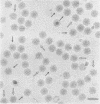
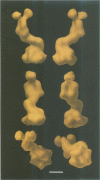
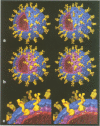
Abstract
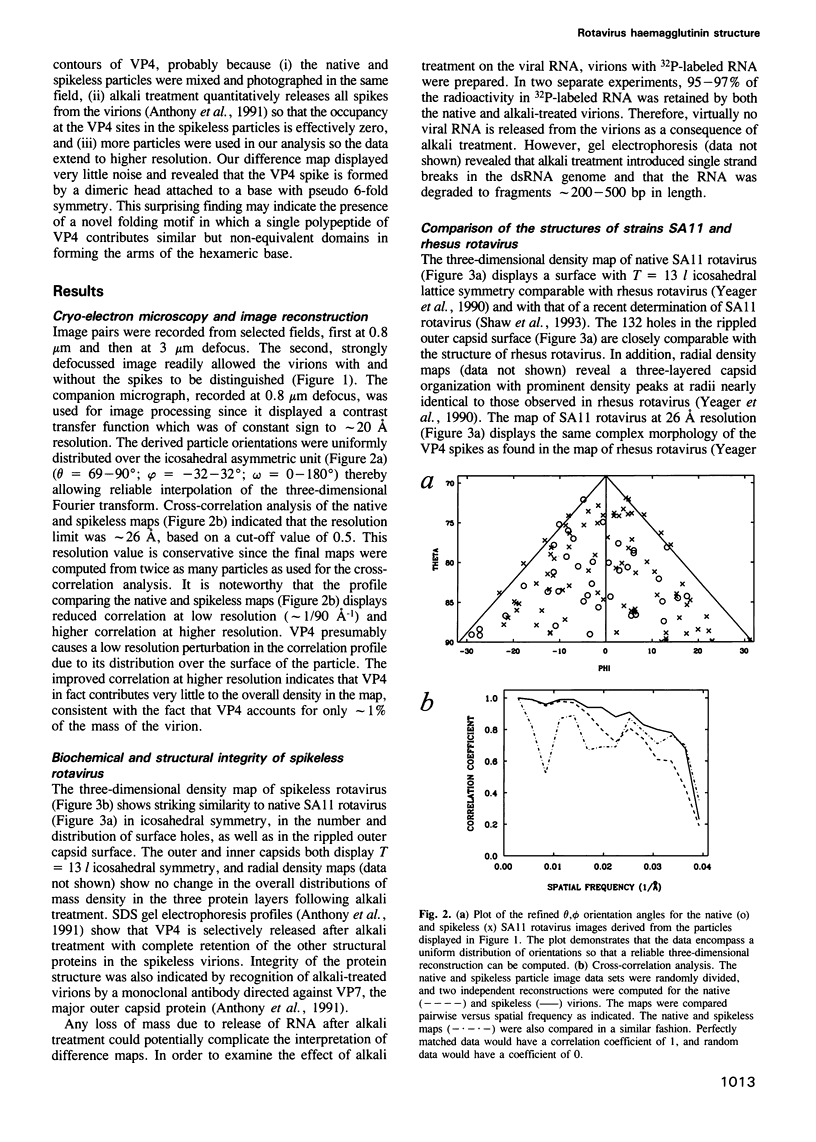
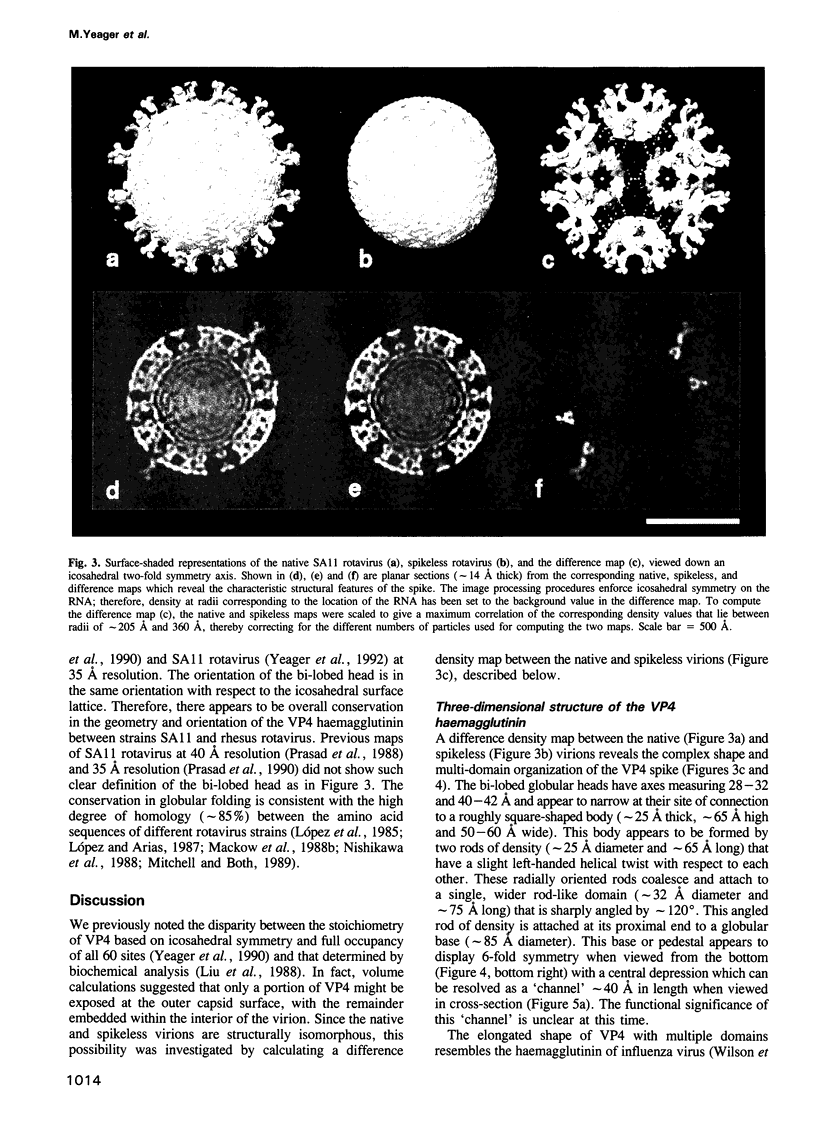
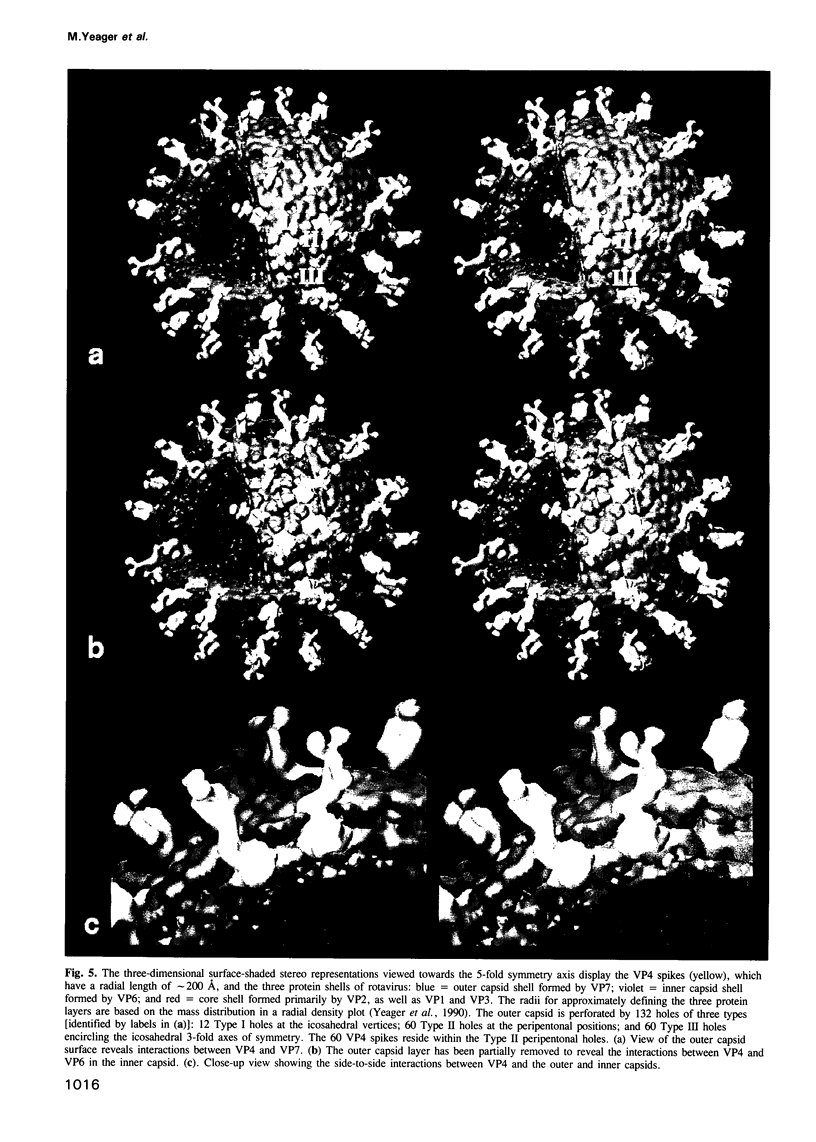
The three-dimensional structure of the rotavirus spike haemagglutinin viral protein 4 (VP4) has been determined to a resolution of 26 A by cryo-electron microscopy and difference analysis of intact virions and smooth (spikeless) particles. Native and spikeless virions were mixed prior to cryo-preservation so that both structures could be determined from the same micrograph, thereby minimizing systematic errors. This mixing strategy was crucial for difference map analysis since VP4 only accounts for approximately 1% of the virion mass. The VP4 spike is multi-domained and has a radial length of approximately 200 A with approximately 110 A projecting from the surface of the virus. Interactions between VP4 and cell surface receptors are facilitated by the bi-lobed head, which allows multi-site interactions, as well as the uniform distribution of the VP4 heads at maximum radius. The bi-lobed head is attached to a square-shaped body formed by two rods that have a slight left-handed helical twist. These rods merge with an angled, rod-like domain connected to a globular base approximately 85 A in diameter. The anchoring base displays pseudo 6-fold symmetry. This surprising finding may represent a novel folding motif in which a single polypeptide of VP4 contributes similar but non-equivalent domains to form the arms of the hexameric base. The VP4 spike penetrates the virion surface approximately 90 A and interacts with both outer (VP7) and inner (VP6) capsid proteins. The extensive VP4-VP7 and VP4-VP6 interactions imply a scaffolding function in which VP4 may participate in maintaining precise geometric register between the inner and outer capsids.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF
