Abstract
The etiology of hypertrophic cardiomyopathy (HCM) has been ascribed to mutations in genes encoding sarcomere proteins. In particular, mutations in MYBPC3, a gene which encodes cardiac myosin binding protein-C (cMyBP-C), have been implicated in over one third of HCM cases. Of these mutations, 70% are predicted to result in C′-truncated protein products, which are undetectable in tissue samples. Heterozygous carriers of these truncation mutations exhibit varying penetrance of HCM, with symptoms often occurring later in life. We hypothesize that heterozygous carriers of MYBPC3 mutations, while seemingly asymptomatic, have subtle functional impairments that precede the development of overt HCM. This study compared heterozygous (+/t) knock-in MYBPC3 truncation mutation mice with wild-type (+/+) littermates to determine whether functional alterations occur at the whole-heart or single-cell level before the onset of hypertrophy. The +/t mice show ∼40% reduction in MYBPC3 transcription, but no changes in cMyBP-C level, phosphorylation status, or cardiac morphology. Nonetheless, +/t mice show significantly decreased maximal force development at sarcomere lengths of 1.9 μm (+/t 68.5 ± 4.1 mN/mm2 vs. +/+ 82.2 ± 3.2) and 2.3 μm (+/t 79.2 ± 3.1 mN/mm2 vs. +/+ 95.5 ± 2.4). In addition, heterozygous mice show significant reductions in vivo in the early/after (E/A) (+/t 1.74 ± 0.12 vs. +/+ 2.58 ± 0.43) and E′/A′ (+/t 1.18 ± 0.05 vs. +/+ 1.52 ± 0.15) ratios, indicating diastolic dysfunction. These results suggest that seemingly asymptomatic heterozygous MYBPC3 carriers do suffer impairments that may presage the onset of HCM.
Keywords: haploinsufficiency, cardiac myosin binding protein-C
hypertrophic cardiomyopathy (HCM) is an inherited form of heart disease that affects 1 in 500 individuals (19). HCM involves pathological myocardial wall thickening and is associated with sudden cardiac death and the development of heart failure. Mutations in genes encoding sarcomere proteins have been identified as the predominant cause of HCM (44), although the mechanisms by which various mutations give rise to a common pathology are poorly understood. Nearly 40% of the identified mutations associated with HCM occur in the MYBPC3 gene, which encodes the sarcomere protein cardiac myosin binding protein-C (cMyBP-C) (3, 4, 46). Of the currently identified mutations in MYBPC3, 70% are predicted to encode for a C′-truncated protein (1, 34). These truncated proteins are often undetectable in samples from human carriers or in mouse models of MYBPC3 mutations, suggesting rapid degradation of the mutant transcript or protein (15, 23, 30, 35). MYBPC3 mutations found in the human population are typically heterozygous and often have a variable penetrance with delayed onset of HCM (6, 9, 24, 25). Protein analysis from heart tissue of symptomatic heterozygous carriers of MYBPC3 mutations has shown a reduction in cMyBP-C levels compared with that of donor hearts (20, 24, 42). This observation is paralleled in certain mouse models of MYBPC3 mutations, where symptomatic heterozygous mice show a decreased amount of cMyBP-C in the heart (5, 7, 8, 45). However, other heterozygous mouse models show no decrease in cMyBP-C levels and display generally asymptomatic phenotypes (15, 23). Therefore, it is currently unclear whether reduction in cMyBP-C content initiates the development of HCM via haploinsufficiency or if other less direct pathways are involved in the early stages of the pathology.
Cardiac MyBP-C is a sarcomeric thick filament protein that incorporates in the C-zone of the cardiac sarcomere via interactions at its COOH-terminus (18, 27, 28). Functionally, cMyBP-C regulates cross-bridge cycling in a phosphorylation-dependent manner (2, 11, 17, 36). When cMyBP-C is dephosphorylated at its N′ domain, it interacts with myosin subfragment 2 (S2) in the interfilament space (14). Upon phosphorylation, the interaction of cMyBP-C with myosin S2 is attenuated, allowing increased cross-bridge cycling kinetics (40). It has been shown that absence of cMyBP-C causes functional deficits in the development of force and impairs proper relaxation (16, 33).
The functional impairments observed in heterozygous carriers of MYBPC3 truncation mutations have been suggested to result from reduced cMyBP-C levels (20, 24, 42). However, transition to symptomatic HCM with reduced cMyBP-C levels remains poorly understood based, in no small part, on the uncertainty of whether such reduced cMyBP-C levels are causative for this transition or a result. Accordingly, this study aims to investigate the physiological consequences exhibited by a mouse model previously described as having preserved cMyBP-C levels, but carrying only one functional allele of MYBPC3. To accomplish this, a MYBPC3 truncation mutant mouse model generated by McConnell et al. was used (22, 23, 30). This model carries a knock-in mutation in MYBPC3 that results in the skipping of exon 30, a frame shift, and inclusion of a premature stop codon. As previously reported, the homozygous (t/t) mouse has no detectable cMyBP-C, increased fibrosis, dilation of the left ventricle, and decreased cardiac function leading to the development of heart failure (23, 29–31). The original characterization of this model focused primarily on the homozygous genotype. However, the heterozygous mouse (+/t) was shown to be asymptomatic, with a level of cMyBP-C equal to that of wild-type (+/+). Also, no hypertrophy was reported in the initial characterization until the animals were over two years of age (22, 23). Information about gene expression, myofilament performance, and in vivo function were not reported for the heterozygote. Because this heterozygous mouse appears to be asymptomatic with normal cMyBP-C levels, it is ideal for testing whether subtle deficits in cellular contractility and cardiac function are present in the heterozygous mouse before the development of overt HCM.
METHODS
Mouse model of MYBPC3 haploinsufficiency.
All +/+, +/t, and t/t mice (23) were in the FVB/N background and between 10 and 12 wk of age when these experiments were performed. The present animal experiments were approved by the Institutional Animal Care and Use Committees at Loyola University Chicago and followed the policies of the Guide for the Use and Care of Laboratory Animals published by the National Institutes of Health (NIH).
Histopathological analyses and gross morphology.
Hearts were excised and either frozen in liquid nitrogen or perfused with formalin and embedded in paraffin for sectioning. Hearts were sectioned into 5-μm-thick slices and stained with hematoxylin and eosin or Masson's Trichrome (36). Gross morphology images were obtained with a light microscope (Zeiss Discovery V8). Histology images were taken using an inverted microscope (Zeiss) with a 40× objective.
Immunohistochemistry.
Isolated hearts were cannulated through the aorta and retrograde perfused with digestion buffer to isolate myocytes as previously described (26). The cells were adhered to chamber slides and fixed with 4% paraformaldehyde and 0.5% Triton X-100 in PBS for 5 min. Antigen retrieval was performed with 0.1 M Glycine, pH 3.5, for 30 min, and the cells were blocked with 1% BSA, 0.1% gelatin, and 0.1% Tween 20, followed by incubation with primary antibodies against cMyBP-C (Cat. No. 137180 clone E7; Santa Cruz) or α-actinin (Sigma A2543), while secondary antibodies were conjugated with Cy3 and Cy5 fluorescent reporters and used for detection. Cells were imaged using a TCS-SP5 confocal microscope (Leica) with a 63× 1.2 numerical aperture water objective. Images were merged using NIH ImageJ software (13).
