Abstract
A locus on human chromosome 11q23 tagged by marker rs3802842 was associated with colorectal cancer (CRC) in a genome-wide association study; this finding has been replicated in case–control studies worldwide. In order to identify biologic factors at this locus that are related to the etiopathology of CRC, we used microarray-based target selection methods, coupled to next-generation sequencing, to study 103 kb at the 11q23 locus. We genotyped 369 putative variants from 1,030 patients with CRC (cases) and 1,061 individuals without CRC (controls) from the Ontario Familial Colorectal Cancer Registry. Two previously uncharacterized genes, COLCA1 and COLCA2, were found to be co-regulated genes that are transcribed from opposite strands. Expression levels of COLCA1 and COLCA2 transcripts correlate with rs3802842 genotypes. In colon tissues, COLCA1 co-localizes with crystalloid granules of eosinophils and granular organelles of mast cells, neutrophils, macrophages, dendritic cells and differentiated myeloid-derived cell lines. COLCA2 is present in the cytoplasm of normal epithelial, immune and other cell lineages, as well as tumor cells. Tissue microarray analysis demonstrates the association of rs3802842 with lymphocyte density in the lamina propria (p = 0.014) and levels of COLCA1 in the lamina propria (p = 0.00016) and COLCA2 (tumor cells, p = 0.0041 and lamina propria, p = 6 × 10–5). In conclusion, genetic, expression and immunohistochemical data implicate COLCA1 and COLCA2 in the pathogenesis of colon cancer. Histologic analyses indicate the involvement of immune pathways.
Keywords: genome-wide association study, genetic risk factors, colon cancer, tumor microenvironment
What’s new? —
Genome-wide association studies have identified genomic loci associated with increased cancer risk, but the precise identification of the genes supporting cancer development within these loci remains challenging. The authors performed high-resolution mapping of a region on chromosome 11q23 previously associated with colorectal cancer predisposition. They identified COLCA1 and COLCA2, two coordinately regulated genes for which transcript and protein expression correlated with the presence or absence of the single-nucleotide polymorphism (SNP) identified in the region. In addition, the presence of the SNP as well as expression levels of the candidate genes correlated with a characteristic histological pattern of lymphocyte infiltration in the lamina propria of the colon tissue. These studies link colon cancer risk with immune pathways providing new opportunities for improved colon cancer diagnostic tools, novel predictive biomarkers, and potential therapeutic strategies modulating the tumor microenvironment.
Genome-wide association studies (GWAS) for over two dozen cancers have identified over 150 loci that individually confer modest increases in cancer risk, although few of these have led to the precise identification of causal genes and alleles.1 Of the 23 published loci associated with colorectal cancer (CRC) predisposition,2–6 functional studies implicate SMAD7,7 MYC,8,9 CDH110 and EIF3H,11 as well as other plausible candidates such as BMP4 and GREM1 that are members or modulators of the transforming growth factor beta superfamily that regulates cell proliferation.
An important challenge in deciphering the molecular basis of a GWAS locus is that the associated marker is a “tag” single nucleotide polymorphism (SNP) that is in linkage disequilibrium (LD) with many nearby SNPs.12 Even when a disease-associated GWAS locus has a plausible candidate gene, the mechanistic basis for the association may be complex. For example, the lung cancer risk-associated variants on chromosome 15q25 are located in a region of strong LD that includes six genes (CHRNB4, CHRNA5, CHRNA3, LOCI123688, PSMA4 and IREB2).13 Although the nicotinic acetylcholine receptor genes are the top candidates due to their involvement in nicotine addiction, IREB2 is being considered, based on its expression levels correlating with risk genotypes and its roles in oxidative stress and inflammation.13 By integrating genome-wide datasets of regulatory variation to specific disease loci, several GWAS loci appear to involve genes whose expression levels correlate with associated variants, including SMAD7 and colon cancer,7 PSCA and bladder cancer,14 TOX3 and breast cancer,5 and genes linked with numerous non-cancer loci.16
Here we report a high-resolution analysis of genetic variants and candidate genes on chromosome 11q23, in the vicinity of GWAS single nucleotide polymorphism (SNP) rs3802842. The associated 11q23 region was first reported in a Scottish study17 and subsequently refined to a 60 kb region using 10,638 cases and 10,457 controls from Europe, North America and Australia.18 The C allele of rs3802842 (global minor allele frequency = 31.3% in the 1,000 Genomes Project19 was shown to predispose to CRC, with odds ratio (OR) = 1.17 per allele, p = 1.08 × 10−12. Replication of the association has been reported in Dutch,20 Chinese,21 European American and Hawaiian22 populations. A recent meta-analysis comprising 38,534 cases and 39,446 controls reported significant association between rs3802842 and CRC risk (OR = 1.45).23
Material and Methods
The study was approved by the research ethics boards of the University of Toronto and Mount Sinai Hospital, Toronto.
Sequenced samples include genomic DNA from 40 sporadic CRC cases and 40 matched controls selected from the 2,380 samples from the Ontario Familial Colorectal Cancer Registry (OFCCR) that were previously genotyped by GWAS24 and 25 probands and 15 affected siblings selected from pedigrees showing autosomal dominant transmission that were selected based on absence of mutations in genes causing familial CRC.
