Abstract
Image file format is often a confusing aspect for someone wishing to process medical images. This article presents a demystifying overview of the major file formats currently used in medical imaging: Analyze, Neuroimaging Informatics Technology Initiative (Nifti), Minc, and Digital Imaging and Communications in Medicine (Dicom). Concepts common to all file formats, such as pixel depth, photometric interpretation, metadata, and pixel data, are first presented. Then, the characteristics and strengths of the various formats are discussed. The review concludes with some predictive considerations about the future trends in medical image file formats.
Keywords: Medical imaging, Image processing, File formats, Dicom, Nifti
Introduction
Image file formats provide a standardized way to store the information describing an image in a computer file. A medical image data set consists typically of one or more images representing the projection of an anatomical volume onto an image plane (projection or planar imaging), a series of images representing thin slices through a volume (tomographic or multislice two-dimensional imaging), a set of data from a volume (volume or three-dimensional imaging), or multiple acquisition of the same tomographic or volume image over time to produce a dynamic series of acquisitions (four-dimensional imaging). The file format describes how the image data are organized inside the image file and how the pixel data should be interpreted by a software for the correct loading and visualization.
The paper is organized as follows. First, some basic concepts common to all image file formats are reviewed. These concepts include: pixel depth, photometric interpretation, metadata, and pixel data. Then, a description of the four major file formats used in medical imaging, Analyze, Neuroimaging Informatics Technology Initiative (Nifti), Minc, and Digital Imaging and Communications in Medicine (Dicom), is provided. Finally, the limits and strengths of the reviewed formats are discussed, with some considerations about the future directions in the field of medical image file formats.
Basic Concepts
A medical image is the representation of the internal structure or function of an anatomic region in the form of an array of picture elements called pixels or voxels. It is a discrete representation resulting from a sampling/reconstruction process that maps numerical values to positions of the space. The number of pixels used to describe the field-of-view of a certain acquisition modality is an expression of the detail with which the anatomy or function can be depicted. What the numerical value of the pixel expresses depends on the imaging modality, the acquisition protocol, the reconstruction, and eventually, the post-processing.
Pixel Depth
Pixel depth is the number of bits used to encode the information of each pixel. Every image is stored in a file and kept in the memory of a computer as group of bytes. Bytes are group of 8 bits and represent the smallest quantity that can be stored in the memory of a computer. This means that if a 256 by 256 pixels image has a pixel depth of 12 or 16 bits, the computer will always store two bytes per pixel and then the pixel data will require 256 × 256 × 2 = 131,072 bytes of memory in both cases. With a pixel depth of 2 bytes per pixels, it is possible to codify and store integer numbers between 0 and 65,535 (216−1); alternatively, it is possible to represent integer numbers between −32,768 and +32,767 using 15 bits to represent the numbers and 1 bit to represent the sign. Image data may also be real numbers. The Institute of Electrical and Electronics Engineers created a standard (IEEE-754) in which defines two basic formats for the encoding in binary of floating-point numbers: the single precision 32-bit and the double precision 64-bit. The standard addresses the problem of the precision with which a finite number of combination obtainable with a sequence of n-bit (2n−1) can represent a continuous range of real numbers. Although unusual, pixels can store complex numbers. Complex data have a real and an imaginary component which are referred as pairs of real numbers. Therefore, complex data typically have a pixel depth twice that used to represent a single real number.
From the previous overview emerges that the pixel depth is a concept related to the memory space necessary to represent in binary the amount of information we want to store in a pixel.
Photometric Interpretation
The photometric interpretation specifies how the pixel data should be interpreted for the correct image display as a monochrome or color image. To specify if color information is or is not stored in the image pixel values, we introduce the concept of samples per pixel (also known as number of channels). Monochrome images have one sample per pixel and no color information stored in the image. A scale of shades of gray from black to white is used to display the images. The number of shades of gray depends clearly from the number of bits used to store the sample that, in this case, coincide with the pixel depth. Clinical radiological images, like x-ray computed tomography (CT) and magnetic resonance (MR) images have a gray scale photometric interpretation. Nuclear medicine images, like positron emission tomography (PET) and single photon emission tomography (SPECT), are typically displayed with a color map or color palette. In this case, each pixel of the image is associated with a color in a predefined color map, but the color regards only the display and is an information associated with and not really stored in the pixel values. The images still have one sample per pixel and are said to be in pseudo-color. To encode color information into pixels, we typically need multiple samples per pixel and to adopt a color model that specifies how to obtain colors combining the samples [1]. Usually, 8-bit are reserved for each sample or color component. The pixel depth is calculated by multiplying the sample depth (number of bits used for each sample) with the number of samples per pixel. Ultrasound images are typically stored employing the red–green–blue color model (briefly, RGB). In this case, the pixel should be intended as a combination of the three primary colors, and three samples per pixel are stored. The images will have a pixel depth of 24 bits and are said to be in true color.