Echocardiography analysis.
Echocardiography was performed under 2% isoflurane anesthesia with a Vevo 2100 (Fujifilm; Visual Sonics, Toronto, Canada) with a MS-550D 22–55 MHz transducer. Wall thickness and contractile function were measured using parasternal long axis M-mode image analysis. Power-Doppler and tissue-Doppler imaging modalities on apical four-chamber view were used to measure early (E) to late (A) ventricular filling velocities (E/A) and early (E′) to late (A′) mitral annulus motion (E′/A′) ratios to assess left ventricular filling and diastolic function. Echocardiography images were measured using the Visual Sonics Vevo 2100 analysis package.
Steady-state force measurements.
Cells were prepared and analyzed for force-pCa relationship as previously described (41). Frozen left ventricular tissue was homogenized and filtered through a 70-μm cell strainer, followed by centrifugation at 120 g for 1 min at 4°C. The cells were skinned by resuspending the pellet in relaxing solution containing (in mM) 97.92 KOH, 6.24 ATP, 10 EGTA, 10 Na2CrP, 47.58 Kprop, 100 BES, and 6.54 MgCl2 with 1% Triton X-100 and then incubated for 10–15 min at room temperature on a rocking table. The Triton was washed out by two centrifugation steps at 120 g for 1 min at 4°C with resuspension in relaxing solution. The skinned myocytes were transferred to a culture dish coated with 0.1% BSA. With the use of an inverted microscope (Leica DM IRB) under bright field at 40× magnification, skinned myocytes were attached to two metal micro-needles using UV-sensitive glue (Norland, Cranbury, NJ). The micro-needles were attached to a high-speed piezo translator (ThorLabs, Newton, NJ) and a force transducer. Myocytes were selected based on uniformity of the cell and clear striation patterns. Myocytes were perfused via a closely placed perfusion pipette with relaxing solution. Subsequent perfusion used a mixture of relaxing and activating solutions with varying calcium concentrations (pCa 10.0-pCa 4.5) to measure force development at sarcomere lengths (SLs) of 1.9 μm and 2.3 μm. Maximal activating calcium solution was administered at the start of the experiment to ensure proper cell attachment. SL was measured using FFT analysis of video images using custom-made LabView software (National Instruments, Austin, TX). Force-pCa curves were fit using a modified Hill equation (P/PO = [Ca2+]n/(kn+[Ca2+]n)), where n is the Hill-slope and k is the pCa50. Cell cross-sectional elliptical area was calculated by buckling the cell, followed by measurement using a calibrated screen monitor. Developed force was measured at both SLs at each activating cycle, with the baseline value of developed force subtracted from subsequent measures. Data were not considered valid if total rundown was greater than 20% after final maximal activation at the end of each activating cycle at both SLs. Six hearts from the +/+ group and seven hearts from the +/t and t/t groups were used, with 3 to 4 cells per heart. All data were acquired by custom-made LabView software and analyzed using Origin Pro 8.0.
Isolation of mRNA and quantitative PCR analysis.
Minced heart tissue was homogenized in 1 ml of TriZol using a bead homogenizer, and mRNA was isolated using the Aurum Total RNA Fatty and Fibrous Tissue Kit (Cat. No. 732–6820; Bio-Rad). Synthesis of cDNA from the mRNA samples was performed using the iScript cDNA synthesis kit (Cat. 170–8891; Bio-Rad) with 1 μg of mRNA template used in each reaction. Templates for quantitative PCR analysis were also generated, but without reverse transcriptase (RT), and these were used as negative controls. Gene expression levels were analyzed using TaqMan primers recognizing MYBPC3 (Cat. No. Mm.PT.53a.2930640, IDT), cardiac β-myosin heavy chain gene (MYH7), NPPA, GAPDH, and calsequestrin 2 gene (CASQ2) (Cat. No. Mm01319006g1; Mm01255748g1; Mm00486742m1 and 4352339E; Applied Biosystems) with iTaq Probes Master Mix (Cat. No. 172–5131; Bio-Rad) and a Bio-Rad CFX96 thermocycler. The cycle protocol was 94°C for 10 min followed by 40 cycles at 94°C for 15 s and 60°C for 60 s. Analysis of quantitative PCR data was performed using the ΔΔCq method, normalized to the geometric mean of GAPDH and CASQ2 Cq values and corrected for primer efficiency as described previously (32).
Protein isolation and analysis.
Myofilament and total heart fractions were isolated as described previously (36). All buffers contained protease inhibitors (Cat. No. 4693159001; Roche) and phosphatase inhibitor cocktails (Cat. Nos. P5726 and P0044; Sigma). For Western blotting, antibodies recognizing the N′ region of cMyBP-C (Cat. No. 137180; Santa Cruz) and antibodies against cardiac sarcomeric actin (Cat. No. A2172; Sigma) and α-tropomyosin (Cat. No. T3605; Sigma) were used to assess protein levels and to normalize loading. Secondary HRP-conjugated antibodies (Cat. No. SC-2004 and SC-2314; Santa Cruz) and ECL Prime Western Blotting Detection Reagent (GE Life Sciences) were used for detection. Myosin isoforms were separated using a Hoefer-format SDS-PAGE gel (6.25% Acrylamide, 99:1 Acrylamide:bis) and run for 21 h at 15 mA at 4°C. Gels were fixed in 7% acetic acid and 40% ethanol for 1 h, stained with SYPRO Ruby overnight, destained in fixing solution for 30 min, followed by overnight destaining in water. All gels and blots were imaged using a Bio-Rad ChemiDoc+, and blots were analyzed using NIH ImageJ software.
Statistical analysis.
All data are represented as means ± SE. Statistical analysis was performed using Graph Pad Prism (version 6.0), and data were analyzed using one-way ANOVA with a Bonferroni post hoc test. For force-pCa data at two sarcomere lengths, two-way ANOVA was performed, followed by a Bonferroni post hoc test. Diastolic echocardiography parameters were compared using an unpaired t-test. Statistical significance was defined as P < 0.05.
RESULTS
Heterozygous mouse hearts show an intermediate level of MYBPC3 transcript but no changes in protein content or phosphorylation levels of cMyBP-C.
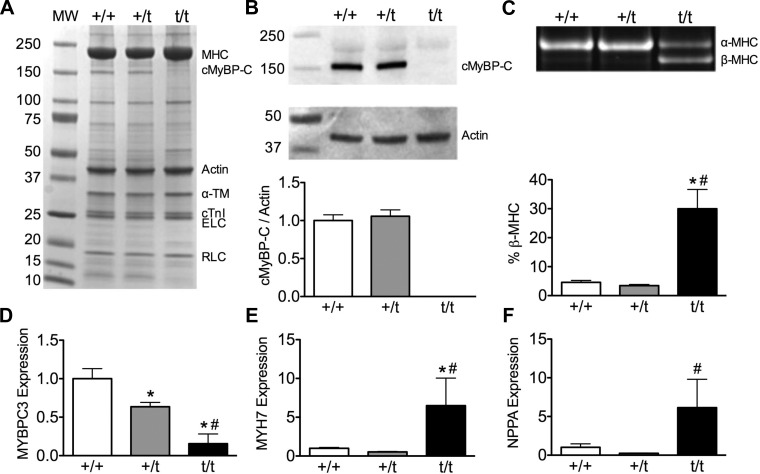
Myofilament protein fractions isolated from +/+ and +/t contained an equal amount of cMyBP-C, whereas no cMyBP-C was present in t/t hearts (Fig. 1A). Furthermore, quantification of cMyBP-C in whole-heart homogenates showed that cMyBP-C levels normalized to actin are unchanged between the +/+ and +/t groups, consistent with previous reports (Fig. 1B) (23, 30). In contrast with the protein data, levels of MYBPC3 transcript measured by quantitative PCR were significantly reduced in the +/t group compared with +/+ (64% of +/+ levels), and t/t levels were significantly reduced compared with +/+ and +/t (16% of +/+ levels) using primers that recognize wild-type and truncated transcript (Fig. 1D).
