Abstract
The fast and reliable detection of bacterial spores is of great importance and still remains a challenge. Here we describe a direct RNA-based diagnostic method for the specific detection of viable bacterial spores which does not depends on an enzymatic amplification step and therefore is directly appropriate for quantification. The procedure includes the following steps: (i) heat activation of spores, (ii) germination and enrichment cultivation, (iii) cell lysis, and (iv) analysis of 16S rRNA in crude cell lysates using a sandwich hybridization assay. The sensitivity of the method is dependent on the cultivation time and the detection limit; it is possible to detect 10 spores per ml when the RNA analysis is performed after 6 h of enrichment cultivation. At spore concentrations above 106 spores per ml the cultivation time can be shortened to 30 min. Total analysis times are in the range of 2–8 h depending on the spore concentration in samples. The developed procedure is optimized at the example of Bacillus subtilis spores but should be applicable to other organisms. The new method can easily be modified for other target RNAs and is suitable for specific detection of spores from known groups of organisms.
Keywords: spore detection, Bacillus subtilis, RNA hybridization
Introduction
Detection of microorganisms in air is challenging due to the very low amount of organisms that generally are collected in air samples and the presence of organisms as dormant spores.
Various biosensors have been developed for the specific detection of harmful microbial spores in air samples (Gooding, 2006). Most methods for the detection of Bacillus spores have been designed for Bacillus anthracis, a causative factor for anthrax and a potential biological threat agent (Edwards et al., 2006; Irenge and Gala, 2012), in order to create sensors for biological warfare agents. Being a causative agent, B. anthracis is difficult to work with, and therefore the closely related species Bacillus subtilis and B. cereus are often used in the development of detection strategies (Arakawa et al., 2003; Stachowiak et al., 2007; Inami et al., 2009; Cheng et al., 2011).
The primary strategies for detection of Bacillus spores include polymerase chain reaction based techniques, immunoassays, spectrometry, chromatography, and protein profiling (Table 1). Recently also some autonomous pathogen detection systems for aerosol collection, sample preparation and detection were developed (Hindson et al., 2005; Stachowiak et al., 2007; Regan et al., 2008; Inami et al., 2009) (Table 1).
Table 1.
Methods for detection of Bacillus spores.
| Method | Sensitivity | Organisms | Comments | References |
|---|---|---|---|---|
| NUCLEIC ACID BASED METHODS | ||||
| Real-time PCR | 1 spore per 100 L of air | B. anthracis | Lee et al., 1999; Makino and Cheun, 2003; Irenge et al., 2010; Wielinga et al., 2011 | |
| Culture-based PCR | 1–10 spores per analysis | B. anthracis | High-throughput | Kane et al., 2009 |
| NASBAa coupled with biosensor | 1–10 spores per analysis | B. anthracis | Analysis after 30 min of germination | Baeumner et al., 2004 |
| FISHb | 103 spores per m3 of air | B. anthracis | Weerasekara et al., 2013 | |
| ICANc DNA detection | 104 spores per analysis | B. subtilis | Does not contain a germination step | Inami et al., 2009 |
| Autonomous pathogen detection system with multiplexed PCR | B. anthracis and others | Regan et al., 2008 | ||
| RAZOR® EX Anthrax Air Detection System | 200 spores per analysis | B. anthracis | DNA extraction and real-time PCR | Spaulding et al., 2012; Hadfield et al., 2013 |
| IMMUNOASSAYS | ||||
| Colorimetric and electrochemilumine-scence immunoassay | 30–100 spores per analysis | B. anthracis | Morel et al., 2012 | |
| ELISAd | B. subtilis | Zhou et al., 2002 | ||
| On-chip ELISA | 105 spores per analysis | B. subtilis | Chemiluminescence method combined with a biochip. Antibodies against surface spore antigen were used | Stratis-Cullum et al., 2003 |
| Luminex assay | 103–104 spores per ml | B. anthracis | Monoclonal antibodies recog-nize anthrose-containing oligosaccharides on the surface of B. anthracis endospores | Tamborrini et al., 2010 |
| Peptide-Function cantilever arrays | 105 spores per ml for analysis | B. subtilis | 1 from 2400 spores was captured | Dhayal et al., 2006; Campbell and Mutharasan, 2007 |
| Chip gel electrophoresis protein profiling (CGE-PP) | 16 particles per liter 100 cells per analysis | Any (adapted for E. coli and B. subtilis) | Autonomous microfluidic system | Pizarro et al., 2007; Stachowiak et al., 2007 |
| Multiplexed Immunoassay with PCR Confirmation | 49 spores per liter of air | B. subtilis | Autonomous Detection of Aerosolized Biological Agents | McBride et al., 2003; Hindson et al., 2004 |
| OTHERS | ||||
| Pyrolysis micromachined differential mobility spectrometry | 103 spores per analysis | B. anthracis | A microfabricated ion mobility spectrometer in combination with a pattern recognition and classification algorithm | Krebs et al., 2006 |
| Microcalorimetric spectroscopy | 100–1000 spores | B. subtilis, B. cereus | Arakawa et al., 2003 | |
| Laser induced breakdown spectroscopy | single particles | B. subtilis | In combination with other detection methods | Hybl et al., 2003, 2006 |
| Mass-spectrometry | 104–105 spores | B. anthracis | In combination with other detection methods | Lasch et al., 2009; Chenau et al., 2011; Li et al., 2012 |
| Raman scattering | 104 spores | Not specific | Based on detection of dipicolinic acid | Cheng et al., 2011, 2012; Cowcher et al., 2013 |
| Optical microchip array biosensor | 5 × 107 spores per ml | B. anthracis and others | Bhatta et al., 2011 | |
NASBA, Nucleic acid sequence based amplification.
FISH, Fluorescence in situ hybridization.
ICAN, Isothermal and chimeric primer-initiated amplification of nucleic acids.
ELISA, Enzyme linked immunosorbent assay.
A number of direct methods are based on DNA or protein detection and cannot distinguish between viable and dead spores. Furthermore, most of them have low sensitivity or require an amplification step such as PCR. RNA-based detection methods have the advantage to specifically analyze only viable spores, and RNA's, especially ribosomal RNAs (rRNAs), are populated in high amounts, making enzyme-based amplifications methods indispensible. Thus it is reasonable to apply RNA detection after the activation of RNA synthesis which takes place already after approximately 10 min of germination (Keijser et al., 2007).