Color is for example used to encode blood flow direction (and velocity) in Doppler ultrasound, to show additional “functional” information onto a gray scale anatomical image as colored overlays, as in the case of fMRI activation sites, to simultaneously display functional and anatomical images as in the PET/CT or PET/MRI, and sometimes in place of gray tones to highlight signal differences.
Metadata
Metadata are information that describe the image. It can seem strange, but in any file format, there is always information associated with the image beyond the pixel data. This information called metadata is typically stored at the beginning of the file as a header and contains at least the image matrix dimensions, the spatial resolution, the pixel depth, and the photometric interpretation. Thanks to metadata, a software application is able to recognize and correctly open an image in a supported file format simply by a double-click or dragging the image icon onto the icon of the application. In the case of medical images, metadata have a wider role due to the nature of the images itself. Images coming from diagnostic modalities typically have information about how the image was produced. For example, a magnetic resonance image will have parameters related to the pulse sequence used, e.g., timing information, flip angle, number of acquisitions, etc. A nuclear medicine image like a PET image will have information about the radiopharmaceutical injected and the weight of the patient. These data allows software like OsiriX [2] to on-the-fly convert pixel values in standardized uptake values (SUV) without the need to really write SUV values into the file. Post-processing file formats have a terser metadata section that essentially describes the pixel data. The different content in the metadata is the main difference between the images produced by a diagnostic modality and post-processed images. Metadata are a powerful tool to annotate and exploit image-related information for clinical and research purposes and to organize and retrieve into archives images and associated data.
Pixel Data
This is the section where the numerical values of the pixels are stored. According to the data type, pixel data are stored as integers or floating-point numbers using the minimum number of bytes required to represent the values (see Table 1). Looking at images generated by tomographic imaging modalities, and sent to a Picture Archiving and Communication System (PACS) or a reading station, radiological images like CT and MR and also modern nuclear medicine modalities, like PET and SPECT, store 16 bits for each pixel as integers. Although integers, eventually with the specification of a scale factor, are adequate for “front-end” images, the use of a float data type is frequent in any post-processing pipeline since it is the most natural representation to address calculations. Image data may also be of complex type even if this data type is not common and can be bypassed by storing the real and imaginary parts as separate images. An example of complex data is provided by arrays that in MRI store acquired data before the reconstruction (the so called k-space) or after the reconstruction if you choose to save both magnitude and phase images.
Table 1.
Summary of file formats characteristics
| Format | Header | Extension | Data types |
|---|---|---|---|
| Analyze | Fixed-length: 348 byte binary format | .img and .hdr | Unsigned integer (8-bit), signed integer (16-, 32-bit), float (32-, 64-bit), complex (64-bit) |
| Nifti | Fixed-length: 352 byte binary formata (348 byte in the case of data stored as .img and .hdr) | .nii | Signed and unsigned integer (from 8- to 64-bit), float (from 32- to 128-bit), complex (from 64- to 256-bit) |
| Minc | Extensible binary format | .mnc | Signed and unsigned integer (from 8- to 32-bit), float (32-, 64-bit), complex (32-, 64-bit) |
| Dicom | Variable length binary format | .dcm | Signed and unsigned integer, (8-, 16-bit; 32-bit only allowed for radiotherapy dose), float not supported |
Not all the software support all the specified data types. Dicom, Analyze, and Nifti support color RGB 24-bit; Nifti also supports RGBA 32-bit (RGB plus an alpha-channel)
aNifti has a mechanism to extend the header
Whenever the value of a pixel is stored using two or more bytes it should be taken into account that the order in which the computer store the bytes is not unique. If we indicate with b1, b2 the two bytes of a 16-bit word, the computer can store the word as (b1:b2) or (b2:b1). The term little endian indicates that the least significant byte is stored first, while big endian which is the most significant byte to be stored first. This issue is typically related to the processor on which the computer hardware is based and regards all the data encoded using more than 8-bit per pixel.