Fig. 1.
Expression of cardiac myosin binding protein-C gene (MYBPC3) and hypertrophic markers. A: myofilament protein fraction resolved with SDS-PAGE and stained with Coomassie blue. B: Western blot showing levels of cMyBP-C and actin. Western blot quantification shows no cMyBP-C present in the homozygous MYBPC3 truncation mutant mouse hearts (t/t) samples and no significant change in cMyBP-C levels between wild-type MYBPC3 mouse hearts (+/+) and heterozygous MYBPC3 truncation mutant mouse hearts (+/t) (n = 5). C: total heart homogenate used for analysis of myosin heavy chain isoforms (α-MHC and β-MHC) resolved on a Hoefer format SDS-PAGE and stained with Sypro-Ruby. The +/+ and +/t hearts show no changes in α-MHC isoform levels, with t/t hearts showing a significant increase in the hypertrophic marker β-MHC (N = 11, 8, 9). D: MYBPC3 transcription is reduced in +/t hearts compared with +/+ hearts, and t/t hearts show an even further reduction in MYBPC3 expression. E and F: transcript levels of hypertrophic markers cardiac β-myosin heavy chain gene (MYH7) and atrial natriuretic factor gene (NPPA) are unchanged in +/+ and +/t hearts, but they show a significant increase in the t/t hearts (N = 6,7, 3). *P < 0.05 vs. +/+; #P < 0.05 vs. +/t. ELC, essential light chain; RLC, regulatory light chain; cTnI, cardiac troponin I protein; α-TM, α-tropomyosin.
Protein levels of the pathological hypertrophy marker β-myosin heavy chain (β-MHC) resolved by SDS-PAGE and stained with Sypro-Ruby showed no change between +/+ and +/t, whereas the t/t hearts showed a significant elevation in β-MHC (Fig. 1C). Analysis of the transcript levels of hypertrophic markers via quantitative PCR showed no difference in either β-MHC (MYH7) or atrial natriuretic factor (NPPA) between +/+ and +/t, with a significant elevation in the t/t mouse hearts (∼6-fold increase in both MYH7 and NPPA; Fig. 1, E and F).
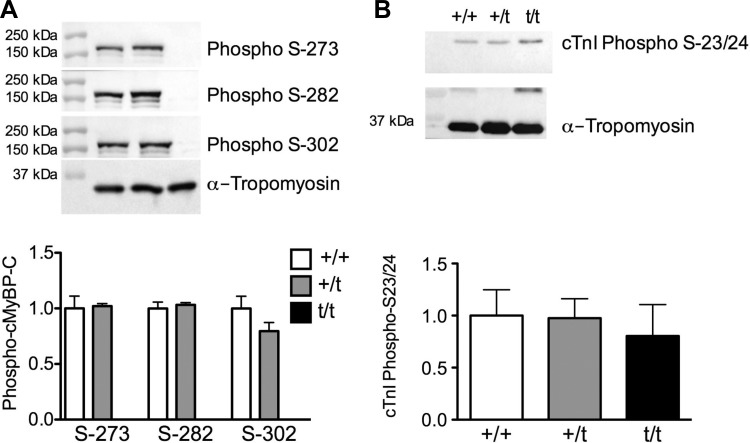
The phosphorylation of three serine residues in the M-domain of cMyBP-C determines its regulatory role in sarcomere function. To determine whether the phosphorylation status of cMyBP-C was altered in the +/t hearts, phosphorylation-dependent, site-specific antibodies to pS273, pS282, and pS302 were used. Phospho-cMyBP-C levels were normalized to total cMyBP-C (as shown in Fig. 1B) and normalized to actin for a loading control. No changes in the phosphorylation status of cMyBP-C at serines 273, 282, or 302 were detected between +/+ and +/t groups (Fig. 2A). Additionally, phosphorylation levels of cardiac troponin I (cTnI) at serines 23 and 24 showed no difference between +/+ and +/t or t/t, although a higher variability of phospho-cTnI was observed compared with phospho-cMyBP-C (Fig. 2B).
Fig. 2.
Phosphorylation of cMyBP-C and cTnI. A: Western blot analysis detecting cMyBP-C phosphorylated at serine-273, -282, and -302. No significant change in phosphorylation of cMyBP-C was observed at any of the phosphorylation sites between the +/+ and +/t groups (n = 5). B: phosphorylation status of cTnI at serine-23 and -24, as determined by Western blot, shows no significant changes in any of the groups (n = 5).
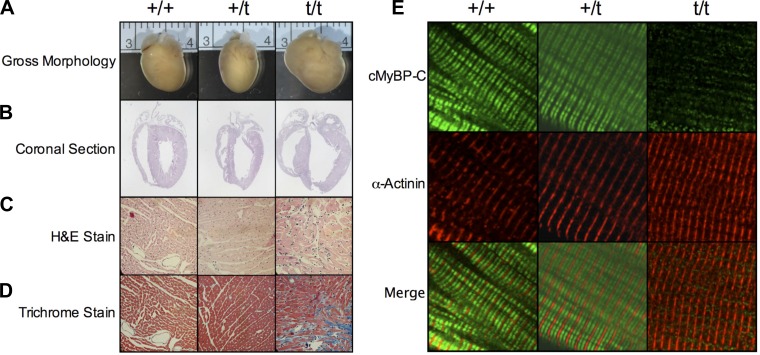
Heterozygous mice do not exhibit gross morphological changes.
Analysis of gross morphology and coronal sections of +/+ and +/t hearts showed no obvious pathology, whereas t/t hearts showed dilation as previously reported (Fig. 3, A and B) (23). Heart weight-to-body weight ratios showed no significant difference between wild-type (4.58 ± 0.32 mg/g heart weight/body weight) and heterozygous (4.62 ± 0.08 mg/g heart weight/body weight) (n = 4). Similarly, hematoxylin and eosin and Trichrome staining revealed no disparities between +/+ and +/t samples (Fig. 3, C and D). Immunostaining of isolated cardiomyocytes showed doublet patterning consistent with properly localized C-band incorporation of cMyBP-C (green) between the α-actinin-stained Z-disks (red) in both the +/+ and +/t groups (Fig. 3E). These observations agree with the original characterization of this model and show that the +/t heart appears to have proper cMyBP-C incorporation with no grossly apparent phenotype.
