0.009), the among-line variance was underlain by a relatively even dispersion of BLUPs, consistent with different putative mutations, each with relatively small effects, in different lines.
Figure 3.
Illustration of mutational variation among MA lines. The best linear unbiased predictors (BLUPs) of each line (i.e., points on the graph) are plotted for six traits with similar mutational heritabilities. Traits are identified by their D. melanogaster gene ortholog annotation, with the exception of EST23134, for which no D. melanogaster ortholog was found. BLUPs are deviations from the population mean, here, on the scale of phenotypic standard deviations, with positive (increased expression) and negative (decreased expression) deviations. Some individual MA lines (10, square; 18, bar; 28, circle; and 39, triangle) are uniquely identified for illustration, and the same symbol is used to indicate the BLUPs of these lines for each gene.
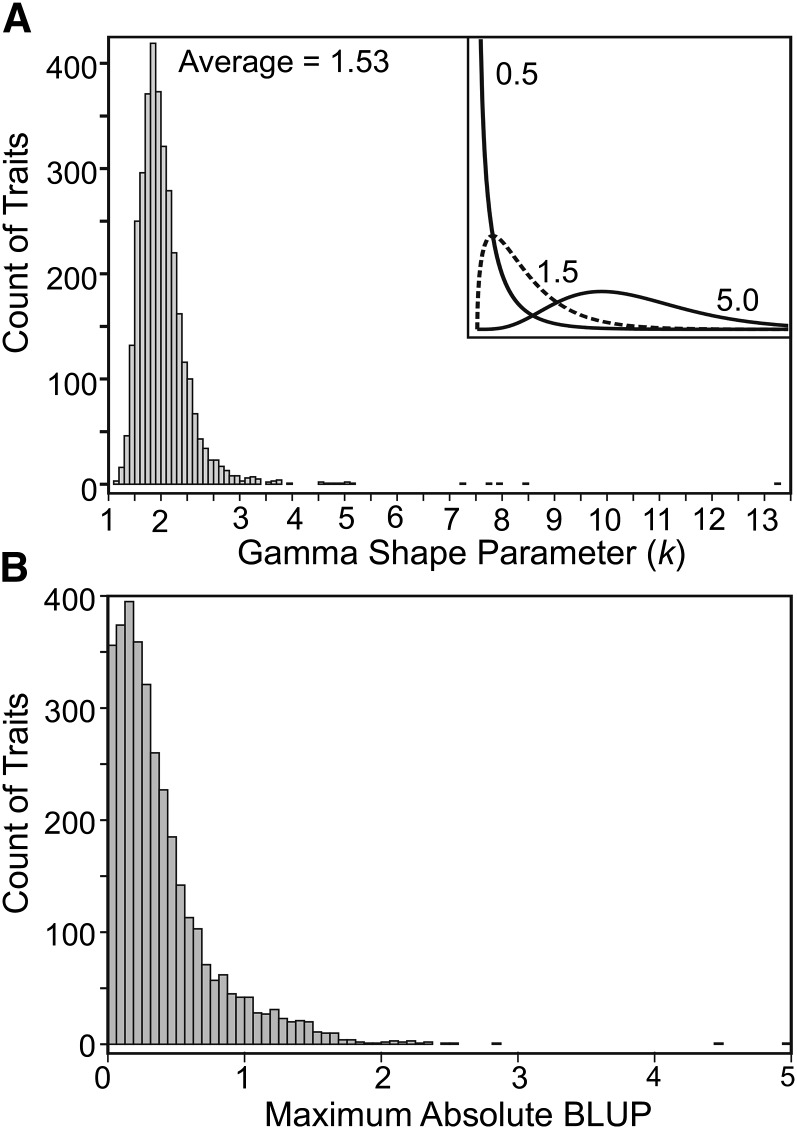
Figure 4 illustrates the general patterns of mutational effects, showing the distribution of the gamma shape parameter (k) (Figure 4A) and the largest individual line BLUP (Figure 4B) for all 3385 variable traits. The observed values of k ranged from 0.66 to 13.29 (Figure 4A). An exponential curve has k = 1 (inset in Figure 4A); fitness is both expected and observed to have a distribution of effects that is more leptokurtic (k < 1) than exponential, where few mutations have large effects while most mutations have small effects (Halligan and Keightley 2009). For the expression traits in this study, the average k was significantly >1 (average = 1.53; t-test, t3,384 = 57.186, P < 0.001), indicating that the distribution of effects was less leptokurtic than observed for fitness. Indeed, only 197 traits (5.8% of variable traits) had k < 1. The inference that the observed mutational variance was typically due to small effect mutations was further supported by the observation that the largest mutational effect sizes were <1 phenotypic standard deviation for most expression traits (3112, corresponding to 91.9% of variable traits) (Figure 4B).