Sandwich hybridization assays (SHA) are suitable for a rapid and quantitative RNA detection (Rautio et al., 2003). These methods are based on the hybridization of a target RNA (or denatured DNA) with two specific oligonucleotide probes. A capture probe is used to immobilize the target on a solid support, such as magnetic microbeads, which provide a large surface area for nucleic acid attachments (Walsh et al., 2001). This binding between the probes and the beads is usually performed by interaction between biotin attached to the oligonucleotide probe and streptavidin coated magnetic beads. The detection probe is labeled with a marker molecule which generates a signal proportional to the amount of target molecules. Oligonucleotide probes required for this assay can be designed for almost any RNA and can easily be modified for other targets. This means that the developed detection system can be applied for different organisms with just some small adaptations. Sandwich hybridization is relatively sensitive (10−16–10−15 moles of a specific target molecule) and can be performed with crude biological samples without any RNA purification. The method has been successfully applied for the detection of 16S rRNA from Legionella sp. in water samples (Leskelä et al., 2005), mycobacteria in soils (Nieminen et al., 2006), Lactobacillus and Pediococcus in brewery yeast slurries (Huhtamella et al., 2007), Salmonella in minced meat (Taskila et al., 2011), Bacillus cereus DNA (Gabig-Ciminska et al., 2004) and for monitoring dynamic changes of different mRNA species in microbial processes (Rautio et al., 2003; Neubauer et al., 2007; Soini et al., 2008; Thieme et al., 2008). The possibility to use various markers makes the method applicable for different read-out systems, such as fluorescence meters (Rautio et al., 2003), chip-based fluorescent biosensors (Wang et al., 2013) or electrical biochip readers (Gabig-Ciminska et al., 2004; Jürgen et al., 2005; Elsholz et al., 2006; Pioch et al., 2008a,b). The SHA coupled with capillary electrophoresis called TRAC (transcript analysis with aid of affinity capture) was developed for multiplex transcript analysis and is commercially available (Rautio et al., 2006, 2008; Rautio, 2010) (PlexPress Oy, Finland).
In the actual study a procedure was developed for detecting bacterial spores, utilizing spore activation, enrichment cultivation and an RNA-based sandwich hybridization assay. In contrast to most assays utilizing DNA and protein detection, the method developed here is specific only for viable organisms since it is based on RNA synthesis.
Materials and methods
Spore preparation
Bacillus subtilis spores were obtained from cells (B. subtilis 6051α, kindly provided by Prof. Dr. Thomas Schweder, Ernst-Moritz-Arndt University of Greifswald, Germany) cultured in Schaeffer's sporulation medium (8 g/l bacto-nutrient broth, 0.1% (w/v) KCl, 0.012% (w/v) MgSO4 × 7H20, 0.5 mM NaOH, 1 mM Ca(NO3)2, 0.01 mM MnCl2, 0.001 mM FeSO4) at 37°C, 200 rpm for 4 days. Cultures were carried out in 3-baffled 1 liter Erlenmeyer flasks with a liquid volume of 100 ml. After harvesting and extensive washing with water at 4°C, spores were inspected by phase-contrast microscopy to show that samples are free of vegetative cells (< 10%). Spores were lyophilized and stored at 4°C. For counting, the spores were diluted in 0.9% NaCl, activated at 70°C for 30 min and plated on nutrient agar plates (Difco, USA).
Activation and growth conditions
B. subtilis spores were diluted in 0.9% NaCl and activated by temperature treatment at 70°C for 30 min. Thereafter the suspensions were transferred into 3-baffled 1 l Erlenmeyer flasks containing 100 ml of a germination medium (20 g/l tryptone, 10 g/l yeast extract, 10 g/l NaCl and 0.5 mM L-alanine) (Moeller et al., 2006). Cultures were carried out at 37°C and 200 rpm on a rotary shaker. Culture growth was monitored by measuring the optical density at 600 nm (OD600, Ultrospec Pro 2100 UV/Visible Spectrophotometer, GE Healthcare, Buckinghamshire, UK). The corresponding colony forming units (cfu/ml) were determined by plating on nutrient agar plates.
Sample preparation
1 ml of the B. subtilis cultures were removed into pre-cooled 1.5 ml microreaction tubes containing 0.125 × sample volume of cold 95:5 ethanol:phenol (v/v) (Thieme et al., 2008). The cells were collected by centrifugation (15,000 × g, + 4°C, 5 min). The pellets were suspended in 500 μl of 1 × TEN buffer (10 mM Tris-HCl, 1 mM EDTA, 100 mM NaCl, pH 8.0) containing 1 μl/ml RNAguard® RNase inhibitor (Amersham Biosciences, New Jersey, USA) and mixed by vortexing for 1 min. Then the cell suspensions were transferred into 2 ml microcentrifuge tubes with a skirt (Greiner Bio-One GmbH, Frickenhausen, Germany) containing 100 mg glass beads (BioSpec Products Inc., Bartlesville, OK, USA) with a diameter of 0.1 mm. Cell disruption was performed with a FastPrep® FP120 Cell Disrupter (Bio-101, Thermo Savant, USA) at 4.5 m/s for 4 × 25 s. The vials were kept on ice for 1 min between the disruption cycles. Cell homogenates were centrifuged (20,000 ×g, + 4°C, 5 min) and supernatants were snap frozen in liquid nitrogen.
Generation of the DNA for the in vitro 16S rRNA transcript
The target gene was amplified by PCR using a colony of B. subtilis cells as a template and 10 pmol of the forward and reverse primers (Weisburg et al., 1991) (Table 2). The forward primer contained at the 5′–end the T7 promoter sequence CTA ATA CGA CTC ACT ATA GGG. The PCR conditions were as follows: 3 min at 95°C; 35 cycles of 15 s at 95°C, 30 s at 50°C and 1 min 45 s at 72°C; 10 min at 72°C and afterwards held at 4°C. PCR products were analyzed by agarose gel electrophoresis and purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany). Purified PCR products were quantified by absorbance measurement at 260 nm (GeneQuant II, Viochrom Ltd., Cambridge, UK).
Table 2.