Fig. 3.
Gross morphology and protein incorporation. Representative whole heart images (A) and coronal sections (B) show no overt hypertrophy in the +/t hearts, whereas t/t hearts display a hypertrophic phenotype. Cross-sectional hematoxylin and eosin (H&E; C) and Trichrome staining (D) reveal deranged muscle arrangement and fibrosis, respectively, in the t/t heart, with no notable differences in +/+ and +/t. E: confocal microcopy immunofluorescence of isolated cardiomyocytes stained for cMyBP-C (green) and α-actinin (red). The cMyBP-C signal shows the characteristic C-zone doublet pattern between each Z-disk, stained for α-actinin in both +/+ and +/t cardiomyocytes. Staining of the t/t cells reveals only minimal cMyBP-C signaling and no structural localization.
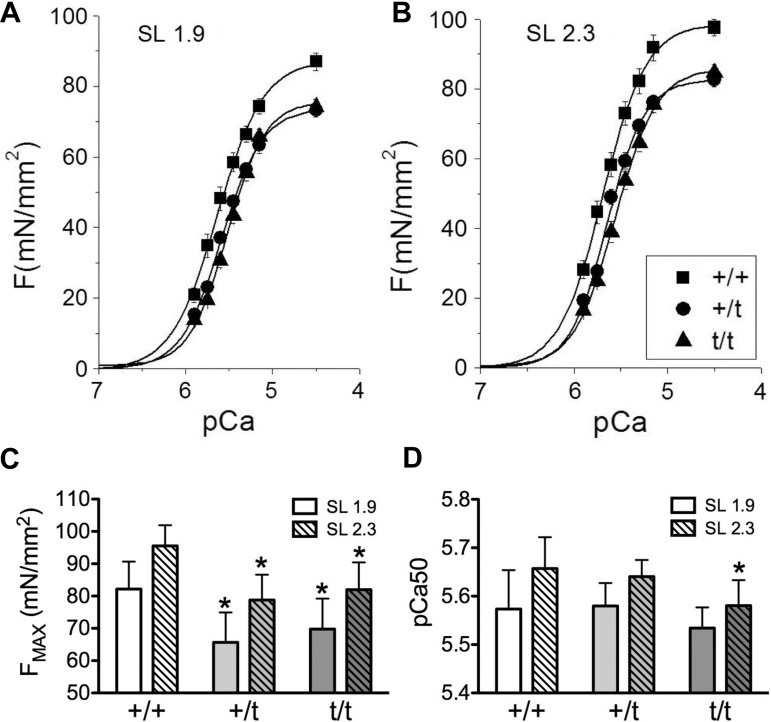
Skinned cardiomyocytes from MYBPC3 heterozygous mutant carriers show a reduction in maximal force generation.
Force-pCa analysis was performed to assess functional disparities in myofilament function between +/+ and +/t myocytes. Skinned myocytes showed a reduction in maximal developed force (FMAX) at pCa 4.5 (3.16 μM Ca2+) in both +/t (68.5 ± 4.1 mN/mm2 at SL 1.9; 79.2 ± 3.1 mN/mm2 at SL 2.3; P = 0.007 SL 1.9, P = 0.004 SL 2.3) and t/t (69.3 ± 3.3 mN/mm2 at SL 1.9; 81.1 ± 3.1 mN/mm2 at SL 2.3; P = 0.0241 SL 1.9, P = 0.009 SL 2.3) groups compared with +/+ (82.2 ± 3.2 mN/mm2 at SL 1.9; 95.5 ± 2.4 mN/mm2 at SL 2.3) (Fig. 4, A–C). We also examined force generation at pCa 5.9 (0.13μM Ca2+) to assess tension development at low calcium concentrations. The force development of the +/+ myocytes (15.5 ± 3.0 mN/mm2 at SL 1.9; 22.8 ± 3.0 mN/mm2 at SL 2.3) was not significantly different than +/t (12.8 ± 1.5 mN/mm2 at SL 1.9; 17.7 ± 1.8 mN/mm2 at SL 2.3), although a significant (P = 0.041) treatment effect was observed between +/+ and t/t (11.8 ± 1.3 mN/mm2 at SL 1.9; 16.1 ± 1.1 mN/mm2 at SL 2.3). Calcium sensitivity of force development, expressed as pCa50, was significantly increased only in t/t hearts at SL 2.3 μm compared with +/+ (Fig. 4D). The +/+ hearts show a significant change in pCa50 (P = 0.01) with changes in sarcomere length, whereas this change is not observed in +/t or t/t (Table 1).
Fig. 4.
Alterations of force development and Ca2+ sensitivity in skinned cardiomyocytes. A–C: maximum development of force (Fmax) is significantly decreased in +/t and t/t skinned cardiomyocytes compared with +/+ at both short [sarcomere length (SL) 1.9 μm] and long (SL 2.3 μm) sarcomere lengths. D: relative force comparisons show a significant decrease in Ca2+ sensitivity of force development in t/t hearts compared with +/+ at SL 2.3 μm. No significant changes in Ca2+ sensitivity were seen between +/t and +/+ (N = 6, 7, and 7 hearts per group, with 3 to 4 cells averaged per heart). *P < 0.05 vs. +/+.
Table 1.
Summary of force-pCa results
| +/+ | +/t | t/t | |
|---|---|---|---|
| ΔFMAX, mN/mm2 | 13.36 ± 1.51 | 12.52 ± 2.33 | 11.72 ± 0.96 |
| ΔpCa50 | 0.084 ± 0.012 | 0.058 ± 0.006 | 0.046 ± 0.008* |
| Sarcomere length 1.9 | |||
| FMAX, mN/mm2 | 82.2 ± 3.2 | 68.5 ± 4.1* | 69.3 ± 3.3* |
| pCa50 | 5.57 ± 0.03 | 5.58 ± 0.02 | 5.53 ± 0.02 |
| nH | 1.91 ± 0.09 | 2.15 ± 0.12 | 2.10 ± 0.04 |
| Sarcomere length 2.3 | |||
| FMAX, mN/mm2 | 95.5 ± 2.4 | 79.2 ± 3.1* | 81.0 ± 3.1* |
| pCa50 | 5.66 ± 0.02 | 5.64 ± 0.01 | 5.58 ± 0.02* |
| nH | 2.02 ± 0.05 | 2.29 ± 0.10 | 2.07 ± 0.08 |
Values are means ± SE and were calculated from force-pCa analysis of skinned cardiomyocytes; n = 3 wild-type MYBPC3 mouse hearts (+/+) and 4 heterozygous MYBPC3 truncation mutant mouse hearts (+/t) and homozygous MYBPC3 truncation mutant mouse hearts (t/t) per group, with 4 cells averaged per heart.
Significant reductions in maximal developed force (FMAX) are observed in +/t and t/t compared with +/+ skinned cardiomyocytes. Significant alteration in calcium sensitivity of force development was only detected in the t/t skinned cardiomyocytes at sarcomere length 2.3 μm. No significant changes in Hill coefficient or length-dependent activation were observed. P < 0.05 vs. +/+.
MYBPC3 heterozygous mice show altered cardiac function in vivo.