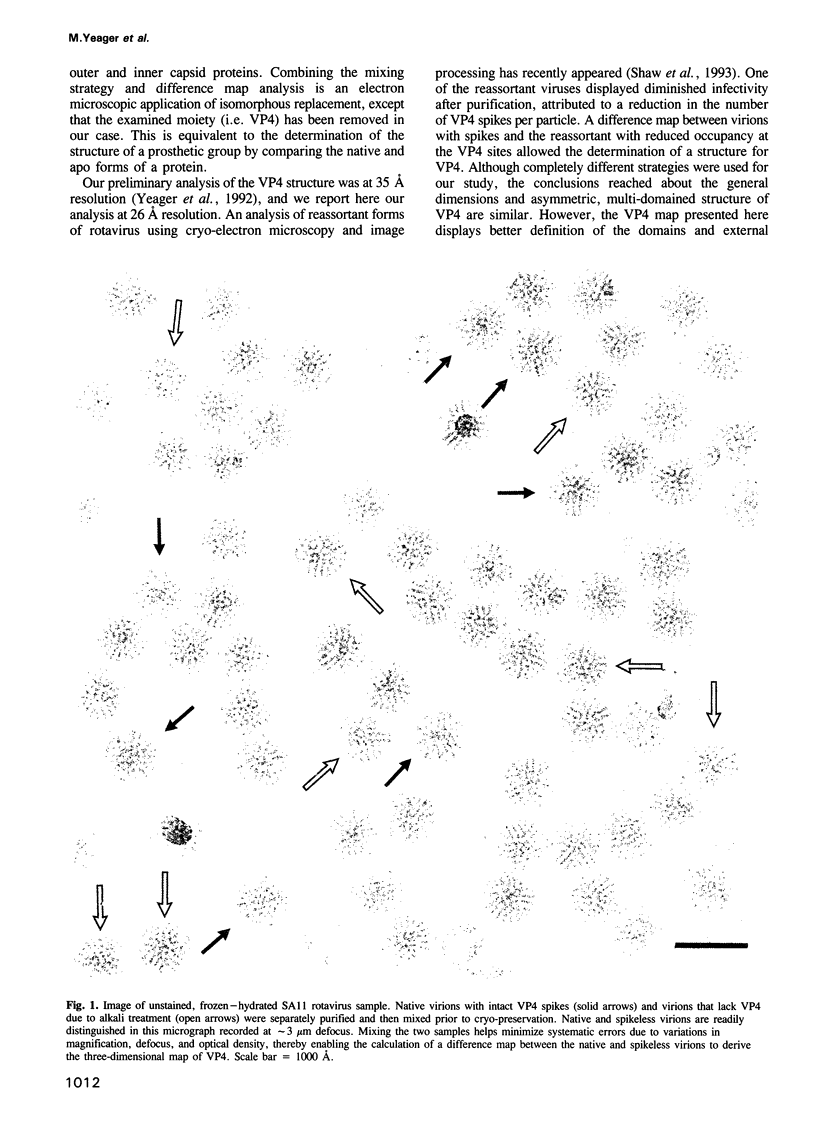



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adrian M., Dubochet J., Lepault J., McDowall A. W. Cryo-electron microscopy of viruses. Nature. 1984 Mar 1;308(5954):32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- Anthony I. D., Bullivant S., Dayal S., Bellamy A. R., Berriman J. A. Rotavirus spike structure and polypeptide composition. J Virol. 1991 Aug;65(8):4334–4340. doi: 10.1128/jvi.65.8.4334-4340.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellamy A. R., Both G. W. Molecular biology of rotaviruses. Adv Virus Res. 1990;38:1–43. doi: 10.1016/s0065-3527(08)60858-1. [DOI] [PubMed] [Google Scholar]
- Bican P., Cohen J., Charpilienne A., Scherrer R. Purification and characterization of bovine rotavirus cores. J Virol. 1982 Sep;43(3):1113–1117. doi: 10.1128/jvi.43.3.1113-1117.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. M., Roth J. R., Clark M. L., Barnett B. B., Spendlove R. S. Trypsin enhancement of rotavirus infectivity: mechanism of enhancement. J Virol. 1981 Sep;39(3):816–822. doi: 10.1128/jvi.39.3.816-822.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther R. A. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Philos Trans R Soc Lond B Biol Sci. 1971 May 27;261(837):221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- Dubochet J., Adrian M., Chang J. J., Homo J. C., Lepault J., McDowall A. W., Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988 May;21(2):129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Esparza J., Gil F. A study on the ultrastructure of human rotavirus. Virology. 1978 Nov;91(1):141–150. doi: 10.1016/0042-6822(78)90362-8. [DOI] [PubMed] [Google Scholar]