Sequences of the oligonucleotide probes used in sandwich hybridization for the detection of B. subtilis 16S rRNA and PCR primers.
| Probe name | Probe specification | Probe sequence 5′–3′ | Location of probe | Probe modification |
|---|---|---|---|---|
| Bsub16Scap | Capture | TGTCTCAGTCCCAGTGTGGa | 319–337 | 5′-Biotin |
| Bsub16Sdet | Detection | CGTAGGAGTCTGGGCCGa | 338–354 | 3′-Digoxigenin |
| Help1 | Helper | CCCCACTGCTGCCTCC | 355–370 | |
| Help2 | Helper | CTGGTCATCCTCTCAGA | 302–318 | |
| Fd-T7 | Forward PCR primer | (CTAATACGACTCACTATAGGG) AGAGTTTGATCCTGGCTCAG | 10–29 | 5′-T7 promoter |
| Rev | Reverse PCR primer | CGGCTACCTTGTTACGACTT | 1502–1521 | |
| NCcap | Negative control capture probe | TGTGAACTTCCATCGGCTTGAGCC | 5′-Biotin | |
| NCdet | Negative control detection probe | GATAGTCCCTCTAAGAAGCCATGTG | 3′-Digoxigenin |
Nucleotides different to general eubacterial probes are underlined.
Target RNA was generated from the purified PCR product by means of T7 RNA polymerase in vitro transcription using the MAXIscript™ T7-kit (Ambion, Austin, TX, USA). The quality of the in vitro transcribed RNA was checked by the Agilent 2100 Bioanalyser and RNA 6000 Nano LabChip Kit (Agilent Technologies, Waldbronn, Germany). RNA was quantified by absorbance measurement at 260 nm (NanoDrop ND 1000, Thermo Fisher Scientific, USA).
Oligonucleotide probes
The oligonucleotide probes for B. subtilis 16S rRNA were designed on the basis of general eubacterial probes with some modifications (Table 2). The oligonucleotide probes were purchased from Oligomer Oy (Helsinki, Finland). The detection probe was labeled with the DIG Oligonucleotide Tailing Kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions.
Sandwich hybridization assay
A quantitative sandwich hybridization assay (SHA) was used for detecting 16S rRNA molecules. The procedure was performed as described by Rautio et al. (2003) and Thieme et al. (2008) with small modifications.
SHAs were carried out in 96-well bright U-shaped microplates (Greiner-Bio-One GmbH, Frickenhausen, Germany) using a Thermomixer Comfort (Eppendorf, Hamburg, Germany). The target RNA was mixed with the 5 × SSC hybridization buffer (0.75 M sodium chloride, 0.075 M sodium citrate, pH 7.0), 20% (v/v) deionized formamide, 3% (w/v) dextran sulphate, 0.2% (v/v) TWEEN20, 0.02% (v/v) Ficoll, 0.02% (w/v) polyvinyl pyrrolidone, 0.02% (w/v) bovine serum albumin, 1% (v/v) blocking reagent (Roche, Mannheim, Germany) in 100 mM maleic acid with 150 mM NaCl (pH 7.5), 5 pmol biotin-labeled capture probe, 1 pmol DIG-labeled detection probe, and 1 pmol of each helper probe in a total volume of 100 μl. The hybridization reactions were performed in three parallels at 750 rpm and 50°C for 30 min. 15 μl of Streptavidin MagneSphere® Paramagnetic Particles (Promega, Madison, WI, USA) were washed as recommended by the manufacturer, and added to each well after the hybridization reaction and incubated at 750 rpm and 50°C for 30 min. Subsequently, beads were washed three times with 120 μl of a washing buffer (1% SSC, 0.1% TWEEN20) at 25°C and 750 rpm for 2 min. During the washing step beads were kept in the wells with a MagnaBot® 96 Magnetic Separation Device (Promega, Madison, WI, USA). 100 μl of anti-DIG alkaline phosphatase Fab-fragments (Roche Diagnostics, Mannheim, Germany) were diluted with 1 × SSC, 0.1% TWEEN20 to a final concentration of 375 U/l, and applied to each well. The 96-well plate was incubated at 450 rpm and 25°C for 30 min. Unbound enzyme was removed by washing three times as described above. Afterwards the whole solutions were transferred into new microplates and washed once more. Finally, 100 μl of 1 mM AttoPhos® fluorescent substrate (Promega, Madison, WI, USA) was added to each well and the enzymatic reaction was performed at 37°C and 750 rpm for 20 min. 90 μl of the contents of well were transferred into a Costar black 96-well assay plate (Corning Inc., Corning, NY, USA) for the fluorescence measurement with a Wallac Victor multilabel counter (PerkinElmer Life Sciences, Turku, Finland) at an excitation wavelength of 430 nm and an emission wavelength of 560 nm.
The detection limit was defined as 3 × SD added to the background fluorescence. Detection limits were calculated separately for each SHA and signals above it were considered as positive. Capture and detection probes which are not complementary to any RNA of B. subtilis (Table 2, NCcap and NCdet, respectively) were used as a negative control for each sample to estimate the background signal.
Results
The aim of this study was the development of a diagnostic method for the detection of bacterial spores at the example of Bacillus subtilis spores. The sequence of the procedure is shown in Figure 1A. First, the spores are activated by a heat treatment (30 min, 70°C) and following transfer into germination medium (Keijser et al., 2007). The duration of the enrichment cultivation was varied depending on the initial amount of spores. After the incubation phase collected samples were disrupted and the supernatant was utilized for the quantification of the 16S ribosomal RNA directly from the cell broth by the use of a sandwich hybridization assay without any further purification.
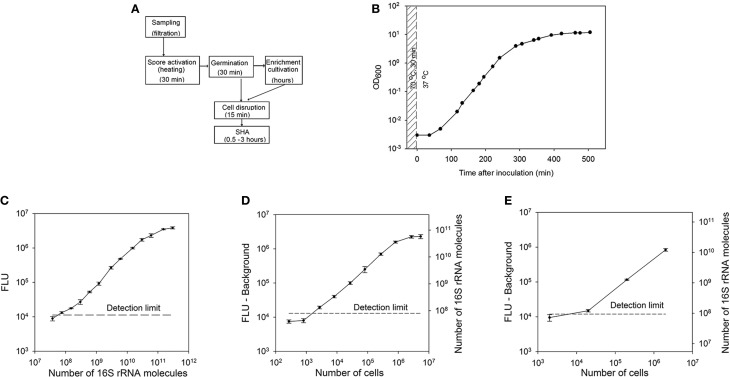
Figure 1.