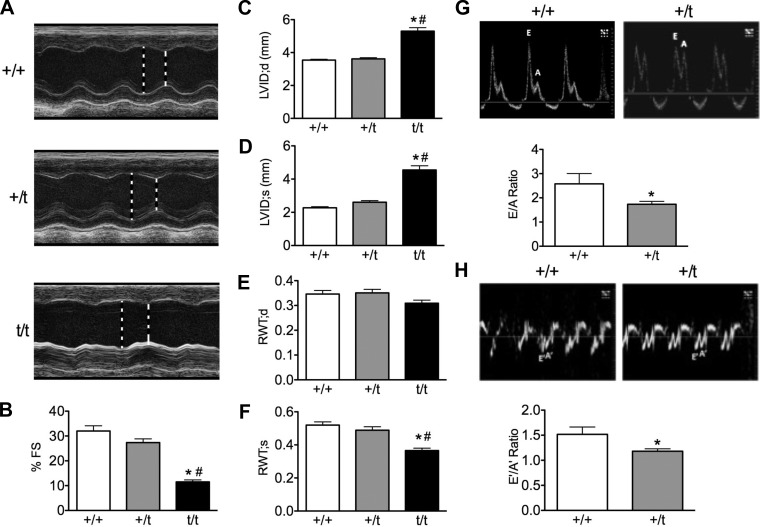
To determine any functional deficits in the heterozygous heart, echocardiography analysis was performed to assess morphology, systolic, and diastolic function. M-mode parasternal long axis echocardiography showed no difference between +/t and +/+ in interventricular septum or posterior wall thickness (Table 2), no dilation of the left ventricle in systole or diastole, and no changes in relative wall thickness compared with total heart thickness (Fig. 5, A and C–F). The +/t mice showed a nonsignificant (P = 0.114) trend toward reduced percent fractional shortening (27.4 ± 1.5%) compared with +/+ (32.0 ± 2.1%), with a significant (P < 0.001) reduction in fractional shortening in the t/t mice (11.53 ± 0.79) (Fig. 5B). Additionally, M mode derived percent ejection fraction followed the same nonsignificant trend between +/+ (60.6% ± 3.0) and +/t (53.6% ± 2.5), with a significant (P < 0.001) reduction in systolic function in the t/t group (24.9% ± 1.6). Power Doppler measurements of left ventricular filling revealed a significant reduction (P = 0.003) in the early/after (E/A) ratio in +/t (1.74 ± 0.12) compared with +/+ (2.58 ± 0.53) (Fig. 5G). Tissue Doppler analysis of mitral valve motion also showed a significant decrease (P = 0.025) in the E′/A′ ratio in the +/t mice (1.18 ± 0.05) compared with +/+ (1.52 ± 0.15) (Fig. 5H). Although these diastolic parameters showed significant impairments in +/t heart relaxation, other parameters, such as the E′ and E velocities, as well as the E/E′ ratio, were not significantly altered (Table 2). Diastolic function was not assessed in t/t mice because that group was not available at the time diastolic parameters were measured.
Table 2.
Summary of echocardiography results
| +/+ | +/t | t/t | |
|---|---|---|---|
| n | 7 | 10 | 9 |
| Fractional shortening, % | 32.02 ± 2.08 | 27.36 ± 1.48 | 11.53 ± 0.79*# |
| E, mm/s | 784 ± 36 | 768 ± 38 | — |
| E/A | 2.58 ± 0.43 | 1.74 ± 0.12* | — |
| E′, mm/s | −22.8 ± 3.2 | −22.7 ± 5.7 | — |
| E′/A' | 1.52 ± 0.15 | 1.18 ± 0.05* | — |
| E/E′ | −35.3 ± 7.2 | −35.8 ± 10.0 | — |
| Interventricular septum, mm | |||
| Diastole | 0.98 ± 0.05 | 1.05 ± 0.05 | 1.09 ± 0.05 |
| Systole | 1.40 ± 0.05 | 1.44 ± 0.07 | 1.27 ± 0.07 |
| Left ventricular internal diameter, mm | |||
| Diastole | 3.75 ± 0.1 | 3.87 ± 0.11 | 4.92 ± 0.16*# |
| Systole | 2.55 ± 0.13 | 2.82 ± 0.13 | 4.36 ± 0.17*# |
| Left ventricular posterior wall, mm | |||
| Diastole | 1.00 ± 0.06 | 1.03 ± 0.05 | 1.10 ± 0.07 |
| Systole | 1.35 ± 0.07 | 1.25 ± 0.06 | 1.23 ± 0.08 |
Values are means ± SE. E/A, ratio of the early (E) to late (A) ventricular filling velocities; E′/A′, ratio of early (E′) to late (A′) mitral annulus motion. Significant reduction in fractional shortening was observed between t/t and +/+. E/A and E′/A′ ratios were significantly reduced in +/t compared with +/+.
P < 0.05 vs. +/+;
#P < 0.05 vs. +/t.
Fig. 5.
In vivo measurements of cardiac morphology and function. A: representative parasternal long-axis M-mode echocardiography tracings from 10- to 12-wk-old +/+, +/t, and t/t mouse hearts. B: percentage of left ventricular fractional shortening (FS) shows that +/t hearts trend toward reduced pump function compared with +/+, whereas t/t hearts show a significant deficit compared with both +/+ and +/t groups. C and D: left ventricular internal diameter (LVID) at peak systole (s) and diastole (d) shows significant dilation in the t/t hearts with no changes between +/+ and +/t. E and F: relative wall thickness (RWT; anterior + posterior wall/total heart thickness) at peak systole and diastole shows no change between +/t and +/+, whereas the wall-to-heart ratio is reduced in t/t. G and H: representative power and tissue Doppler images depicting the early (E) and late (A) blood filling of the left ventricle and mitral valve motion. E/A and E′/A′ ratios were significantly reduced in +/t hearts compared with +/+, with t/t hearts showing a significant deficit compared with both groups (N = 7, 10, 9). *P < 0.05 vs. +/+; #P < 0.05 vs. +/t.
DISCUSSION
This study aimed to detect functional deficits in a MYBPC3 heterozygous mutant mouse with preserved cMyBP-C levels. Although this heterozygous model has normal cMyBP-C levels, proper protein incorporation, and normal morphology, the results of these experiments demonstrate that functional deficits do exist at the whole heart and single myocyte levels. Here we show a 36% decrease in MYBPC3 transcription in the heterozygote, whereas cMyBP-C protein levels remain normal. The preservation of cMyBP-C level in the heterozygote could be explained by reduced myofilament degradation or a longer half-life of cMyBP-C in the sarcomere. The wild-type levels of transcript are either in excess under normal conditions or compensatory mechanisms in the heterozygote compensate, such as with reduced cMyBP-C turnover. However, despite proper cMyBP-C stoichiometry, maximal force development is reduced in heterozygous myocytes, indicating that these mice have impairments in contractility. In addition to these deficits, the heterozygous mice demonstrate a trend toward reduced fractional shortening, which is consistently measured on the low-normal side, as well as significantly reduced E/A and E′/A′ ratios, indicating the development of diastolic dysfunction. On the other hand, force development in skinned myocytes did not show any significant difference at diastolic calcium concentrations, leaving the cause of diastolic dysfunction unclear. The myofilament abnormalities and alterations in organ function do not appear to manifest morphologically, since the heterozygote does not show any signs of hypertrophy at the whole organ level nor does the heterozygote display an increase in levels of hypertrophic markers. These findings suggest that seemingly asymptomatic heterozygous MYBPC3 mutant carriers do have functional impairments, which may cause the development of HCM with increased age or when subjected to additional cardiac stress.