Figure 4.
The distribution of the mutational effects for individual traits. (A) The distribution of gamma shape parameters for each trait, including an illustration of curve shapes for different shape parameter values (inset). (B) The distribution of the largest absolute best linear unbiased predictors (i.e., deviations from the population mean breeding value), for each trait, plotted as phenotypic standard deviations.
Mutational pleiotropy
We began our characterization of the extent of pleiotropy with its simplest form: pleiotropy affecting just two traits. Sampling 6768 (0.1%) of the possible >5.7 × 106 bivariate pairs of variable traits, we found 35 significant (FDR < 0.05) covariances. This suggests that pleiotropy might be uncommon. However, a problem with inferring the presence of pleiotropy is the generally low statistical power to detect contributions from alleles of small phenotypic effect (Wagner and Zhang 2011; Hill and Zhang 2012b; Paaby and Rockman 2013), such as those observed in this experiment (Figure 4).
Multivariate mixed modeling of the genetic covariance among multiple traits substantially improved the detectability of pleiotropy over bivariate analyses. We generated 677 five-trait sets and implemented factor analytic modeling, estimating specific variances for each trait, and a single common factor. The LRT analysis supported the presence of a common factor, indicative of mutational covariance and pleiotropy, for 245 (36.2%) of the 677 random five-trait matrices, 145 (21.4%) of which remained significant at 5% FDR (Figure 2B). In contrast, when we shuffled the data, breaking covariances, only 3.2% of the same five-trait matrices had significant covariance, and none remained significant at FDR < 0.05. Furthermore, the mutational variance in the common factor, mmax, was significantly lower in the shuffled than in the observed data (paired t-test: t = 9.801, d.f. = 676, P < 0.0001).
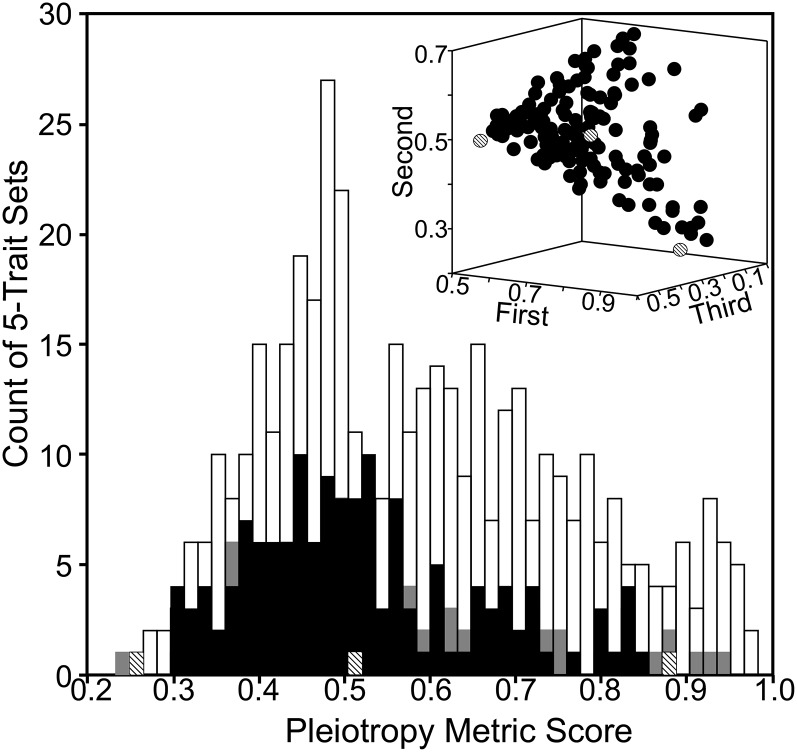