Spore detection using a sandwich hybridization assay. (A) Principle of airborne spore detection using a sandwich hybridization assay. Spores collected from air are activated and cultivated in a germination medium. The first sample for analysis can be collected after approximately 30 min of germination. If the amount of spores is too low to be detected at this step, the incubation can be continued to allow a multiplication of the cells. (B) A Growth curve of B. subtilis cells. After heat activation (70°C for 30 min) B. subtilis spores were inoculated into a germination medium to a final concentration of 106 spores/ml. (C) Standard curve for the B. subtilis 16S rRNA sandwich hybridization assay. (D) SHA dilution curve for B. subtilis 16S rRNA. The dilutions of the crude extracts of B. subtilis cells collected at the exponential growth phase (OD600 = 1.4) were used as a target. The corresponding numbers of 16S rRNA molecules were calculated according to the standard curve of in vitro transcribed 16SrRNA at 260 nm. (E) 16S rRNA measured with SHA in B. subtilis spores after their activation and germination for 30 min. Corresponding numbers of 16S rRNA molecules were calculated according to the standard curve (Figure 1C). The error bars show the ±SD of three parallel experiments and the detection limit is shown as a dashed horizontal line.
RNA-based detection of spores is feasible only after the activation of RNA synthesis, that takes place during the spore germination. In order to activate B. subtilis spores and initiate the germination process, dormant spores were exposed to heat (70°C for 30 min) and afterwards cultured in the germination medium at 37°C. Cell germination and growth were monitored by measuring the OD600 (Figure 1B). An increase of the cell density after 1 h of cultivation indicated that the germination and outgrowth were completed and proliferation began.
For this feasibility study oligonucleotide probes for the detection of B. subtilis 16S rRNA with SHA were designed on the basis of general eubacterial probes with some modifications to detect the sequences specific for B. subtilis (Table 2). The probes were tested using an in vitro transcribed fragment of B. subtilis 16S rRNA as target molecule, which also is applied as a quantitative standard. 6 × 107 (0.1 fmol) target molecules in the hybridization solution gave a signal that was significantly above the detection limit. The reaction was linear over a range of nearly 3 orders of magnitude up to approximately 3 × 1010 i.e., 50 fmol of target molecules (Figure 1C).
In order to determine the detection limit and the range of the SHA for crude cell extracts, different dilutions of B. subtilis cell extracts were analyzed with the sandwich hybridization assay. The cells were collected at the end of the exponential growth phase (OD600 = 1.4), disrupted, and 2.7 × 102 to 5.4 × 106 cells were used as targets for a dilution curve (Figure 1D). The detection limit was about 1.5 × 103 cells per assay for exponentially growing B. subtilis cells (7.5 × 104 cells per ml of culture medium). The calculated amount of 16S rRNA was 4.8 (±0.6) × 104 molecules per cell at this growth phase. Consequently, the detection limit in crude cell extracts corresponded to about 7.2 × 107 16S rRNA molecules. The linear range of the assay was between 103 and 106 cells in a hybridization solution.
In order to establish the analysis of cells from enrichment cultures the germination medium was inoculated with 105–108 activated B. subtilis spores per mL. After 30 min of cultivation (before the first proliferation), the cells were disrupted and analyzed with SHA (Figure 1E). It was possible to detect about 20,000 cells per assay (106 cells per ml of culture medium). This number of cells corresponds to approximately 108 16S rRNA molecules. According to the measurements at this stage of germination one B. subtilis cell contained 5.6 (±0.9) × 103 16S rRNA molecules.
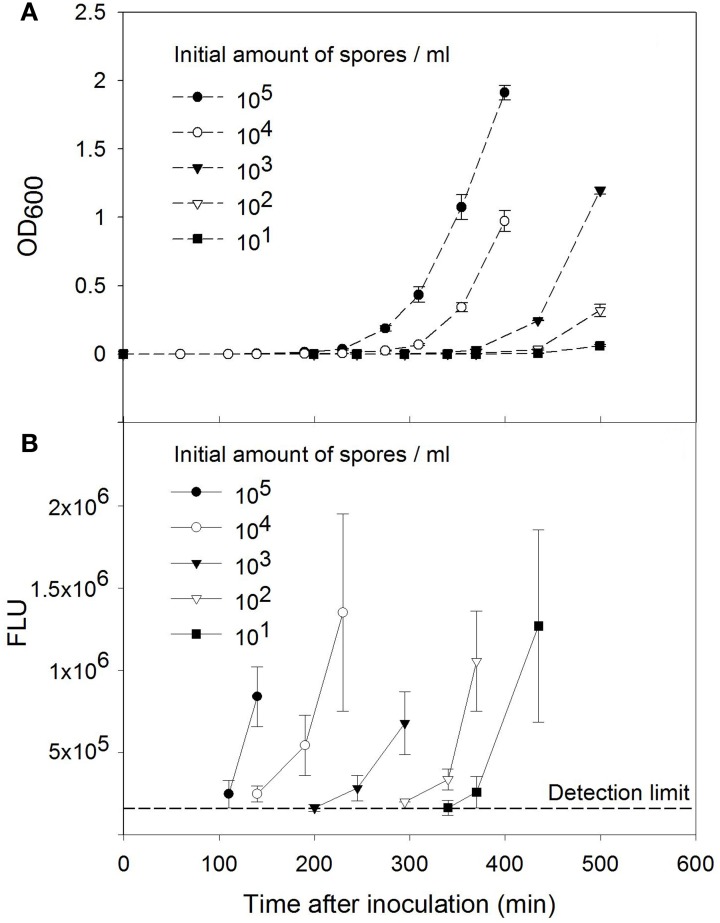
Next, the sensitivity of the method including activation, enrichment cultivation, and 16S rRNA analysis of B. subtilis spores was studied. Therefore the germination medium was inoculated with 101–105 activated B. subtilis spores per ml. Germination and growth of the cells were monitored by measuring the OD600 (Figure 2A). The cells were collected, disrupted and analyzed with SHA using the probes against the16S rRNA (Figure 2B). Detectable signals were observed after 110, 140, 245, 340, and 370 min of cultivation for the samples with initial number of 105, 104, 103, 102, and 101 spores per ml of culture medium, respectively.
Figure 2.
Sensitivity of the B. subtilis spore detection method. (A) Growth curves of B. subtilis cells after spore activation. The initial number of the spores was 101–105 spores per ml of germination medium. (B) Detection of B. subtilis 16S rRNA by sandwich hybridization after enrichment cultivation. The error bars show ±SD of three independent cultivations and measurements.