The mechanism by which heterozygous truncation mutations of MYBPC3 initiate the development of HCM is unclear. Human carriers of these mutations exhibit variable disease penetrance, often with no expression of the mutant protein (35). Heterozygous carriers of various MYBPC3 mutations are typically asymptomatic until adulthood and often develop a mild phenotype compared with homozygous MYBPC3 mutant carriers (6, 9, 24, 25). The variable penetrance of these mutations is evident in pedigree analysis of mutation-carrying families, where the same mutation can result in asymptomatic or symptomatic phenotypes (6, 9, 24, 25). The role that MYBPC3 haploinsufficiency plays in these cases is also unclear. In symptomatic heterozygous mutation carriers, a reduced level of cMyBP-C has been identified (21, 24, 42). This reduction in protein has been linked to a reduction in force development, although this may be attributed to the HCM phenotype, since these alterations are also observed in HCM patients with non-MYBPC3 mutations and normal cMyBP-C levels (39, 43). However, because samples from asymptomatic mutation carriers are unavailable, it is unclear whether reduction of cMyBP-C initiates the development of HCM or merely accompanies it, as the result of an unknown primary effect. A better understanding of the early changes in these pathways would provide additional opportunity for diagnosis and intervention (12).
Several cMyBP-C knockout mouse models have been developed over the last 15 years. The model developed by the Seidman laboratory in 1999, which was used for this work, was generated with a knock-in mutation resulting in a gene encoding a predicted truncated protein, which is undetectable in the heart. Heterozygous mice with this mutation do not show a hypertrophic phenotype, whereas the homozygous mouse develops dilated cardiomyopathy, decreased cardiac function, and a reduction in force development (22, 23, 29–31). The MYBPC3 knockout “null” mouse model was generated in the Moss laboratory in 2002. This model has exons 3–10 removed, resulting in the absence of cMyBP-C expression in the homozygous mouse (15). In initial reports, the heterozygous genotype of this model does not exhibit any reported deficits, with normal levels of cMyBP-C, despite reduced mRNA levels (15). However, more recent evaluation of this heterozygous model does show a 32% decrease in cMyBP-C levels, as well as mild functional impairments consistent with the onset of HCM (7, 8). Two MYBPC3 mutant mouse models were developed in the Carrier lab (5, 45), one of which is a knockout mouse lacking exon 1 and 2, resulting in a null allele (5). In contrast with the Seidman model, this heterozygous mouse model does show a reduction in cMyBP-C levels (25% of wild-type), with a reduction in MYBPC3 mRNA (from ∼80% to ∼50% of wild-type levels from 6 to 11 mo of age), and develops HCM symptoms at 10 to 11 mo of age (5). The other model is a knock-in mouse carrying a common human mutation that results in several mRNA splice products. This causes the expression of one potential poison polypeptide and an overall 50% reduction in mRNA, in turn resulting in a 20% reduction of cMyBP-C in the heterozygote (45). Although this model does not appear to develop HCM, these mice do show increased calcium sensitivity at the skinned fiber level, and alterations in calcium handling were observed in intact cardiomyocytes (10).
A consistent theme found in all these models is that homozygous mutant carriers show a severe dilated or eccentric hypertrophy phenotype, whereas the heterozygous models are more variable in their pathology, typically showing no hypertrophy or mild hypertrophy, with the development of age-dependent HCM. These observations parallel those made in human carriers of MYBPC3 mutations that are linked to HCM. The study of these various heterozygous mouse models underscores the subtle mechanistic differences among specific MYBPC3 mutations, illustrating the complexity of the heterozygous condition in mouse models and human patients. The mechanism that drives the development of dysfunction in heterozygous mutant mice is currently unclear. A haploinsufficiency model suggests that reduction of cMyBP-C levels from insufficient expression from one wild-type allele causes the initial insult for the development of HCM. The poison polypeptide model proposes that mutant MYBPC3 genes express a protein that is potentially unable to incorporate properly into the sarcomere, causing disruption of cellular processes. However, in cases where cMyBP-C levels are preserved and little mutant MYBPC3 transcript is present, other mechanisms may be involved. For example, it has been suggested that impairment of the ubiquitin-proteasome system or the nonsense-mediated mRNA decay pathway over an individual's lifetime may account for the late onset of hypertrophy (37, 38, 45).
In this study, it was observed that heterozygous MYBPC3 truncation mutations in mice results in reduced mRNA levels, a full cMyBP-C complement, and only minor functional disparities. Thus, even in nonhypertrophic carriers, some perturbations may be occurring. The identity of these alterations and whether they are compensatory processes or the result of decompensation remain to be clarified. Elucidating the mechanism by which lower levels of gene expression can lead to a disease phenotype, while protein levels are maintained, is an interesting problem. One line of inquiry into this problem might examine MYBPC3 gene dosage effect relative to normal cMyBP-C protein turnover in the sarcomere such that lower gene expression could lead to an increased half-life of cMyBP-C. Another potential explanation previously reported implicates haploinsufficiency of MYBPC3 (37, 38, 45). This notion holds that an increased need to degrade aberrant mRNA via nonsense-mediated decay or misfolded protein via proteasome or autophagic pathways exerts a stress on the cardiomyocytes, leading to dysfunction. It is also not absolutely clear if the truncated gene is able to express small amounts of mutant cMyBP-C, which could result in a functional impairment of the sarcomere.
Because the results of this study indicate that the this heterozygous mouse is already functionally compromised, it would be beneficial to determine whether these animals have an increased risk for hypertrophy following cardiovascular stress, since this may provide direct insight into the development of HCM and heart failure in human heterozygous MYBPC3 mutant carriers.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute Grants R01HL-105826 and K02HL-114749 (to S. Sadayappan), HL-101297, HL-75494, and HL-62426 (to P. de Tombe) and the American Heart Association Grant 11PRE7240022 (to D. Barefield).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: D.B., P.P.d.T., and S.S. conception and design of research; D.B. and M.K. performed experiments; D.B., M.K., P.P.d.T., and S.S. analyzed data; D.B., P.P.d.T., and S.S. interpreted results of experiments; D.B., M.K., and S.S. prepared figures; D.B. drafted manuscript; D.B., P.P.d.T., and S.S. edited and revised manuscript; D.B., M.K., P.P.d.T., and S.S. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Suresh Govindan, Xiang Ji, and Beth Mayer in the Department of Cell and Molecular Physiology at Loyola for support of this work. We also thank Jonathan Seidman and Christine Seidman in the Department of Genetics, Harvard Medical School, for the t/t mouse model and critique of the study.