We characterized the extent of pleiotropy within each five-trait matrix as the squared maximum contribution of any trait to the normalized mmax, a metric that ranges from 1.0 (no pleiotropy) to 0.2 (all five traits shared mutational covariance to the same extent). The distribution of this pleiotropy metric suggested that pleiotropy, although manifest in many matrices, typically did not simultaneously affect all five traits (Figure 5). The average pleiotropy metric score, 0.57 (0.52 for matrices significant at FDR), was significantly lower (more pleiotropic) than for the shuffled data (paired t-test: t = 5.083, d.f. = 676, P < 0.0001). Both positive and negative mutational covariances occurred among traits (Figure 6), although positive covariance between the two traits with the greatest contribution to mmax was observed significantly more frequently than negative covariances (95 vs. 50 cases; binomial test: P = 0.0002).
Figure 5.
Multivariate mutational covariance among expression traits. The pleiotropy metric (squared maximum contribution to normalized mmax vector) is plotted for each of the 677 five-trait matrices. Open, shaded, and solid bars indicate significance, as in Figure 2. The inset illustrates the three largest absolute loadings on mmax for the 145 matrices with significant pleiotropy at the FDR (solid bars in main plot); the shaded circles illustrate (left to right) the trait loadings of matrices with the smallest (0.26), average (0.52), and largest (0.89) pleiotropy metric scores (as indicated by shaded bars in the main plot).
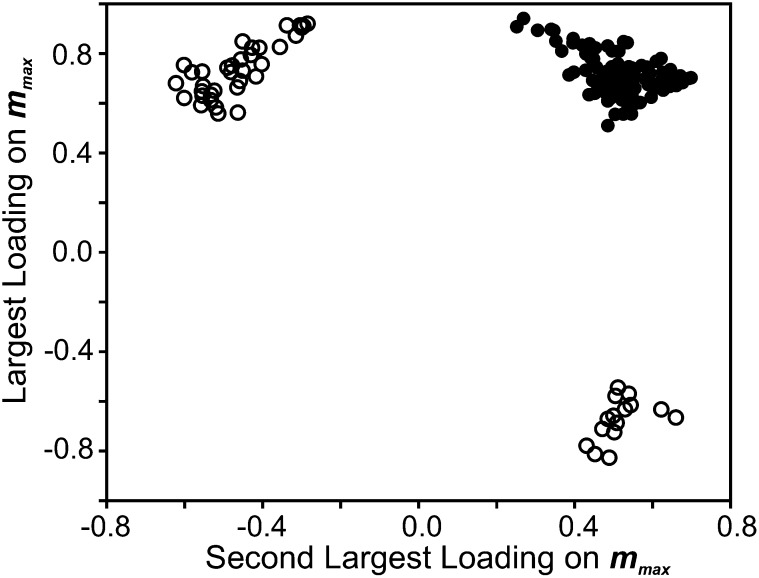
Figure 6.
The distribution of the direction of mutational covariances. Plotted are the largest (y-axis) and the second largest (x-axis) contributions of traits to the normalized mmax eigenvectors of the 145 matrices that were significant at 5% FDR. Negative and positive covariances are revealed by opposing directions of contribution (open circles) and coincident directions of contribution (solid circles), respectively. Directions of trait loading on mmax do not correspond to up- vs. downregulation of gene expression. Eigenvector direction can be arbitrary, such that the largest loading is typically positive. Thus, negative loadings from the two traits with the largest two contributions were not observed in this subset of the data, but would reveal the same positive mutational covariance as positive loadings of two traits with the largest contributions to mmax (i.e., solid circles).