- Flores J., Midthun K., Hoshino Y., Green K., Gorziglia M., Kapikian A. Z., Chanock R. M. Conservation of the fourth gene among rotaviruses recovered from asymptomatic newborn infants and its possible role in attenuation. J Virol. 1986 Dec;60(3):972–979. doi: 10.1128/jvi.60.3.972-979.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N., Yoshie O., Kitaoka S., Konno T., Ishida N. Evidence for endocytosis-independent infection by human rotavirus. Arch Virol. 1987;97(1-2):93–99. doi: 10.1007/BF01310737. [DOI] [PubMed] [Google Scholar]
- Fuller S. D. The T=4 envelope of Sindbis virus is organized by interactions with a complementary T=3 capsid. Cell. 1987 Mar 27;48(6):923–934. doi: 10.1016/0092-8674(87)90701-x. [DOI] [PubMed] [Google Scholar]
- Gorziglia M., Hoshino Y., Buckler-White A., Blumentals I., Glass R., Flores J., Kapikian A. Z., Chanock R. M. Conservation of amino acid sequence of VP8 and cleavage region of 84-kDa outer capsid protein among rotaviruses recovered from asymptomatic neonatal infection. Proc Natl Acad Sci U S A. 1986 Sep;83(18):7039–7043. doi: 10.1073/pnas.83.18.7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorziglia M., Larrea C., Liprandi F., Esparza J. Biochemical evidence for the oligomeric (possibly trimeric) structure of the major inner capsid polypeptide (45K) of rotaviruses. J Gen Virol. 1985 Sep;66(Pt 9):1889–1900. doi: 10.1099/0022-1317-66-9-1889. [DOI] [PubMed] [Google Scholar]
- Kabcenell A. K., Atkinson P. H. Processing of the rough endoplasmic reticulum membrane glycoproteins of rotavirus SA11. J Cell Biol. 1985 Oct;101(4):1270–1280. doi: 10.1083/jcb.101.4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalica A. R., Flores J., Greenberg H. B. Identification of the rotaviral gene that codes for hemagglutination and protease-enhanced plaque formation. Virology. 1983 Feb;125(1):194–205. doi: 10.1016/0042-6822(83)90073-9. [DOI] [PubMed] [Google Scholar]