REFERENCES
- 1.Alcalai R, Seidman J, Seidman C. Genetic basis of hypertrophic cardiomyopathy: from bench to the clinics. J Cardiovasc Electrophysiol 19: 104–110, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Barefield D, Sadayappan S. Phosphorylation and function of cardiac myosin binding protein-C in health and disease. J Mol Cell Cardiol 48: 866–875, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonne G, Carrier L, Bercovici J, Cruaud C, Richard P, Hainque B, Gautel M, Labeit S, James M, Beckmann J, Weissenbach J, Vosberg HP, Fiszman M, Komajda M, Schwartz K. Cardiac myosin binding protein-C gene splice acceptor site mutation is associated with familial hypertrophic cardiomyopathy. Nat Genet 11: 438–440, 1995 [DOI] [PubMed] [Google Scholar]
- 4.Carrier L, Bonne G, Bahrend E, Yu B, Richard P, Niel F, Hainque B, Cruaud C, Gary F, Labeit S, Bouhour J, Dubourg O, Desnos M, Hagege A, Trent R, Komajda M, Fiszman M, Schwartz K. Organization and sequence of human cardiac myosin binding protein C gene (MYBPC3) and identification of mutations predicted to produce truncated proteins in familial hypertrophic cardiomyopathy. Circ Res 80: 427–434, 1997 [PubMed] [Google Scholar]
- 5.Carrier L, Knoll R, Vignier N, Keller D, Bausero P, Prudhon B, Isnard R, Ambroisine M, Fiszman M, Ross J, Schwartz K, Chien K. Asymmetric septal hypertrophy in heterozygous cMyBP-C null mice. Cardiovasc Res 63: 293–304, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Charron P, Dubourg O, Desnos M, Bennaceur M, Carrier L, Camproux A, Isnard R, Hagege A, Langlard J, Bonne G, Richard P, Hainque B, Bouhour J, Schwartz K, Komajda M. Clinical features and prognostic implications of familial hypertrophic cardiomyopathy related to the cardiac myosin-binding protein c gene. Circulation 97: 2230–2236, 1998 [DOI] [PubMed] [Google Scholar]
- 7.Cheng Y, Wan X, McElfresh T, Chen X, Gresham K, Rosenbaum D, Chandler M, Stelzer J. Impaired contractile function due to decreased cardiac myosin binding protein C content in the sarcomere. Am J Physiol Heart Circ Physiol 305: H52–H65, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desjardins C, Chen Y, Coulton A, Hoit B, Yu X, Stelzer J. Cardiac myosin binding protein C insufficency leads to early onset of mechanical dysfunction. Circ Cardiovsc Imaging 5: 127–136, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhandapany PS, Sadayappan S, Xue Y, Powell GT, Rani DS, Nallari P, Rai TS, Khullar M, Soares P, Bahl A, Tharkan JM, Vaideeswar P, Rathinavel A, Narasimhan C, Ayapati DR, Ayub Q, Mehdi SQ, Oppenheimer S, Richards MB, Price AL, Patterson N, Reich D, Singh L, Tyler-Smith C, Thangaraj K. A common MYBPC3 (cardiac myosin binding protein C) variant associated with cardiomyopathies in South Asia. Nat Genet 41: 187–191, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraysse B, Weinberger F, Bardswell S, Cuello F, Vignier N, Geertz B, Starbatty J, Kramer E, Coirault C, Eschenhagen T, Kentish J, Avkiran MC, L Increased myofilament Ca2+ sensitivity and diastolic dysfunction as early consequences of Mybpc3 mutation in heterozygous knock-in mice. J Mol Cell Cardiol 52: 1299–1307, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gautel M, Zuffardi O, Freiburg A, Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: a modulator of cardiac contraction? Embo J 14: 1952–1960, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Germans T, Russel I, Gotte M, Spreeuwenberg M, Doevendans P, Pinto Y, van der Geest R, van der Velden J, Wilde A, van Rossum A. How do hypertrophic cardiomyopathy mutations affect myocardial function in carriers with normal wall thickness? Assessment with cardiovascular magnetic resonance. J Cardiov Magn Reson 12: 13, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Govindan S, Sarkey J, Ji X, Sundaresan NR, Gupta MP, de Tombe PP, Sadayappan S. Pathogenic properties of the N-terminal region of cardiac myosin binding protein-C in vitro. J Muscle Res Cell Motil 33: 17–30, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruen M, Prinz H, Gautel M. cAPK-phosphorylation controls the interaction of the regulatory domain of cardiac myosin binding protein C with myosin-S2 in an on-off fashion. FEBS Lett 453: 254–259, 1999 [DOI] [PubMed] [Google Scholar]
- 15.Harris S, Bartley C, Hacker T, McDonald K, Douglas P, Greaser M, Powers P, Moss R. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res 90: 594–601, 2002 [DOI] [PubMed] [Google Scholar]
- 16.Korte F, McDonald K, Harris S, Moss R. Loaded shortening, power output, and rate of force redevelopment are increased with knockout of cardiac myosin binding protein-C. Circ Res 93: 752–758, 2003 [DOI] [PubMed] [Google Scholar]
- 17.Kuster DW, Sequeira V, Najafi A, Boontje NM, Wijnker PJ, Witjas-Paalberends ER, Marston SB, Dos Remedios CG, Carrier L, Demmers JA, Redwood C, Sadayappan S, van der Velden J. GSK3beta phosphorylates newly identified site in the proline-alanine-rich region of cardiac myosin-binding protein C and alters cross-bridge cycling kinetics in human: short communication. Circ Res 112: 633–639, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luther P, Bennett P, Knupp C, Craig R, Padron R, Harris S, Patel J, Moss R. Understanding the organisation and role of myosin binding protein C in normal striated muscle by comparison with MyBP-C knockout cardiac muscle. J Mol Biol 384: 60–72, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maron B, Gardin J, Flack J, Gidding S, Kurosaki T, Bild D. Prevalence of hypertrophic cardiomyopathy in a general population of young adults: echocardiographic analysis of 4111 subjects in the CARDIA study. Circulation 92: 785–789, 1995 [DOI] [PubMed] [Google Scholar]
- 20.Marston S, Copeland O, Gehmlich K, Schlossarek S, Carrier L. How do MYBPC3 mutations cause hypertrophic cardiomyopathy? J Muscle Res Cell Motil 33: 75–80, 2012 [DOI] [PubMed] [Google Scholar]
- 21.Marston S, Copeland O, Jacques A, Livesey K, Tsang V, McKenna W, Jalilzadeh S, Carballo S, Redwood C, Watkins H. Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circ Res 105: 219–222, 2009 [DOI] [PubMed] [Google Scholar]
- 22.McConnell B, Fatkin D, Semsarian C, Jones K, Georgakopoulous D, Maguire C, Healey M, Mudd J, Moskowitz I, Conner D, Giewat M, Wakimoto H, Berul C, Schoen F, Kass D, Seidman C, Seidman J. Comparison of two murine models of familial hypertrophic cardiomyopathy. Circ Res 88: 383–389, 2001 [DOI] [PubMed] [Google Scholar]