Positive associations between the magnitude of effects on an individual trait and the number of traits affected by a gene have been observed in gene knockout data (Wang et al. 2010) and for QTL underlying divergence among populations (Wagner et al. 2008). We did not observe a relationship between the magnitude of a mutation’s effect on a trait (characterized by the trait’s mutational effect kurtosis: Figure 4A) and the pleiotropic variance in that trait (characterized by the loading of the trait on the normalized pleiotropy eigenvector mmax) (r = −0.028, d.f. = 3385, P = 0.105).
Number of traits affected by a pleiotropic mutation
Based on the observations of FDR significant covariances between traits in 0.52% of our bivariate analyses and in 21% of multivariate analyses, we implemented probability calculations to predict a range of values for the number of traits affected by a single putative pleiotropic mutation. Such sets of coaffected expression traits correspond to variational modules (Wagner et al. 2007). Our goal here was not to identify and characterize specific variational modules in our data, but to provide some general indication of how many traits might typically be affected by a pleiotropic mutation, under some simplifying assumptions.
Assuming any trait was affected by only a single mutation and that each pleiotropic mutation affected equal numbers of traits (i.e., all modules are the same size), let m be the number of variational modules and n be the number of traits in a module, so that m × n = 3384 (one less than the number of variable traits to allow for bivariate sampling). In a bivariate sample, the probability of sampling two traits from the same module is then n(n – 1)/33842, which scales across all variational modules to m[n(n – 1)/33842]. Substituting for m and solving for n, given the observed frequency of 1 in 193 significant bivariate covariances, results in an estimate of 18.5 traits in each module.
The multivariate modeling indicated that 21% (1 in every 4.8) of five-trait matrices displayed significant mutational pleiotropy. With five traits, there are 10 potential pleiotropic bivariate covariances that could have generated the observed statistically significant mutational pleiotropy. A minimum of one bivariate covariance must be present for a significant mmax vector to be found, and therefore there must be covariance between at least 1 in 48 bivariate pairs of traits. Again solving m[n(n – 1)/33842], we obtain an estimate of 70.5 traits per variational module. As we discuss further below, both the bivariate and the multivariate estimates of average variational module size will be downwardly biased.
Discussion
Although the vast majority of expression traits were heritable in an outbred population of our study species, mutations did not affect the expression of more than two-thirds of our traits, lending no support to universal pleiotropy across the transcriptome, consistent with recent analyses of gene knockout data and QTL mapping of crosses between divergent populations (Wagner and Zhang 2011). Nonetheless, our data suggest that pleiotropic mutations might often generate genetic covariance among traits. An overall assessment of the extent of pleiotropy generated by new mutations can be gained by contrasting the results of the univariate, bivariate, and multivariate analyses. First, the average mutational heritability of the five-trait mmax was 2.6 times higher than for individual traits analyzed in isolation (Figure 2), and 58.8% of the total mutational variance, summed across all 3385 variable traits, was accounted for by the sum of the pleiotropic mutational variances (i.e., the eigenvalues of mmax). These summary measures, which are not affected by detection limits imposed by statistical significance thresholds (Hill and Zhang 2012b), suggest that pleiotropy is widespread. Second, at 5% FDR, we inferred pleiotropy in 21% of five-trait matrices, but only in 0.52% of bivariate covariances. This suggests bivariate covariances that do not pass stringent FDR often cumulatively contribute to significant variance in mmax, as reflected in the distribution of trait loadings on mmax vectors (inset in Figure 5).
Taking the observed frequency of mutational covariance, and assuming that equal numbers of traits are affected by different pleiotropic mutations, we use a simple probabilistic argument to infer the number of pleiotropically related traits. Based only on the frequency with which FDR significant bivariate covariances were observed, we estimate that the average module size is 19; based on the frequency of FDR significant mmax (in the multivariate analyses), we predict an average module size of 70. Because these predictions are based only on the FDR significant results, they are likely to be underestimates, a problem that the bivariate analysis was particularly sensitive to. Further, for the multivariate analysis, we assumed that only 1 of the 10 bivariate covariances in the five-trait matrices contributed to the FDR significance of mmax; if multiple traits in a five-trait set covary, module size will have been underestimated.