- Kalica A. R., Greenberg H. B., Wyatt R. G., Flores J., Sereno M. M., Kapikian A. Z., Chanock R. M. Genes of human (strain Wa) and bovine (strain UK) rotaviruses that code for neutralization and subgroup antigens. Virology. 1981 Jul 30;112(2):385–390. doi: 10.1016/0042-6822(81)90285-3. [DOI] [PubMed] [Google Scholar]
- Kaljot K. T., Shaw R. D., Rubin D. H., Greenberg H. B. Infectious rotavirus enters cells by direct cell membrane penetration, not by endocytosis. J Virol. 1988 Apr;62(4):1136–1144. doi: 10.1128/jvi.62.4.1136-1144.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé M., Charpilienne A., Crawford S. E., Estes M. K., Cohen J. Expression of rotavirus VP2 produces empty corelike particles. J Virol. 1991 Jun;65(6):2946–2952. doi: 10.1128/jvi.65.6.2946-2952.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Offit P. A., Estes M. K. Identification of the simian rotavirus SA11 genome segment 3 product. Virology. 1988 Mar;163(1):26–32. doi: 10.1016/0042-6822(88)90230-9. [DOI] [PubMed] [Google Scholar]
- López S., Arias C. F., Bell J. R., Strauss J. H., Espejo R. T. Primary structure of the cleavage site associated with trypsin enhancement of rotavirus SA11 infectivity. Virology. 1985 Jul 15;144(1):11–19. doi: 10.1016/0042-6822(85)90300-9. [DOI] [PubMed] [Google Scholar]
- López S., Arias C. F., Méndez E., Espejo R. T. Conservation in rotaviruses of the protein region containing the two sites associated with trypsin enhancement of infectivity. Virology. 1986 Oct 15;154(1):224–227. doi: 10.1016/0042-6822(86)90445-9. [DOI] [PubMed] [Google Scholar]
- López S., Arias C. F. The nucleotide sequence of the 5' and 3' ends of rotavirus SA11 gene 4. Nucleic Acids Res. 1987 Jun 11;15(11):4691–4691. doi: 10.1093/nar/15.11.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López S., López I., Romero P., Méndez E., Soberón X., Arias C. F. Rotavirus YM gene 4: analysis of its deduced amino acid sequence and prediction of the secondary structure of the VP4 protein. J Virol. 