In formats that adopt a fixed-size header, the pixel data start at a fixed position after skipping the header length. In the case of variable length header, the starting location of the pixel data is marked by a tag or a pointer. In any case, to calculate the pixel data size, we have to do:
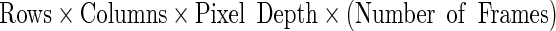 |
where the pixel depth is expressed in bytes. The image file size will be given by:
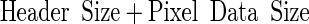 |
Both the expressions are valid in the case of uncompressed data. Image data may also be compressed to reduce requirements for storage and transmission, in which case the file size is reduced by a factor that depends on the compression technique adopted. Generally speaking, compression may be reversible (lossless) or irreversible (lossy). Lossless compression techniques allow a moderate gain in terms of image storage. Lossy techniques allow a greater advantage at the cost of information loss but, for this reason, their use in the world of medical imaging is controversial. It is not clear under which conditions the reading of the images and/or the quantitative post-processing procedures are not influenced by information loss. On the other hand, the adoption of lossy compression schemes with a low or moderate loss of information in place of lossless ones might appear not justified [3].
File Formats
Medical image file formats can be divided in two categories. The first is formats intended to standardize the images generated by diagnostic modalities, e.g., Dicom [4]. The second is formats born with the aim to facilitate and strengthen post-processing analysis, e.g., Analyze [5], Nifti [6], and Minc [7]. Medical image files are typically stored using one of the following two possible configurations. One in which a single file contains both the metadata and image data, with the metadata stored at the beginning of the file. This paradigm is used by Dicom, Minc, and Nifti file formats, even if it is allowed by other formats. The second configuration stores the metadata in one file and the image data in a second one. The Analyze file format uses the two-files paradigm (.hdr and .img).
In this section, we describe some of the most popular formats currently used: Analyze, Nifti, Minc, and Dicom. Table 1 summarizes the characteristics of the described file formats.
Historically, the one of the first projects aimed to create standardized file formats in the field of medical imaging was the Interfile format [8]. It was created in the 1980s and has been used for many years for the exchange of nuclear medicine images. An Interfile image consists of a pair of files, one containing metadata information in ASCII format, that the standard calls administrative information, and one containing the image data. The Interfile header can be viewed and edited with a normal text editor.
Analyze
Analyze 7.5 was created at the end of 1980s as format employed by the commercial software Analyze developed at the Mayo Clinic in Rochester, MN, USA. For more than a decade, the format was the standard “de facto” for the medical imaging post-processing. The big insight of the Analyze format was that it has been designed for multidimensional data (volume). Indeed, it is possible to store in one file 3D or 4D data (the fourth dimension being typically the temporal information). An Analyze 7.5 volume consists of two binary files: an image file with extension “.img” that contains the voxel raw data and a header file with extension “.hdr” that contains the metadata, such as number of pixels in the x, y, and z directions, voxel size, and data type. The header has a fixed size of 348 bytes and is described as a structure in the C programming language. The reading and the editing of the header require a software utility. The format is today considered “old” but it is still widely used and supported by many processing software packages, viewers, and conversion utilities. A new version of the format (AnalyzeAVW) used in the latest versions of the Analyze software is not discussed here since it is not widespread.
As summarized in Table 1, Analyze 7.5 does not support certain basic data types including the unsigned 16 bits, and this can be sometimes a limitation forcing users to use a scale factor or to switch to a pixel depth of 32-bit. Moreover, the format does not store enough information to unambiguously establish the image orientation.
Nifti
Nifti is a file format created at the beginning of 2000s by a committee based at the National Institutes of Health with the intent to create a format for neuroimaging maintaining the advantages of the Analyze format, but solving the weaknesses. The Nifti can in fact be thought as a revised Analyze format. The format fills some of the unused/little used fields present in the Analyze 7.5 header to store new information like image orientation with the intent to avoid the left–right ambiguity in brain study. Moreover, Nifti include support for data type not contemplated in the Analyze format like the unsigned 16-bit. Although the format also allows the storage of the header and pixel data in separate files, images are typically saved as a single “.nii” file in which the header and the pixel data are merged. The header has a size of 348 bytes in the case of “.hdr” and “.img” data storage, and a size of 352 bytes in the case of a single “.nii” file for the presence of four additional bytes at the end, essentially to make the size a multiple of 16, and also to provide a way to store additional-metadata, in which case these 4 bytes are nonzero. A practical implementation of an extended Nifti format for the processing of diffusion-weighted magnetic resonance data is described in [9].