In reality, there is likely to be considerable variation among pleiotropic mutations in the number of traits affected (Wang et al. 2010; Runcie and Mukherjee 2013). To explain the observed frequency of pleiotropy among D. serrata expression traits, at least some mutations must affect a substantial number of traits (>70); we cannot distinguish between a distribution with few very large modules and many relatively small ones vs. a more normal distribution with module sizes distributed about a fairly large mean (∼70). Mapping studies have identified individual trans eQTL with pleiotropic effects on many expression traits [e.g., up to 2528 (West et al. 2007)]. Analyses of null mutations have inferred that the average proportion of traits affected by a null allele is 2–11% (Wang et al. 2010). For the expression trait set analyzed here, an average variational module size of 70 corresponds to effects on 0.6% of all expression traits, or 2.1% of variable traits, at the lower range reported for null mutations.
Cis-acting regulatory factors might be expected to have lower pleiotropy than trans-acting factors, with effects restricted to neighboring genes. However, a mutation in the cis-regulatory region of a gene that acted as a trans-regulatory factor for other genes could potentially affect many traits. Analysis of mutational contribution to gene expression in Caenorhabditis elegans suggested the observed variation in expression of 660 genes might be due to few trans-acting factors (Denver et al. 2005). Our data do not allow us to determine the regulatory nature of the mutations generating the observed covariances.
Mutations affecting gene expression were predominantly of small phenotypic effect, with only ∼8% of mutationally variable traits affected by a mutation with an effect size of >1 phenotypic standard deviation. Alleles with individually small effects have been implicated as contributing much of the standing genetic variance in other quantitative traits (Yang et al. 2010). It is important to remember, however, that our experimental design cannot exclude the possibility that mutations with larger effects on expression, and which also had detrimental pleiotropic effects on fitness, were removed by natural selection before we could observe them. Although MA experiments aim to minimize selection, mutations causing a >10% decline in fitness are unlikely to accumulate (Lynch et al. 1999). Viability selection operating on our set of MA lines caused divergence in wing size and shape between lines that went extinct vs. those that remained extant at the end of the experiment (and were the subset analyzed for expression trait variation here) (McGuigan and Blows 2013). Mutations associated with very low fitness might differ from surveyed mutations in their distribution of allelic effects, including the extent of pleiotropy. We did not observe any association between the effect size of a mutation and the extent of pleiotropy within each five-trait set; however, our ability to determine whether a relationship exists was limited by our analytical approach.
The range of mutational heritabilities observed in the current study corresponds well to the range reported for life-history and morphological traits [0.0005–0.005 (Lynch et al. 1999)], but was two orders of magnitude larger than previously reported for gene expression traits (Rifkin et al. 2005; Landry et al. 2007). Using a similar approach to that of our study, Rifkin et al. (2005) investigated mutational heritability for two early life-history stages in 12 D. melanogaster MA lines that had accumulated mutations for ∼200 generations. Although the number of traits with significant mutational variance (32% of ESTs) was greater than what we observed for adult male D. serrata (4.6%), as might be expected given the longer time frame over which mutations accumulated, the median mutational heritabilities were much lower: 0.000023 and 0.000025 for the two life-history stages. Rifkin et al. (2005) scaled their estimate of within-line (residual) variance by the number of individual flies from which RNA was extracted for each hybridization; when we applied this scaling approach, our estimate of median mutational heritability (0.000309) was still an order of magnitude greater than that reported by Rifkin et al. (2005). Given the similarity in experimental design, it is not clear to us why the mutational heritability of expression traits in D. melanogaster is lower than that reported here for D. serrata.