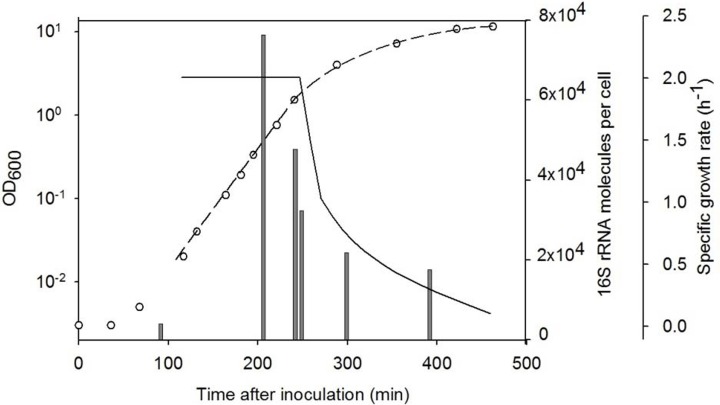
Since the SHA sensitivity depends on the number of target RNA molecules in one cell, the amount of 16S rRNA was measured for different growth phases of B. subtilis cells (Figure 3). As it was shown earlier one B. subtilis cell contained 5.6 (±0.9) × 103 16S rRNA molecules already after 30 min of germination. At the early exponential growth phase the amount of 16S rRNA molecules per cell was maximal (about 9 × 104 rRNA molecules per cell for an OD600 of 0.27) and the 16S rRNA content decreased in later growth stages. Therefore the analysis is most sensitive if the OD600 is approximately 0.3.
Figure 3.
Level of 16S rRNA molecules per cell (gray bars) in different growth phases of B. subtilis. Quantity of RNA was measured with SHA. The growth was followed by measuring the optical density at 600 nm (◦) and the specific growth rate was calculated (black line). The dashed line shows fitting of the OD curve.
Discussion
Detection of microorganisms in bioaerosols is an important issue in determining bio-warfare agents during biological attacks and for the monitoring of indoor air quality. A fast and sensitive method for the detection of bacterial spores was developed in this study. The procedure includes activation of spores, their germination, enrichment cultivation and RNA detection using a sandwich hybridization assay, thus only viable spores are detected, which is advantageous over standard detection methods as listed in Table 1. There are no sensitivity limitations as the method is adaptable by the enrichment cultivation time. The RNA analysis can be performed directly with crude cell extracts avoiding laborious RNA purification. The method was developed for B. subtilis, a model organism capable of spore formation and quite abundant in aerosols.
Since spores only contain low amounts of RNA molecules, the idea of this study was to initiate RNA synthesis to increase the detection system sensitivity. At first, the spores are activated at 70°C for 30 min and placed into a medium containing L-alanine as a germination factor. During germination and outgrowth the activated spores synthesize RNA, proteins, and ATP. Chromosomal replication is initiated after 0.5 h (Garrick-Silversmith and Torriani, 1973) and the first cell division takes place after approximately 70 min (Keijser et al., 2007). Depending on the initial amount of spores, cells are disrupted either directly after the germination or after the enrichment cultivation required for the accumulation of necessary amount of RNA molecules. The presented method allows the use of crude cells lysates as sample material for the RNA detection, and no further RNA purification is needed (Rautio et al., 2003; Thieme et al., 2008). We believe that this is a strong advantage for using the sandwich hybridization assay for the RNA detection compared to reverse transcriptase real time PCR.
The assay is based on the hybridization event of the target RNA with two oligonucleotide probes. Alkaline phosphatase attached to one of the probes generates a fluorescent signal used for quantification. The SHA has a potential to be automated, e.g., by the use of an electrical chip reader (Gabig-Ciminska et al., 2004).
After the activation the spores germinated very quickly. The first increase of the OD600 was noticed already after 1 h for the cultures with an initial amount of 106 spores per ml. The detectable SHA signal for these cultures was observed after 0.5 h of germination. Since the RNA synthesis in germinating B. subtilis spores starts within about 5 min after spore activation (Matsuda and Kameyama, 1986), the cells have accumulated a detectable amount of RNA molecules already after 30 min. Furthermore, at this moment the spore core is rehydrated making cells more accessible to disruption (Sloma and Smith, 1979; Moeller et al., 2006).
The sensitivity of the developed method is not limited due to enrichment cultivation for which the time can be adapted. Very low initial spore amounts in the sample could be detected after about 5 h of enrichment cultivation. Consequently, the maximum duration of the assay for a very low amount of spores including spore activation, enrichment cultivation, and detection with the SHA is about 8 h. The detection limit of the SHA was estimated as 6 × 107 target molecules, which in case of using rRNA as a target corresponds to about 103–104 cells in a sample depending on their growth stage.
By the use of in vitro synthesized RNA probes as a standard with the SHA it was possible to estimate the amount of 16S rRNA molecules per cell. As expected, the number of molecules increased during the germination and the outgrowth, reached the maximum at the beginning of the exponential phase of growth, and then slowly decreased. After 0.5 h of germination one cell contained approximately 5.6 (±0.9) × 103 16S rRNA molecules. Exponentially growing cells contained about 9 × 104 16S rRNA molecules. These numbers correlate well with the known amount of ribosomes in Escherichia coli cells, that is 6700–71,000 depending on the specific growth rate (Bremer and Dennis, 1996).
The 16S rRNA oligonucleotide probes used in this study are capable of detecting most bacteria and are relevant only for method development. Specific probes for the SHA can be designed for almost any organism or groups of organisms if the gene sequence is available. Furthermore, it would be possible to apply the procedure for the analysis of mRNAs and their dynamics during germination. However, due to numerical advantage of ribosomes compared to a specific mRNA species, the method sensitivity would be lower when mRNA is used as a target for the SHA instead of rRNA and additionally the analytical error eventually would be higher due to the low stability of mRNA molecules. For selecting target mRNAs for specific identification of B. subtilis a transcription profile of germinated spores (Keijser et al., 2007) and the most abundant proteins of growing Bacillus cells have to be reviewed. Protein abundance during the exponential growth of Bacillus subtilis has been studied in great detail (Büttner et al., 2001; Eymann et al., 2004). The most abundant proteins in B. subtilis cytosolic extracts perform mainly housekeeping functions as components of the translational apparatus (e.g., translation elongation factors, ribosomal proteins), the glycolytic pathways, the tricarboxylic acid cycle, the metabolism of amino and nucleic acids, and protein quality control (e.g., chaperons). The mRNAs of these proteins can be used as a target for the specific detection of B. subtilis spores using the presented method.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Dr. Pekka Belt, Dr. Janne Härkönen, and Dr. Matti Möttönen are acknowledged for critical editing of this manuscript. This study was supported by the Finnish Funding Agency for Technology and Innovation (TEKES) project ENRICH (40254/07).
Glossary
Abbreviations
- SHA
sandwich hybridization assay.