The Nifti format allows a double way to store the orientation of the image volume in the space. The first, comprising a rotation plus a translation, to be used to map voxel coordinates to the scanner frame of reference; this “rigid body” transformation is encoded using a “quaternion” [10]. The second method is used to save the 12 parameters of a more general linear transformation which defines the alignment of the image volume to a standard or template-based coordinate system. This spatial normalization task is common in brain functional image analysis [11].
The Nifti format has rapidly replaced the Analyze in neuroimaging research, being adopted as the default format by some of the most widespread public domain software packages, as, FSL [12], SPM [13], and AFNI [14]. The format is supported by many viewers and image analysis software like 3D Slicer [15], ImageJ [16], and OsiriX, as well as other emerging software like R [17] and Nibabel [18], besides various conversion utilities.
An update version of the standard, the Nifti-2, developed to manage larger data set has been defined in the 2011. This new version encode each of the dimensions of an image matrix with a 64-bit integer instead of a 16-bit as in the Nifti-1, eliminating the restriction of having a size limit of 32,767. This updated version maintains almost all the characteristics of the Nifti-1 but, as reserve for some header fields the double precision, comes with a header of 544 bytes [19].
Minc
The Minc file format was developed at the Montreal Neurological Institute (MNI) starting from 1992 to provide a flexible data format for medical imaging. The first version of Minc format (Minc1) was based on the standard Network Common Data Format (NetCDF). Subsequently, to overcame the limit in supporting large data files and to provide other new features, the Minc development team chose to switch from NetCDF to Hierarchical Data Format version 5 (HDF5). This new release, not compatible with the previous one, was called Minc2. The format is mainly used by software tools developed by the MNI Brain Imaging Center, i.e., a viewer and a processing software library [7]. A set of utilities which allow the conversion to and from Dicom and Nifti formats, and between Minc1 and Minc2 have been made available by the same group.
Dicom
The Dicom standard was established by the American College of Radiology and the National Electric Manufacturers Association. Despite of its 1993 date of birth, the real introduction of the Dicom standard into imaging departments takes place at the end of 1990s. Today, the Dicom standard is the backbone of every medical imaging department. The added value of its adoption in terms of access, exchange, and usability of diagnostic medical images is, in general, huge. Dicom is not only a file format but also a network communication protocol, and although the two aspects cannot be completely separated, here, we will discuss only Dicom as a file format.
The innovation of Dicom as a file format has been to establish that the pixel data cannot be separated from the description of the medical procedure which led to the formation in the image itself. In other words, the standard stressed the concept that an image that is separate from its metadata becomes “meaningless” as medical image. Metadata and pixel data are merged in a unique file, and the Dicom header, in addition to the information about the image matrix, contains the most complete description of the entire procedure used to generate the image ever conceived in terms of acquisition protocol and scanning parameters. The header also contains patient information such as name, gender, age, weight, and height. For these reasons, the Dicom header is modality-dependent and varies in size. In practice, the header allows the image to be self-descriptive. In order to easily understand the power of this approach, just think to the software which Siemens first introduced for its MRI systems to replicate an acquisition protocol. The software, known as “Phoenix”, is able to extract from a Dicom image series dragged into the acquisition window the protocol and to replicate it for a new acquisition. There are similar tools for all the major manufacturers.
Regarding the pixel data, Dicom can only store pixel values as integers. Dicom cannot currently save pixel data in floating-point while it supports various data types, including floats, to store metadata. Whenever the values stored in each voxel have to be scaled to different units, Dicom makes use of a scale factor using two fields into the header defining the slope and the intercept of the linear transformation to be used to convert pixel values to real world values.
Dicom supports compressed image data through a mechanism that allow a non-Dicom-formatted document to be encapsulated in a Dicom file. Compression schemes supported by Dicom include the following: JPEG, run-length encoding (RLE), JPEG-LS, JPEG-2000, MPEG2/MPEG4, and Deflated, as described in Part 5 of the standard [20]. The newly JPEG-XR compression standard has been proposed to be adopted by Dicom. An encapsulated Dicom file includes metadata related to the native document plus the metadata necessary to create the Dicom shell.
Discussion
The existence of multiple file formats is clearly a limiting factor for the access, exchange and, usability of medical images and associated information.
At present, the clinical and research domains still appear well separated. There is sometimes a contraposition between the two approaches that is not completely justified in practice. People who work with images from a post-processing point of view have the opinion that formats like Dicom are too complicated or unnecessarily detailed, while in practice, it is simply an answer to a different need. On the other hand, post-processing file formats are more simple and terse but on careful analysis are always somewhat incomplete because much of the information other than the numerical values of the pixels is lost. The creation of a format well suited both for clinical and research is difficult, but the creation of a unique format for the post-processing is desirable. The format should be flexible enough to prevent that the development and adoption of a new protocol or image analysis procedure which needs a new set of metadata leads to the creation of a new format.