- 23.McConnell B, Jones K, Farkin D, Arroyo L, Lee R, Aristizabal O, Turnbull D, DG, Kass D, Bond M, Niimura H, Schoen F, Conner D, Fischman D, Seidman C, Seidman J. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J Clin Invest 104: 1235–1244, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moolman J, Reith S, Uhl K, Bailey S, Gautel M, Jeschke B, Fischer C, Ochs J, McKenna W, Klues H, Vosberg H. A newly created splice donor site in econ 25 of the MyBP-C gene is responsible for inherited hypertrophic cardiomyopathy with incomplete disease penetrance. Circulation 101: 1396–1402, 2000 [DOI] [PubMed] [Google Scholar]
- 25.Niimura H, Bachinski L, Sangwatanaroj S, Watkins H, Chudley A, McKenna W, Kristinsson A, Roberts R, Sole M, Maron B, Seidman J, Seidman C. Mutations in the gene for cardiac myosin-binding protein c and late-onset familial hypertrophic cardiomyopathy. N Engl J Med 338: 1248–1257, 1998 [DOI] [PubMed] [Google Scholar]
- 26.O′Connell T, Rodrigo M, Simpson P. Isolation and culture of adult mouse cardiac myocytes. Methods Mol Biol 357: 271–296, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Offer G, Moos C, Starr R. A new protein of the thick filaments of vertebrate skeletal myofibrils. Extractions, purification and characterization. J Mol Bio 74: 653–676, 1973 [DOI] [PubMed] [Google Scholar]
- 28.Okagaki T, Weber F, Fischman D, Vaughan K, Mikawa T, Reinach F. The major myosin-binding domain of skeletal myscle MyBP-C (C protein) resides in the COOH-terminal, immunoglobulin C2 motif. J Cell Biol 123: 619–626, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer B, Georgakopoulous D, Janssen P, Wang Y, Alpert N, Belardi D, Harris S, Moss R, Burgon P, Seidman C, Seidman J, Maughan D, Kass D. Role of cardiac myosin binding protein C in sustaining left ventricular systolic stiffening. Circ Res 94: 1249–1255, 2004 [DOI] [PubMed] [Google Scholar]
- 30.Palmer B, McConnell B, Li G, Seidman C, Seidman J, Irving T, Alpert N, Maughan D. Reduced cross-bridge dependent stiffness of skinned myocardium from mice lacking cardiac myosin binding protein-C. Mol Cell Biochem 263: 73–80, 2004 [DOI] [PubMed] [Google Scholar]
- 31.Palmer B, Noguchi T, Wang Y, Heim J, Alpert N, Burgon P, Seidman C, Seidman J, Maughan D, LeWinter M. Effect of cardiac myosin binding protein-C on mechanoenergetics in mouse myocardium. Circ Res 94: 1615–1622, 2004 [DOI] [PubMed] [Google Scholar]
- 32.Pfaffl M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: 2002–2007, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pohlmann L, Kroger I, Vignier N, Schlossarek S, Kramer E, Coirault C, Sultan K, El-Armouche A, Winegrad S, Eschenhagen T, Carrier L. Cardiac myosin-binding protein C is required for complete relaxation in intact myocytes. Circ Res 101: 928–938, 2007 [DOI] [PubMed] [Google Scholar]
- 34.Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, Benaiche A, Isnard R, Dubourg O, Burban M, Gueffet J, Millaire A, Desnos M, Schwartz K, Hainque B, Komajda M. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations and implications for molecular diagnosis stategy. Circulation 107: 2227–2232, 2003 [DOI] [PubMed] [Google Scholar]
- 35.Rottbauer W, Gautel M, Zehelein J, Labeit S, Franz W, Fischer C, Vollrath B, Mall G, Dietz R, Kubler W, Katus H. Novel splice donor site mutation in the cardiac myosin-binding protein-C gene in familial hypertrophic cardiomyopathy. Characterization of cardiac transcript and protein. J Clin Invest 100: 475–482, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadayappan S, Gulick J, Osinska H, Martin LA, Hahn HS, Dorn GW, 2nd, Klevitsky R, Seidman CE, Seidman JG, Robbins J. Cardiac myosin binding protein-C phosphorylation and cardiac function. Circ Res 97: 1156–1163, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlossarek S, Englmann D, Sultan K, Sauer M, Eschenhagen T, Carrier L. Defective proteolytic systems in Mybpc3-targeted mice with cardiac hypertrophy. Basic Res Cardiol 107: 235, 2012 [DOI] [PubMed] [Google Scholar]
- 38.Schlossarek S, Schuermann F, Geertz B, Mearini G, Eschenhagen T, Carrier L. Adrenergic stress reveals septal hypertrophy and proteasome impairment in heterozygous Mybpc3-targeted knock-in mice. J Muscle Res Cell Motil 33: 5–15, 2012 [DOI] [PubMed] [Google Scholar]
- 39.Sequeira V, Wijnker P, Nijenkamp L, Kuster D, Najafi A, Witjas-Paalberends E, Regan J, Boontje N, ten Cate F, Germans T, Carrier L, Sadayappan S, van Slegtenhorst M, Zaremba R, Foster D, Murphy A, Poggesi C, dos Remedios C, Stienen G, Ho C, Michels M, van der Velden J. Purterbed length-dependent activation in human hypertrophic cardiomyopathy with missense sarcomeric gene mutations. Circ Res 112: 1491–1505, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong C, Stelzer J, Greaser M, Powers P, Moss R. Acceleration of crossbridge kinetics by protein kinase A phosphorylation of cardiac myosin binding protein C modulates cardiac function. Circ Res 103: 974–982, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Velden J, Papp Z, Boontje N, Zaremba R, de Jong J, Janssen P, Hasenfuss G, Stienen G. The effect of myosin light chain 2 dephosphorylation on Ca2+-sensitivity of force is enchanced in failing human hearts. Cardiovasc Res 57: 505–514, 2003 [DOI] [PubMed] [Google Scholar]
- 42.van Dijk S, Dooijes D, dos Remedios C, Michels M, Lamers J, Winegrad S, Schlossarek S, Carrier L, ten Cate F, Stienen G, van der Velden J. Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation 119: 1473–1483, 2009 [DOI] [PubMed] [Google Scholar]
- 43.van Dijk S, Paalberends E, Najafi A, Michels M, Sadayappan S, Carrier L, Boontje N, Kuster D, van Slegtenhorst M, Dooijes D, dos Remedios C, ten Cate F, Stienen G, van der Velden J. Contractile dysfunction irrespective of the mutant protein in human hypertrophic cardiomyopathy with normal systolic function. Circ Heart Fail 5: 36–46, 2012 [DOI] [PubMed] [Google Scholar]
- 44.Van Driest S, Ommen S, Tajik A, Gersh B, Ackerman M. Sarcomeric genotyping in hypertrophic cardiomyopathy. Mayo Clin Proc 80: 463–469, 2005 [DOI] [PubMed] [Google Scholar]
- 45.Vignier N, Schlossarek S, Fraysse B, Mearini G, Kramer E, Herve P, Mougenot N, Guiard J, Reimer R, Hohenberg H, Schwartz K, Vernet M, Eschenhagen T, Carrier L. Nonsense-mediated mRNA decay and ubiquitin-proteasome system regulate cardiac myosin-binding protein C mutant levels in cardiomyopathic mice. Circ Res 105): 239–248, 2009 [DOI] [PubMed] [Google Scholar]
- 46.Watkins H, Conner D, Thierfelder L, Jarcho JA, MacRae C, McKenna WJ, Maron BJ, Seidman JG, Seidman CE. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet 11: 434–437, 1995 [DOI] [PubMed] [Google Scholar]