References
- Arakawa E. T., Lavrik N. V., Rajic S., Datskos P. G. (2003). Detection and differentiation of biological species using microcalorimetric spectroscopy. Ultramicroscopy 97, 459–465 10.1016/S0304-3991(03)00074-3 [DOI] [PubMed] [Google Scholar]
- Baeumner A. J., Leonard B., McElwee J., Montagna R. A. (2004). A rapid biosensor for viable B. anthracis spores. Anal. Bioanal. Chem. 380, 15–23 10.1007/s00216-004-2726-7 [DOI] [PubMed] [Google Scholar]
- Bhatta D., Michel A. A., Marti Villalba M., Emmerson G. D., Sparrow I. L. G., Perkins E. A., et al. (2011). Optical microchip array biosensor for multiplexed detection of bio-hazardous agents. Biosens. Bioelectron. 30, 78–86 10.1016/j.bios.2011.08.031 [DOI] [PubMed] [Google Scholar]
- Bremer H., Dennis P. P. (1996). Modulation of chemical composition and other parameters of the cell by growth rate, in Escherichia coli and Salmonella typhimurium Cellular and Molecular Biology, ed Neidhardt F. C. (Washington, DC: American Society of Microbiology), 1553–1569 [Google Scholar]
- Büttner K., Bernhardt J., Scharf C., Schmid R., Mäder U., Eymann C., et al. (2001). A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 22, 2908–2935 [DOI] [PubMed] [Google Scholar]
- Campbell G. A., Mutharasan R. (2007). Method of measuring Bacillus anthracis spores in the presence of copious amounts of Bacillus thuringiensis and Bacillus cereus. Anal. Chem. 79, 1145–1152 10.1021/ac060982b [DOI] [PubMed] [Google Scholar]
- Chenau J., Fenaille F., Ezan E., Morel N., Lamourette P., Goossens P. L., et al. (2011). Sensitive detection of Bacillus anthracis spores by immunocapture and liquid chromatography-tandem mass spectrometry. Anal. Chem. 83, 8675–8682 10.1021/ac2020992 [DOI] [PubMed] [Google Scholar]
- Cheng H. W., Chen Y. Y., Lin X. X., Huan S. Y., Wu H. L., Shen G. L., et al. (2011). Surface-enhanced Raman spectroscopic detection of Bacillus subtilis spores using gold nanoparticle based substrates. Anal. Chim. Acta 707, 155–163 10.1016/j.aca.2011.09.007 [DOI] [PubMed] [Google Scholar]
- Cheng H. W., Huan S. Y., Yu R. Q. (2012). Nanoparticle-based substrates for surface-enhanced Raman scattering detection of bacterial spores. Analyst 137, 3601–3608 10.1039/c2an35448a [DOI] [PubMed] [Google Scholar]
- Cowcher D. P., Xu Y., Goodacre R. (2013). Portable, quantitative detection of Bacillus bacterial spores using surface-enhanced Raman scattering. Anal. Chem. 85, 3297–3302 10.1021/ac303657k [DOI] [PubMed] [Google Scholar]
- Dhayal B., Henne W. A., Doorneweerd D. D., Reifenberger R. G., Low P. S. (2006). Detection of Bacillus subtilis spores using peptide-functionalized cantilever arrays. J. Am. Chem. Soc. 128, 3716–3721 10.1021/ja0570887 [DOI] [PubMed] [Google Scholar]
- Edwards K. A., Clancy H. A., Baeumner A. J. (2006). Bacillus anthracis: toxicology, epidemiology and current rapid-detection methods. Anal. Bioanal. Chem. 384, 73–84 10.1007/s00216-005-0090-x [DOI] [PubMed] [Google Scholar]
- Elsholz B., Worl R., Blohm L., Albers J., Feucht H., Grunwald T., et al. (2006). Automated detection and quantitation of bacterial RNA by using electrical microarrays. Anal. Chem. 78, 4794–4802 10.1021/ac0600914 [DOI] [PubMed] [Google Scholar]
- Eymann C., Dreisbach A., Albrecht D., Bernhardt J., Becher D., Gentner S., et al. (2004). A comprehensive proteome map of growing Bacillus subtilis cells. Proteomics 4, 2849–2876 10.1002/pmic.200400907 [DOI] [PubMed] [Google Scholar]
- Gabig-Ciminska M., Holmgren A., Andresen H., Bundvig B. K., Wümpelmann M., Albers J., et al. (2004). Electric chips for rapid detection and quantification of nucleic acids. Biosens. Bioelectron. 19, 537–546 10.1016/S0956-5663(03)00273-2 [DOI] [PubMed] [Google Scholar]
- Garrick-Silversmith L., Torriani A. (1973). Macromolecular syntheses during germination and outgrowth of Bacillus subtilis spores. J. Bacteriol. 114, 507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding J. J. (2006). Biosensor technology for detecting biological warfare agents: recent progress and future trends. Anal. Chim. Acta 559, 137–151 10.1016/j.aca.2005.12.020 [DOI] [Google Scholar]
- Hadfield T., Ryan V., Spaulding U. K., Clemens K. M., Ota I. M., Brunelle S. L. (2013). RAZOR EX Anthrax Air Detection System for detection of Bacillus anthracis spores from aerosol collection samples: collaborative study. J. AOAC Int. 96, 392–398 10.5740/jaoacint.CS2012-06 [DOI] [PubMed] [Google Scholar]
- Hindson B. J., Brown S. B., Marshall G. D., McBride M. T., Makarewicz A. J., Gutierrez D. M., et al. (2004). Development of an automated sample preparation module for environmental monitoring of biowarfare agents. Anal. Chem. 76, 3492–3497 10.1021/ac035365r [DOI] [PubMed] [Google Scholar]
- Hindson B. J., McBride M. T., Makarewicz A. J., Henderer B. D., Setlur U. S., Smith S. M., et al. (2005). Autonomous detection of aerosolized biological agents by multiplexed immunoassay with polymerase chain reaction confirmation. Anal. Chem. 77, 284–289 10.1021/ac0489014 [DOI] [PubMed] [Google Scholar]
- Huhtamella S., Leinonen M., Nieminen T., Fahnert B., Myllykoski L., Breitenstein A., et al. (2007). RNA-based sandwich hybridisation method for detection of lactic acid bacteria in brewery samples. J. Microbiol. Methods 68, 543–553 10.1016/j.mimet.2006.10.009 [DOI] [PubMed] [Google Scholar]
- Hybl J. D., Lithgow G. A., Buckley S. G. (2003). Laser-induced breakdown spectroscopy detection and classification of biological aerosols. Appl. Spectrosc. 57, 1207–1215 10.1366/000370203769699054 [DOI] [PubMed] [Google Scholar]