1991 Jul;65(7):3738–3745. doi: 10.1128/jvi.65.7.3738-3745.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackow E. R., Shaw R. D., Matsui S. M., Vo P. T., Benfield D. A., Greenberg H. B. Characterization of homotypic and heterotypic VP7 neutralization sites of rhesus rotavirus. Virology. 1988 Aug;165(2):511–517. doi: 10.1016/0042-6822(88)90595-8. [DOI] [PubMed] [Google Scholar]
- Mackow E. R., Shaw R. D., Matsui S. M., Vo P. T., Dang M. N., Greenberg H. B. The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc Natl Acad Sci U S A. 1988 Feb;85(3):645–649. doi: 10.1073/pnas.85.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. L., Palmer E. L., Middleton P. J. Ultrastructure of infantile gastroenteritis virus. Virology. 1975 Nov;68(1):146–153. doi: 10.1016/0042-6822(75)90156-7. [DOI] [PubMed] [Google Scholar]
- Mitchell D. B., Both G. W. Complete nucleotide sequence of the simian rotavirus SA11 VP4 gene. Nucleic Acids Res. 1989 Mar 11;17(5):2122–2122. doi: 10.1093/nar/17.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa K., Taniguchi K., Torres A., Hoshino Y., Green K., Kapikian A. Z., Chanock R. M., Gorziglia M. Comparative analysis of the VP3 gene of divergent strains of the rotaviruses simian SA11 and bovine Nebraska calf diarrhea virus. J Virol. 1988 Nov;62(11):4022–4026. doi: 10.1128/jvi.62.11.4022-4026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offit P. A., Blavat G., Greenberg H. B., Clark H. F. Molecular basis of rotavirus virulence: role of gene segment 4. J Virol. 1986 Jan;57(1):46–49. doi: 10.1128/jvi.57.1.46-49.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poruchynsky M. S., Atkinson P. H. Rotavirus protein rearrangements in purified membrane-enveloped intermediate particles. J Virol. 1991 Sep;65(9):4720–4727. doi: 10.1128/jvi.65.9.4720-4727.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B. V., Burns J. W., Marietta E., Estes M. K., Chiu W. Localization of VP4 neutralization sites in rotavirus by three-dimensional cryo-electron microscopy. Nature. 1990 Feb 1;343(6257):476–479. doi: 10.1038/343476a0. [DOI] [PubMed] [Google Scholar]