In a clinical context, the Dicom format has been widely accepted and successfully used. The standard specifies rules not only for the encoding of image and image-related information but also for their transmission across a network. In the following, we will discuss some of the characteristics of the standard and also try to answer the question of why the Dicom format is not used for post-processing.
The documentation about Dicom is extensive also because it is not possible to completely separate the communication protocol aspects of the standard from the data format aspects. This presents a barrier for anyone who wants to gain a deep knowledge of the standard. The unavailability of the documentation in a format such as the HTML certainly does not help. People who are only interested into file format may limit the studies to the Parts 3 and 5 of the standard [20]. The format is born basically as a 2D image file format. A 3D volume is described by a series of files containing single slices. This approach is today considered “anachronistic” since many tomographic imaging modalities generate several thousand of images for each volume, and in dynamic studies multiple volumes in a single session. The multiframe Dicom specification arrived late and even today it is not widespread. The header of a multiframe Dicom file is structured to store attributes shared by all frames as well as per-frame attributes, maintaining the possibility to specify parameters on a slice-by-slice base. In such cases, this aspect comes in handy, e.g., in modern CT where, to optimize the dose, the x-ray tube current is modulated as a function of the anatomical area irradiated so its value varies with the slice location [21], or in the case of PET images where sometimes the scale factor or the acquisition time are specified for single slice or bed position, respectively.
In addition to standard information, Dicom metadata can also contain manufacturer-specific attributes (private fields), for which there is not a description and that are not included in the Dicom data dictionary. Private data have the same structure of standard data elements and are identified with tags belonging to specific group numbers. In case post-processing analysis requires some of these vendor-specific data, the user needs to parse them trying to find out the desired information or refer to the manufacturer.
For storing images having non-integer values as integers Dicom makes use of a scale factor. Generally speaking, the purpose of the scale factor is to allow values in images that cannot be stored using the supported data types. It is a mechanism used by many others file formats. For example, the Analyze format uses a scale factor to have unsigned integers storing signed ones. It is clearly possible to use scaling within the same data type, for example, to “compress” a 16-bit integer to 8-bit (or a 32-bit integer to 16-bit).
A format that provides a full support to the different data types is the Nifti. This feature allows to minimize, or even eliminate, the use of the scale factor, that the standard anyway admits for backwards compatibility.
The byte order is almost never a problem. Some formats specify in the metadata the byte order. In the case the metadata does not explicitly contain any indication, it is the software that will do a check typically on one of the metadata field that has a known value to determine the order. In both cases, the recognition of the byte order with which the pixel data have been recorded is transparent to the end user.
Any post-processing pipeline typically starts with the conversion of an image data set from Dicom to the format chosen for the processing. Sometimes, it is necessary to convert data from a format to another when, for example, it is necessary to use multiple software modules or third part data. It is a good practice to accurately check the output of each format conversion utility in order to avoid unwanted actions. For example, the aforementioned scale factor is one possible pitfall for a format conversion operation. The correct application of scale factors is particularly important when voxel values are expression of quantities or parameters in a certain measurement units. This is, for example, the case of radiotracer activity per unit volume in PET images or perfusion measurements with MR arterial-spin labeling. Another issue that can be potentially affected by a format conversion operation is image orientation.
At the end of a post-processing pipeline, a certain number of results are produced. These data remain typically confined in a research environment so that no specific needs about the “format” are required. But in the case of a quantitative imaging procedure that is moving from research-only applications into clinical use, it could be interesting to find a way that does not leave post-processing results “separated” from the context of a hospital information system. It is in this context that the Annotation and Image Markup project was started by the US National Cancer Institute for the creation of a software tool to associate quantitative and descriptive information to images, with the intent to facilitate the use of imaging as biomarker in cancer studies [22].
There is a growing interest in better understanding how to save or integrate, for example using the Dicom standard, some of the results obtained by image analysis methods so that they could be sent to a Picture Archiving and Communication System and archived for documentation or subsequent analysis or reuse. The Dicom standard should encourage and clarify the encoding of post-processing results to give them the chance to be effectively integrated into the rest of the Dicom world.