- Hybl J. D., Tysk S. M., Berry S. R., Jordan M. P. (2006). Laser-induced fluorescence-cued, laser-induced breakdown spectroscopy biological-agent detection. Appl. Opt. 45, 8806–8814 10.1364/AO.45.008806 [DOI] [PubMed] [Google Scholar]
- Inami H., Tsuge K., Matsuzawa M., Sasaki Y., Togashi S., Komano A., et al. (2009). Semi-automated bacterial spore detection system with micro-fluidic chips for aerosol collection, spore treatment and ICAN DNA detection. Biosens. Bioelectron. 24, 3299–3305 10.1016/j.bios.2009.04.025 [DOI] [PubMed] [Google Scholar]
- Irenge L. M., Durant J. F., Tomaso H., Pilo P., Olsen J. S., Ramisse V., et al. (2010). Development and validation of a real-time quantitative PCR assay for rapid identification of Bacillus anthracis in environmental samples. Appl. Microbiol. Biotechnol. 88, 1179–1192 10.1007/s00253-010-2848-0 [DOI] [PubMed] [Google Scholar]
- Irenge L. M., Gala J. L. (2012). Rapid detection methods for Bacillus anthracis in environmental samples: a review. Appl. Microbiol. Biotechnol. 93, 1411–1422 10.1007/s00253-011-3845-7 [DOI] [PubMed] [Google Scholar]
- Jürgen B., Pioch D., Hoi L. T., Albers J., Hintsche R., Schweder T. (2005). At-line monitoring of bioprocess-relevant marker genes using an electric DNA-chip. J. Biotechnol. 118:S36 10.1002/bit.20578 [DOI] [PubMed] [Google Scholar]
- Kane S. R., Létant S. E., Murphy G. A., Alfaro T. M., Krauter P. W., Mahnke R., et al. (2009). Rapid, high-throughput, culture-based PCR methods to analyze samples for viable spores of Bacillus anthracis and its surrogates. J. Microbiol. Methods 76, 278–284 10.1016/j.mimet.2008.12.005 [DOI] [PubMed] [Google Scholar]
- Keijser B. J., Ter Beek A., Rauwerda H., Schuren F., Montijn R., van der Spek H., et al. (2007). Analysis of temporal gene expression during Bacillus subtilis spore germination and outgrowth. J. Bacteriol. 189, 3624–3634 10.1128/JB.01736-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M. D., Mansfield B., Yip P., Cohen S. J., Sonenshein A. L., Hitt B. A., et al. (2006). Novel technology for rapid species-specific detection of Bacillus spores. Biomol. Eng. 23, 119–127 10.1016/j.bioeng.2005.12.003 [DOI] [PubMed] [Google Scholar]
- Lasch P., Beyer W., Nattermann H., Stämmler M., Siegbrecht E., Grunow R., et al. (2009). Identification of Bacillus anthracis by using matrix-assisted laser desorption ionization-time of flight mass spectrometry and artificial neural networks. Appl. Environ. Microbiol. 75, 7229–7242 10.1128/AEM.00857-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. A., Brightwell G., Leslie D., Bird H., Hamilton A. (1999). Fluorescent detection techniques for real-time multiplex strand specific detection of Bacillus anthracis using rapid PCR. J. Appl. Microbiol. 87, 218–223 10.1046/j.1365-2672.1999.00908.x [DOI] [PubMed] [Google Scholar]
- Leskelä T., Tilsala-Timisjärvi A., Kusnetsov J., Neubauer P., Breitenstein A. (2005). Sensitive genus-specific detection of Legionella by a 16S rRNA based sandwich hybridization assay. J. Microbiol. Methods 62, 167–179 10.1016/j.mimet.2005.02.008 [DOI] [PubMed] [Google Scholar]
- Li D., Truong T. V., Bills T. M., Holt B. C., VanDerwerken D. N., Williams J. R., et al. (2012). GC/MS method for positive detection of Bacillus anthracis endospores. Anal. Chem. 84, 1637–1644 10.1021/ac202606x [DOI] [PubMed] [Google Scholar]
- Makino S., Cheun H. (2003). Application of the real-time PCR for the detection of airborne microbial pathogens in reference to the anthrax spores. J. Microbiol. Methods 53, 141–147 10.1016/S0167-7012(03)00019-8 [DOI] [PubMed] [Google Scholar]
- Matsuda M., Kameyama T. (1986). Fractionation of ribonucleic-acid transcripts synthesized during spore germination in Bacillus subtilis. J. Basic Microbiol. 26, 91–99 10.1002/jobm.3620260209 [DOI] [PubMed] [Google Scholar]
- McBride M. T., Masquelier D., Hindson B. J., Makarewicz A. J., Brown S., Burris K., et al. (2003). Autonomous detection of aerosolized Bacillus anthracis and Yersinia pestis. Anal. Chem. 75, 5293–5299 10.1021/ac034722v [DOI] [PubMed] [Google Scholar]
- Moeller R., Horneck G., Rettberg P., Mollenkopf H. J., Stackebrandt E., Nicholson W. L. (2006). A method for extracting RNA from dormant and germinating Bacillus subtilis strain 168 endospores. Curr. Microbiol. 53, 227–231 10.1007/s00284-006-0099-1 [DOI] [PubMed] [Google Scholar]
- Morel N., Volland H., Dano J., Lamourette P., Sylvestre P., Mock M., et al. (2012). Fast and sensitive detection of Bacillus anthracis spores by immunoassay. Appl. Environ. Microbiol. 78, 6491–6498 10.1128/AEM.01282-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer A., Soini J., Bollok M., Zenker M., Sandqvist J., Myllyharju J., et al. (2007). Fermentation process for tetrameric human collagen prolyl 4-hydroxylase in Escherichia coli: improvement by gene optimisation of the PDI/beta subunit and repeated addition of the inducer anhydrotetracycline. J. Biotechnol. 128, 308–321 10.1016/j.jbiotec.2006.10.017 [DOI] [PubMed] [Google Scholar]
- Nieminen T., Pakarinen J., Tsitko I., Salkinoja-Salonen M., Breitenstein A., Ali-Vehmas T., et al. (2006). 16S rRNA targeted sandwich hybridization method for direct quantification of mycobacteria in soils. J. Microbiol. Methods 67, 44–55 10.1016/j.mimet.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Pioch D., Jürgen B., Evers S., Maurer K. H., Hecker M., Schweder T. (2008a). Improved sandwich-hybridization assay for an electrical DNA-chip-based monitoring of bioprocess-relevant marker genes. Appl. Microbiol. Biotechnol. 78, 719–728 10.1007/s00253-008-1347-z [DOI] [PubMed] [Google Scholar]