Estimates of mutational variance from mutation accumulation studies are rarely matched with simultaneous estimates of the standing genetic variance in the same traits, measured under the same conditions (Houle et al. 1996). It is well established that expression traits are heritable (Skelly et al. 2009). However, heritability inferences typically come from experimental designs such as QTL mapping of crosses between phenotypically divergent strains (Schadt et al. 2003; Skelly et al. 2009), rather than from classical quantitative genetic designs (but see Ayroles et al. 2009), such as the inbred lines design typically used to characterize mutational variance (Lynch and Walsh 1998; Halligan and Keightley 2009). In this study, we have quantified gene expression using a microarray platform that was specifically designed for the study species, employing well-matched quantitative genetic breeding designs in two populations, one capturing outbred, standing genetic variation in a population and the other capturing the new mutations that contribute to that variation. The measurement of the same large set of expression traits in the MA and outbred lines provides the opportunity for a formal comparison between patterns of mutational (VM) and standing genetic (VG) variance. The strength of selection acting on mutations affecting quantitative traits can be estimated as s = VM/VG, where s is the selection coefficient of the mutation in heterozygous form (Barton 1990; Houle et al. 1996). Selection coefficients can be estimated both for individual traits and for the multivariate trait combinations that have been established here to be affected by pleiotropic mutations. A detailed analysis of the strength of selection acting on these mutations, and how this selection is influenced by the extent of pleiotropy, will be presented in a future article.
In conclusion, phenome-wide assessment of mutational variance did not provide support for universal pleiotropy, but did establish that pleiotropy generated by new, naturally occurring mutations of relatively small effect is widespread. Pleiotropy can be expected to increase among traits such as morphological and life-history traits, which will aggregate the effects of numerous underlying gene expression traits and therefore have larger mutational targets (Houle 1998). The frequency of mutational pleiotropy among small sets of randomly associated expression traits is consistent with recent findings that the vast majority of the multivariate genetic variance of quantitative traits is often restricted to a limited number of phenotypic dimensions (Kirkpatrick 2009; Walsh and Blows 2009).
Supplementary Material
Acknowledgments
We acknowledge the dedication and hard work of people who maintained the experimental populations and collected RNA samples, particularly J. Thompson, C. Oesch-Lawson, D. Petfield, F. Frentiu, O. Sissa-Zubiate, and T. Gosden.
Footnotes
Communicating editor: N. H. Barton
Literature Cited
- Allen S. L., Bonduriansky R., Chenoweth S. F., 2013. The genomic distribution of sex-biased genes in Drosophila serrata: X-chromosome demasculanisation, feminisation, and hyper-expression in both sexes. Genome Biol. Evol. 5: 1986–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles J. F., Gibson G., 2006. Analysis of variance of microarray data, pp. 214–233 in DNA Microarrays, Part B: Databases and Statistics, edited by Kimmel A., Oliver B. Elsevier Academic Press Inc., San Diego, CA. [Google Scholar]
- Ayroles J. F., Carbone M. A., Stone E. A., Jordan K. W., Lyman R. F., et al. , 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Troup D. B., Wilhite S. E., Ledoux P., Evangelista C., et al. , 2011. NCBI GEO: archive for functional genomics data sets-10 years on. Nucleic Acids Res. 39: D1005–D1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., 1990. Pleiotropic models of quantitative variation. Genetics 124: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 2000. On the adaptive control of the false discovery fate in multiple testing with independent statistics. J. Educ. Behav. Stat. 25: 60–83. [Google Scholar]
- Britten R. J., Davidson E. H., 1971. Repetitive and non-repetitive DNA sequences and a speculation on origins of evolutionary novelty. Q. Rev. Biol. 46: 111–138. [DOI] [PubMed] [Google Scholar]
- Carroll S. B., 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. [DOI] [PubMed] [Google Scholar]