- Prasad B. V., Wang G. J., Clerx J. P., Chiu W. Three-dimensional structure of rotavirus. J Mol Biol. 1988 Jan 20;199(2):269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- Shaw A. L., Rothnagel R., Chen D., Ramig R. F., Chiu W., Prasad B. V. Three-dimensional visualization of the rotavirus hemagglutinin structure. Cell. 1993 Aug 27;74(4):693–701. doi: 10.1016/0092-8674(93)90516-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirzaker S. C., Whitfeld P. L., Christie D. L., Bellamy A. R., Both G. W. Processing of rotavirus glycoprotein VP7: implications for the retention of the protein in the endoplasmic reticulum. J Cell Biol. 1987 Dec;105(6 Pt 2):2897–2903. doi: 10.1083/jcb.105.6.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street J. E., Croxson M. C., Chadderton W. F., Bellamy A. R. Sequence diversity of human rotavirus strains investigated by northern blot hybridization analysis. J Virol. 1982 Aug;43(2):369–378. doi: 10.1128/jvi.43.2.369-378.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin N. The use of cryoelectron microscopy in elucidating molecular design and mechanisms. Ann N Y Acad Sci. 1986;483:1–4. doi: 10.1111/j.1749-6632.1986.tb34485.x. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Skehel J. J., Wiley D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981 Jan 29;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Yeager M., Dryden K. A., Olson N. H., Greenberg H. B., Baker T. S. Three-dimensional structure of rhesus rotavirus by cryoelectron microscopy and image reconstruction. J Cell Biol. 1990 Jun;110(6):2133–2144. doi: 10.1083/jcb.110.6.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]