- Pioch D., Schweder T., Jürgen B. (2008b). Novel developments for improved detection of specific mRNAs by DNA chips. Appl. Microbiol. Biotechnol. 80, 953–963 10.1007/s00253-008-1680-2 [DOI] [PubMed] [Google Scholar]
- Pizarro S. A., Lane P., Lane T. W., Cruz E., Haroldsen B., VanderNoot V. A. (2007). Bacterial characterization using protein profiling in a microchip separations platform. Electrophoresis 28, 4697–4704 10.1002/elps.200700005 [DOI] [PubMed] [Google Scholar]
- Rautio J., Barken K. B., Lahdenperä J., Breitenstein A., Molin S., Neubauer P. (2003). Sandwich hybridisation assay for quantitative detection of yeast RNAs in crude cell lysates. Microb. Cell Fact. 2, 4 10.1186/1475-2859-2-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautio J. J. (2010). Multiplex gene expression analysis by TRAC in fungal cultures. Methods Mol. Biol. 638, 165–173 10.1007/978-1-60761-611-5_12 [DOI] [PubMed] [Google Scholar]
- Rautio J. J., Kataja K., Satokani R., Penttilä M., Söderlund H., Saloheimo M. (2006). Rapid and multiplexed transcript analysis of microbial cultures using capillary electophoresis-detectable oligonucleotide probe pools. J. Microbiol. Methods 65, 404–416 10.1016/j.mimet.2005.08.010 [DOI] [PubMed] [Google Scholar]
- Rautio J. J., Satokari R., Vehmaan-Kreula P., Serkkola E., Söderlund H. (2008). TRAC in high-content gene expression analysis: applications in microbial population studies, process biotechnology and biomedical research. Expert Rev. Mol. Diagn. 8, 379–385 10.1586/14737159.8.4.379 [DOI] [PubMed] [Google Scholar]
- Regan J. F., Makarewicz A. J., Hindson B. J., Metz T. R., Gutierrez D. M., Corzett T. H., et al. (2008). Environmental monitoring for biological threat agents using the autonomous pathogen detection system with multiplexed polymerase chain reaction. Anal. Chem. 80, 7422–7429 10.1021/ac801125x [DOI] [PubMed] [Google Scholar]
- Sloma A., Smith I. (1979). RNA synthesis during spore germination in Bacillus subtilis. Mol. Gen. Genet. 175, 113–120 10.1007/BF00425526 [DOI] [PubMed] [Google Scholar]
- Soini J., Falschlehner C., Liedert C., Bernhard J., Vuoristo J., Neubauer P. (2008). Norvaline is accumulated after a down-shift of oxygen in Escherichia coli W3110. Microb. Cell Fact. 7:30 10.1186/1475-2859-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaulding U. K., Christensen C. J., Crisp R. J., Vaughn M. B., Trauscht R. C., Gardner J. R., et al. (2012). RAZOR EX anthrax air detection system. J. AOAC Int. 95, 860–891 10.5740/jaoacint.11-521 [DOI] [PubMed] [Google Scholar]
- Stachowiak J. C., Shugard E. E., Mosier B. P., Renzi R. F., Caton P. F., Ferko S. M., et al. (2007). Autonomous microfluidic sample preparation system for protein profile-based detection of aerosolized bacterial cells and spores. Anal. Chem. 79, 5763–5770 10.1021/ac070567z [DOI] [PubMed] [Google Scholar]
- Stratis-Cullum D. N., Griffin G. D., Mobley J., Vass A. A., Vo-Dinh T. (2003). A miniature biochip system for detection of aerosolized Bacillus globigii spores. Anal. Chem. 75, 275–280 10.1021/ac026068+ [DOI] [PubMed] [Google Scholar]
- Tamborrini M., Holzer M., Seeberger P. H., Schürch N., Pluschke G. (2010). Anthrax spore detection by a luminex assay based on monoclonal antibodies that recognize anthrose-containing oligosaccharides. Clin. Vaccine Immunol. 17, 1446–1451 10.1128/CVI.00205-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taskila S., Osmekhina E., Tuomola M., Ruuska J., Neubauer P. (2011). Modification of buffered peptone water for improved recovery of heat-injured Salmonella Typhimurium. J. Food Sci. 76, M157–M162 10.1111/j.1750-3841.2010.02050.x [DOI] [PubMed] [Google Scholar]
- Thieme D., Neubauer P., Nies D. H., Grass G. (2008). Sandwich hybridization assay for sensitive detection of dynamic changes in mRNA transcript levels in crude Escherichia coli cell extracts in response to copper ions. Appl. Environ. Microbiol. 74, 7463–7470 10.1128/AEM.01370-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh M. K., Wang X., Weimer B. C. (2001). Optimizing the immobilization of single-stranded DNA onto glass beads. J. Biochem. Biophys. Methods 47, 221–231 10.1016/S0165-022X(00)00146-9 [DOI] [PubMed] [Google Scholar]
- Wang Z., Fan Y., Chen J., Guo Y., Wu W., He Y., et al. (2013). A micro-fluidic chip-based fluorescent biosensor for the sensitive and specific detection of label-free single-base mismatch via magnetic beads-based “sandwich” hybridization strategy. Electrophoresis 34, 2177–2184 10.1002/elps.201300131 [DOI] [PubMed] [Google Scholar]
- Weerasekara M. L., Ryuda N., Miyamoto H., Okumura T., Ueno D., Inoue K., et al. (2013). Double-color fluorescence in situ hybridization (FISH) for the detection of Bacillus anthracis spores in environmental samples with a novel permeabilization protocol. J. Microbiol. Methods 93, 177–184 10.1016/j.mimet.2013.03.007 [DOI] [PubMed] [Google Scholar]
- Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. (1991). 16S Ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielinga P. R., Hamidjaja R. A., Agren J., Knutsson R., Segerman B., Fricker M., et al. (2011). A multiplex real-time PCR for identifying and differentiating B. anthracis virulent types. Int. J. Food Microbiol. 145 (Suppl. 1), S137–S144 10.1016/j.ijfoodmicro.2010.07.039 [DOI] [PubMed] [Google Scholar]
- Zhou B., Wirsching P., Janda K. D. (2002). Human antibodies against spores of the genus Bacillus: a model study for detection of and protection against anthrax and the bioterrorist threat. Proc. Natl. Acad. Sci. U.S.A. 99, 5241–5246 10.1073/pnas.082121599 [DOI] [PMC free article] [PubMed] [Google Scholar]