Granulicatella adiacens is a catalase-negative, oxidase-negative, facultatively anaerobic, Gram-positive coccus [1]. This bacterium is considered nutritionally variant streptococci (NVS) because of its requirement for pyridoxal, L-cysteine, or additional nutrients, such as sulfhydryl compounds, for growth [2]. G. adiacens can grow well on culture medium containing these required nutrients. G. adiacens colonies can grow as satellite colonies around other bacteria, such as Staphylococcus aureus [3]. However, most clinical microbiologists have overlooked the fact that G. adiacens isolates can show discrepant satellite testing results based on the different kinds of nutrients contained in blood agar plates (BAPs) produced by different manufacturers. Here, to the best of our knowledge, we firstly report a discrepant satellite testing result of G. adiacens isolates on BAPs produced by different manufacturers.
A 62-yr-old man was admitted to our hospital after 2 days of fever. He had no history of cardiac disease. There was no vegetation on his cardiac valves on the basis of echocardiography results.
Blood samples were collected from peripheral veins at two other sites and inoculated into broth media in aerobic and anaerobic blood culture bottles. The BacT/Alert 3D system (BioMérieux Inc, Durham, NC, USA) was used to incubate and monitor bacterial growth. After blood culture tests were performed, the patient received empirical antibiotic treatment with 2 g intravenous cefotaxime every 8 hr.
After 3 days of incubation, two sets of aerobic blood culture bottles appeared positive on the basis of visual inspection. Gram staining of the positive smear preparations revealed Gram-positive pleomorphic cocci in pairs and short chains. The cultured blood specimens were inoculated onto BAP, MacConkey agar (MAC), and chocolate agar plates (ASAN Pharmaceutical, Hwaseong, Korea). After 72 hr, pin-point sized, grayish-white colored colonies grew on the BAP and chocolate agar. No colony growth was observed on MAC, and catalase and oxidase tests were negative.
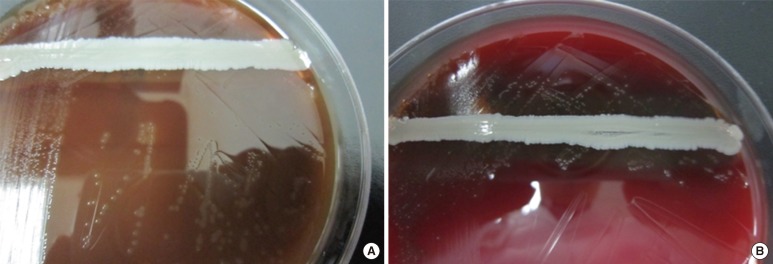
We performed a satellite test to identify the organism because the Gram staining and biochemical testing results suggested NVS. A streak of S. aureus ATCC 25923 was made across the BAP surface for satellite testing. These microorganisms exhibited no satellite effect on BAPs. We used the same method to perform a satellite test using another BAP (MICROMEDIA Corp., Busan, Korea), and found that the microorganism exhibited a satellite effect (Fig. 1). The isolate was identified as G. adiacens with 98.7% probability using the VITEK 2 Gram-Positive Identification Card (BioMérieux, Marcy-l'Etoile, France).
Fig. 1.
Growth of Granulicatella adiacens colonies after 3 days of incubation. (A) No satellite effect on blood agar plate (BAP) manufactured by ASAN. (B) Satellite effect on BAP manufactured by MICROMEDIA.
We performed 16S ribosomal RNA (rRNA) gene sequencing analysis to confirm the identity of the isolate. PCR was used to amplify the first 500 bp of the 5' end of the 16S rRNA gene using the MicroSeq 500 16S rDNA Bacterial Identification PCR kit (Applied Biosystems, Foster City, CA, USA). Cycle sequencing reactions were performed with a MicroSeq 500 16S rDNA Bacterial Identification Sequencing kit (Applied Biosystems). The purified sequencing products were analyzed on a 3130 Genetic Analyzer (Applied Biosystems) according to the manufacturer's instructions.
The resulting sequence was compared with sequences in GenBank (http://www.ncbi.nlm.nih.gov). A BLAST search revealed that the 16S rRNA gene sequence of the isolate was 100% homologous with G. adiacens strain CCUG 60768 (GenBank accession number, FR822389.1). This strain showed percent identity with >0.8% separation from other species that were identified in the search [4]. Therefore, the 16S rRNA gene sequencing analysis confirmed that the microorganism was G. adiacens.
Antimicrobial susceptibility testing was performed by using microdilution methods with 0.001% pyridoxal hydrochloride and cation-adjusted Mueller-Hinton broth supplemented with 5% lysed horse blood [5]. The isolate was susceptible to clindamycin, rifampin, and vancomycin, and it was resistant to penicillin, cefotaxime, ceftriaxone, and meropenem. Because the isolate was resistant to cefotaxime, the patient received antibiotic therapy with intravenous injection of 1 g vancomycin every 12 hr. After the third day of vancomycin therapy, the patient's fever subsided, and subsequent blood cultures were negative for G. adiacens and any other microorganisms.