- Carvalho B. S., Irizarry R. A., 2010. A framework for oligonucleotide microarray preprocessing. Bioinformatics 26: 2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver D. R., Morris K., Streelman J. T., Kim S. K., Lynch M., et al. , 2005. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat. Genet. 37: 544–548. [DOI] [PubMed] [Google Scholar]
- Draghici, S., 2012 Statistics and Data Analysis for Microarrays Using R and Bioconductor. CRC Press, Taylor and Francis Group/Chapman & Hall, Boca Raton, FL. [Google Scholar]
- Edgar R., Domrachev M., Lash A. E., 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., 2010. Genetic architecture of a complex trait and its implications for fitness and genome-wide association studies. Proc. Natl. Acad. Sci. USA 107: 1752–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618. [DOI] [PubMed] [Google Scholar]
- Frentiu F. D., Adamski M., McGraw E. A., Blows M. W., Chenoweth S. F., 2009. An expressed sequence tag (EST) library for Drosophila serrata, a model system for sexual selection and climatic adaptation studies. BMC Genomics 10: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., et al. , 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y., Rifkin S. A., Pritchard J. K., 2008. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 24: 408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory T. R., Johnston J. S., 2008. Genome size diversity in the family Drosophilidae. Heredity 101: 228–238. [DOI] [PubMed] [Google Scholar]
- Grundberg E., Small K. S., Hedman A. K., Nica A. C., Buil A., et al. , 2012. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 44: 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag-Liautard C., Dorris M., Maside X., Macaskill S., Halligan D. L., et al. , 2007. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445: 82–85. [DOI] [PubMed] [Google Scholar]
- Halligan D. L., Keightley P. D., 2009. Spontaneous mutation accumulation studies in evolutionary genetics. Annu. Rev. Ecol. Evol. Syst. 40: 151–172. [Google Scholar]
- He X. L., Zhang J. Z., 2006. Toward a molecular understanding of pleiotropy. Genetics 173: 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., 2010. Understanding and using quantitative genetic variation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Zhang X. S., 2012a Assessing pleiotropy and its evolutionary consequences: pleiotropy is not necessarily limited, nor need it hinder the evolution of complexity. Nat. Rev. Genet. 13: 296. [DOI] [PubMed] [Google Scholar]
- Hill W. G., Zhang X. S., 2012b On the pleiotropic structure of the genotype-phenotype map and the evolvability of complex organisms. Genetics 190: 1131–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D., 1998. How should we explain variation in the genetic variance of traits? Genetica 103: 241–253. [PubMed] [Google Scholar]
- Houle D., Morikawa B., Lynch M., 1996. Comparing mutational variabilities. Genetics 143: 1467–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T., Barton N., 2005. Theoretical models of selection and mutation on quantitative traits. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360: 1411–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Halligan D. L., 2009. Analysis and implications of mutational variation. Genetica 136: 359–369. [DOI] [PubMed] [Google Scholar]
- Keightley P. D., Trivedi U., Thomson M., Oliver F., Kumar S., et al. , 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19: 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M. C., Wilson A. C., 1975. Evolution at two levels in humans and chimpanzees. Science 188: 107–116. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M., 2009. Patterns of quantitative genetic variation in multiple dimensions. Genetica 136: 271–284. [DOI] [PubMed] [Google Scholar]
- Landry C. R., Lemos B., Rifkin S. A., Dickinson W. J., Hartl D. L., 2007. Genetic properties influencing the evolvability of gene expression. Science 317: 118–121. [DOI] [PubMed] [Google Scholar]
- Lawson H. A., Cady J. E., Partridge C., Wolf J. B., Semenkovich C. F., et al. , 2011. Genetic effects at pleiotropic loci are context-dependent with consequences for the maintenance of genetic variation in populations. PLoS Genet. 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littell R. C., Milliken G. A., Stroup W. W., Wolfinger R. D., Schabenberger O., 2006. SAS System for Mixed Models. SAS Institute, Cary, NC. [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Lynch M., Blanchard J., Houle D., Kibota T., Schultz S., et al. , 1999. Spontaneous deleterious mutation. Evolution 53: 645–663. [DOI] [PubMed] [Google Scholar]