Conclusion
The field of image file formats is constantly evolving. Regarding the encoding of images generated by diagnostic modalities, Dicom is the preferred standard and it will be for many years to come thanks to a great flexibility to follow the technological development and medical imaging advances and the integration with a network infrastructure. On the other hand, in the near future, we will probably see the proposal and the adoption of new file formats for the post-processing, even if the idea that these formats are a convenient way to only retain “essential information” will lose strength. The weight of image-specific and image-related information is growing in the world of image processing, and the potential integration of results obtained by quantitative analysis in a clinical context is an open issue.
Acknowledgments
The authors acknowledge the support of the Italian Ministry of Instruction, University and Research—Progetti MERIT RBNE08E8CZ and PRIN 2010XE5L2R.
References
- 1.The Encyclopedia of Graphics File Formats. http://www.fileformat.info/mirror/egff/ (accessed in June 2013)
- 2.Rosset A, Spadola L, Ratib O. OsiriX: an open-source software for navigating in multidimensional DICOM images. J Digit Imaging. 2004;17(3):205–16. doi: 10.1007/s10278-004-1014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clunie DA. Lossless compression of grayscale medical images: effectiveness of traditional and state-of-the-art approaches. Imaging: SPIE-Medical; 2000. pp. 74–84. [Google Scholar]
- 4.Bidgood WD, Jr, Horii SC, Prior FW, Van Syckle DE. Understanding and using DICOM, the data interchange standard for biomedical imaging. J Am Med Inform Assoc. 1997;4(3):199–212. doi: 10.1136/jamia.1997.0040199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robb RA, Hanson DP, Karwoski RA, Larson AG, Workman EL, Stacy MC. Analyze: a comprehensive, operator-interactive software package for multidimensional medical image display and analysis. Comput Med Imaging Graph. 1989;13(6):433–54. doi: 10.1016/0895-6111(89)90285-1. [DOI] [PubMed] [Google Scholar]
- 6.NIFTI documentation. http://nifti.nimh.nih.gov/nifti-1/documentation (accessed in June 2013)
- 7.MINC software library and tools. http://www.bic.mni.mcgill.ca/ServicesSoftware/MINC (accessed in June 2013)
- 8.Todd-Pokropek A, Cradduck TD, Deconinck F. A file format for the exchange of nuclear medicine image data: a specification of Interfile version 3.3. Nucl Med Commun. 1992;13(9):673–99. doi: 10.1097/00006231-199209000-00007. [DOI] [PubMed] [Google Scholar]
- 9.Patel V, Dinov ID, Van Horn JD, Thompson PM, Toga AW. LONI MiND: metadata in NIfTI for DWI. Neuroimage. 2010;51(2):665–76. doi: 10.1016/j.neuroimage.2010.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrew J. Hanson. Visualizing Quaternions. Morgan Kaufmann Pub, 2006
- 11.Brett M, Johnsrude IS, Owen AM. The problem of functional localization in the human brain. Nat Rev Neurosci. 2002;3(3):243–9. doi: 10.1038/nrn756. [DOI] [PubMed] [Google Scholar]
- 12.Jenkinson M, Beckmann CF, Behrens TE. Woolrich MW, and Smith SM. FSL. NeuroImage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Ashburner J. SPM: a history. Neuroimage. 2012;62(2):791–800. doi: 10.1016/j.neuroimage.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox RW. AFNI: what a long strange trip it's been. Neuroimage. 2012;62(2):743–7. doi: 10.1016/j.neuroimage.2011.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012;30(9):1323–41. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ImageJ—Image processing and analysis in Java. http://rsbweb.nih.gov/ij/ (accessed in June 2013)
- 17.Whitcher B, Schmid VJ, Thornton A. Working with the DICOM and NIfTI Data Standards in R. Journal of Statistical Software. 2011;44(6):1–29. [Google Scholar]
- 18.Neuroimaging in Python (Nipy project). http://nipy.sourceforge.net/nibabel/ (accessed in June 2013)
- 19.The NIFTI-2 header. http://brainmap.wustl.edu/wiki/index.php/Nifti2.h (accessed in June 2013)
- 20.Digital Imaging and Communications in Medicine (DICOM) documentation. http://medical.nema.org/standard.html (accessed in June 2013)
- 21.Tsalafoutas IA, Metallidis SI. A method for calculating the dose length product from CT DICOM images. Br J Radiol. 2011;84(999):236–43. doi: 10.1259/bjr/37630380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Channin DS, Mongkolwat P, Kleper V, Rubin DL. The Annotation and Image Mark-up project. Radiology. 2009;253(3):590–2. doi: 10.1148/radiol.2533090135. [DOI] [PubMed] [Google Scholar]