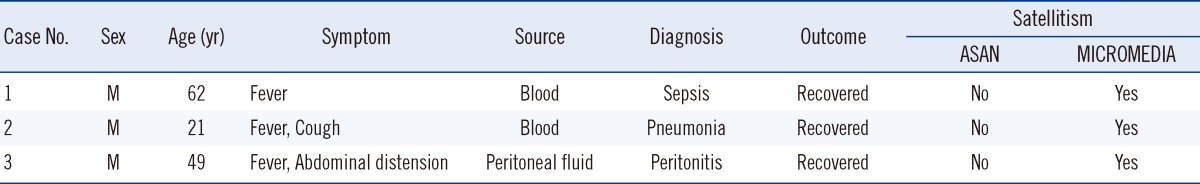
We reviewed the medical records of all patients who had visited our hospital, and found two other patients with G. adiacens identified from clinical specimens. One patient was a 21-yr-old man who had a persistent cough and fever for 7 days. The condition of the patient was diagnosed with pneumonia. An isolate from blood culture tests was identified as G. adiacens. We inoculated this isolate onto two kinds of BAPs produced by ASAN and MICROMEDIA manufacturers, and satellite testing was performed as described above. This isolate showed no satellitism on the BAP manufactured by ASAN, but was positive for satellitism on the BAP from MICROMEDIA. The second patient was a 49-yr-old woman who had abdominal distension and fever. Her condition was diagnosed with peritonitis and advanced gastric cancer. An isolate from the peritoneal fluid was identified as G. adiacens. The isolate also showed no satellitism on the BAP manufactured by ASAN, but did show satellites on the BAP from MICROMEDIA (Table 1).
Table 1.
Three cases of Granulicatella adiacens infection
Abbreviation: M, male.
Granulicatella spp. should be suspected when Gram positive pleomorphic cocci are revealed in pairs and short chains that fail to grow in subculture. Once a suspected Granulicatella isolate is cultured, its identity should be confirmed by establishing its requirement for pyridoxal. Organisms can be cultured on media supplemented with filter-sterilized 0.01% pyridoxal hydrochloride at a final concentration of 0.001%. Pyridoxal disks may also be used in the satellite test. When pyridoxal-supplemented blood agar is not available, Granulicatella spp. can also be identified by the demonstration of satellitism around a S. aureus streak on BAP [6]. Because S. aureus provides nutrients, such as pyridoxal, to Granulicatella spp. by hemolyzing erythrocytes on BAP, Granulicatella species can grow well around S. aureus.
Chocolate agar and brucella agar media contain pyridoxal hydrochloride; thus, Granulicatella can grow well independently on these media. Granulicatella may not grow on conventional BAPs without pyridoxal hydrochloride or L-cysteine. However, Granulicatella may grow on blood agar independently, if the isolation medium containing pyridoxal hydrochloride or L-cysteine offers enough enrichment for growth [7].
In our three cases, we performed satellite tests of the G. adiacens isolates. These isolates exhibited a satellite effect on BAPs manufactured by MICROMEDIA but not on BAPs obtained from ASAN. We found that the ASAN BAPs contained thioketone, which is one of the required nutrients for G. adiacens growth. This result indicates that this isolate may grow well independently and show no satellite effect on BAPs containing thioketone without L-cysteine or pyridoxal hydrochloride. Thus, we suggest that clinical microbiologists should be aware of all nutrients contained in media used to culture Granulicatella spp. We also recognize that streaking S. aureus for satellite testing may not be needed to inoculate BAPs containing nutrients required for G. adiacens growth.
G. adiacens isolates can be identified by biochemical testing with or without molecular confirmation. However, biochemical testing has been reported to lead to incorrect identification of this organism. G. adiacens can be misidentified as other species of Granulicatella, Abiotrophia, or Gemella [8]. It can be difficult for clinical microbiologists to separate these genera by only using standard biochemical tests. When biochemical tests show discrepant or conflicting results, molecular methods, such as 16S rRNA gene sequencing analysis, can be used to confirm the identity of the microorganism.
In conclusion, we showed that streaking S. aureus for satellite testing may not be necessary when BAPs contain the required nutrients for G. adiacens growth. Furthermore, we demonstrated that16S rRNA gene sequencing analysis can be used to confirm the identity of G. adiacens when biochemical tests do not provide clear results.
Acknowledgments
This work was supported by the year 2013 clinical research grant from Pusan National University Hospital.
Footnotes
No potential conflicts of interest relevant to this article were reported.
References
- 1.Collins MD, Lawson PA. The genus Abiotrophia is not monophyletic: proposal of Granulicatella gen. nov., Granulicatella adiacens comb. nov., Granulicatella elegans comb. nov., and Granulicatella balaenopterae comb. nov. Int J Syst Evol Microbiol. 2000;50:365–369. doi: 10.1099/00207713-50-1-365. [DOI] [PubMed] [Google Scholar]
- 2.Ruoff KL. Nutritionally variant streptococci. Clin Microbiol Rev. 1991;4:184–190. doi: 10.1128/cmr.4.2.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.George RH. The isolation of symbiotic streptococci. J Med Microbiol. 1974;7:77–83. doi: 10.1099/00222615-7-1-77. [DOI] [PubMed] [Google Scholar]
- 4.CLSI. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing: approved guideline. Wayne, PA: Clinical and Laboratory Standards Institute; 2008. CLSI document MM18-A. [Google Scholar]
- 5.CLSI. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria: approved guideline. 2nd ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2010. CLSI document MM45-A2. [Google Scholar]
- 6.Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, Warnock DW, editors. Manual of clinical microbiology. 10th ed. Washington, DC: ASM Press; 2011. pp. 365–376. [Google Scholar]
- 7.Christensen JJ, Facklam RR. Granulicatella and Abiotrophia species from human clinical specimens. J Clin Microbiol. 2001;39:3520–3523. doi: 10.1128/JCM.39.10.3520-3523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cargill JS, Scott KS, Gascoyne-Binzi D, Sandoe JA. Granulicatella infection: diagnosis and management. J Med Microbiol. 2012;61:755–761. doi: 10.1099/jmm.0.039693-0. [DOI] [PubMed] [Google Scholar]