- McGuigan K., Blows M. W., 2010. Evolvability of individual traits in a multivariate context: partitioning the additive genetic variance into common and specific components. Evolution 64: 1899–1911. [DOI] [PubMed] [Google Scholar]
- McGuigan K., Blows M. W., 2013. Joint allelic effects on fitness and metric traits. Evolution 67: 1131–1142. [DOI] [PubMed] [Google Scholar]
- McGuigan K., Petfield D., Blows M. W., 2011. Reducing mutation load through sexual selection on males. Evolution 65: 2816–2829. [DOI] [PubMed] [Google Scholar]
- Meyer K., 2009. Factor-analytic models for genotype x environment type problems and structured covariance matrices. Genet. Sel. Evol. 41: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nica A. C., Dermitzakis E. T., 2013. Expression quantitative trait loci: present and future. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368: 20120362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby A. B., Rockman M. V., 2013. The many faces of pleiotropy. Trends Genet. 29: 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin S. A., Houle D., Kim J., White K. P., 2005. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature 438: 220–223. [DOI] [PubMed] [Google Scholar]
- Roche NimbleGen, Inc. 2011 NimbleGen Software: Guide to Interpreting the Experimental Metrics Report. Roche, Madison, Wisconsin. [Google Scholar]
- Runcie D. E., Mukherjee S., 2013. Dissecting high-dimensional phenotypes with Bayesian sparse factor analysis of genetic covariance matrices. Genetics 194: 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E. E., Monks S. A., Drake T. A., Lusis A. J., Che N., et al. , 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302. [DOI] [PubMed] [Google Scholar]
- Self S. G., Liang K.-Y., 1987. Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J. Am. Stat. Assoc. 82: 605–610. [Google Scholar]
- Sivakumaran S., Agakov F., Theodoratou E., Prendergast J. G., Zgaga L., et al. , 2011. Abundant pleiotropy in human complex diseases and traits. Am. J. Hum. Genet. 89: 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelly D. A., Ronald J., Akey J. M., 2009. Inherited variation in gene expression. Annu. Rev. Genom. Hum. Genet. 10: 313–332. [DOI] [PubMed] [Google Scholar]
- Stearns F. W., 2010. One hundred years of pleiotropy: a retrospective. Genetics 186: 767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. L., 2000. Evolutionary developmental biology and the problem of variation. Evolution 54: 1079–1091. [DOI] [PubMed] [Google Scholar]
- Storey J. D., Tibshirani R., 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G. P., Zhang J. Z., 2011. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat. Rev. Genet. 12: 204–213. [DOI] [PubMed] [Google Scholar]
- Wagner G. P., Pavlicev M., Cheverud J. M., 2007. The road to modularity. Nat. Rev. Genet. 8: 921–931. [DOI] [PubMed] [Google Scholar]
- Wagner G. P., Kenney-Hunt J. P., Pavlicev M., Peck J. R., Waxman D., et al. , 2008. Pleiotropic scaling of gene effects and the ’cost of complexity’. Nature 452: 470–473. [DOI] [PubMed] [Google Scholar]
- Walsh B., Blows M. W., 2009. Abundant genetic variation + strong selection = multivariate genetic constraints: a geometric view of adaptation. Annu. Rev. Ecol. Evol. Syst. 40: 41–59. [Google Scholar]
- Wang Z., Liao B. Y., Zhang J. Z., 2010. Genomic patterns of pleiotropy and the evolution of complexity. Proc. Natl. Acad. Sci. USA 107: 18034–18039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M. A. L., Kim K., Kliebenstein D. J., van Leeuwen H., Michelmore R. W., et al. , 2007. Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics 175: 1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp P. J., Kalay G., 2012. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13: 59–69. [DOI] [PubMed] [Google Scholar]
- Wray G. A., 2007. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8: 206–216. [DOI] [PubMed] [Google Scholar]
- Wyman M. J., Agrawal A. F., Rowe L., 2010. Condition-dependence of the sexually dimorphic transcriptome in Drosophila melanogaster. Evolution 64: 1836–1848. [DOI] [PubMed] [Google Scholar]
- Yang J. A., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. J., Graze R. M., Walts B. M., Lopez C. M., Baker H. V., et al. , 2011. Partitioning transcript variation in Drosophila: abundance, isoforms, and alleles. G3 1: 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. S., Hill W. G., 2005. Genetic variability under mutation selection balance. Trends Ecol. Evol. 20: 468–470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.