Genotyping of 11q23 SNPs was performed using the iSelect array from Illumina. Novel SNPs that have been successfully genotyped and validated were submitted to dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) under the submission handle OICR_HUDSON.
RNA expression analyses, luciferase reported assays, protein expression and histochemical studies followed common laboratory protocols.
Further methods: Detailed methods and associated tables, figures and references are available in Supporting Information materials.
Results
High-resolution mapping of the 11q23 CRC locus
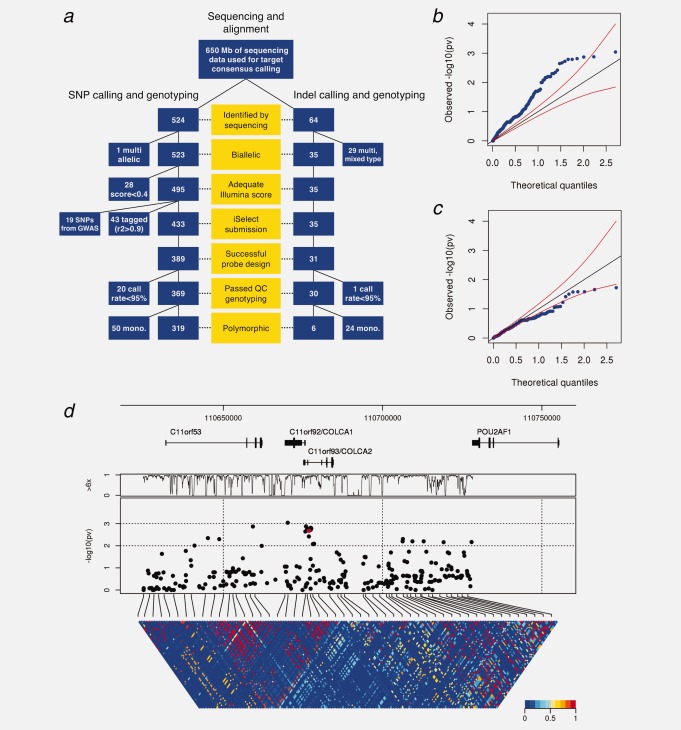
In a panel of 120 individuals recruited in the OFCCR and one CEPH sample, we used microarray-based target selection coupled to next-generation sequencing,25 to interrogate 103,418 bp of DNA including exonic, intronic and intergenic intervals at the 11q23 CRC locus. The selected region was defined as the largest interval that includes SNPs in LD with rs3802842 (r2 > 0.20), based on release 23a of the CEU HapMap data. This defines a broad region that should contain the causal SNP/haplotype picked up by the tag SNP. We generated 650 Mb of usable sequencing data in the target regions, corresponding to an average of 53 reads per base of genomic target and 72 reads per exonic base. We identified 524 putative SNPs and 64 putative insertion/deletions (indels) (Fig. 1a and Supporting Information Table 1).
Figure 1.
Association analysis of the CRC locus tagged by GWAS SNP rs3802842. (a) Polymorphism discovery, quality filters and genotyping flow-chart for 11q23. (b) Quantile–quantile plot of significance levels against theoretical quantiles for unconditional tests of association. Red lines represent 95% confidence bands. (c) Same plot as in (b), but with tests of association conditional on rs3802842. (d) Architecture of the 11q23 locus. From top to bottom: base position and known genes; percentage of samples with at least 6× sequence coverage as a function of base position; significance of tests of association, on the negative log scale, with the red dot indicating rs3802842; LD structure between all variants, with color shading showing the squared correlation coefficient r2.
We genotyped 369 putative variants in 1,030 cases and 1,061 controls from the OFCCR, in addition to the sequenced samples. The data revealed a high overall concordance (98.6%) between sequence-based genotypes and array-based genotypes, although there were many false-positives indels (80%) and a moderate rate of false-positives for SNPs (13.5%), which generally corresponded to putative variants detected at low coverage and/or misalignment at the codon level. We confirmed two coding non-synonymous SNPs in C11orf92 in the original sequenced samples: (1) p.Gly22Arg for which only one additional instance of the alternative allele was seen in an independent set of 2,091 genotyped samples and (2) p.Ala7Thr, which has an allele frequency of 1.5% in cases and 1.8% in controls. Only one coding non-synonymous SNP was discovered in C11orf93; SNP rs201868314 was present in a subject with sporadic CRC, but not observed in the subsequent panel of 2,091 samples.
There are signals of association at 11q23 for many SNPs in the interval. A quantile–quantile (Q–Q) plot illustrating all significance levels shows deviation of the weight of the distribution toward greater levels of associations (Fig. 1b). This shift in weight is caused by LD with rs3802842. Performing tests of association by conditioning on the presence or the absence of the GWAS risk allele on the same haplotypes as the test alleles shifts the Q–Q plot distribution back towards the null (Fig. 1c).
Figure 1d shows the location of all SNPs with minimum allele frequencies above 1% in cases and controls combined and risk of CRC in 1,030 cases and 1,061 controls, the significance levels of tests of association and comprehensive LD maps among common variants. GWAS SNP rs3802842 reaches a significance level of p < 0.002 in this sample set. It is noteworthy that extensive sequencing and genotyping of the 11q23 locus in the OFCCR did not reveal other variants that are more significant than rs3802842; several 11q23 variants show similar association and are in strong LD with rs3802842.
Expression of two transcripts at 11q23 show association with rs3802842
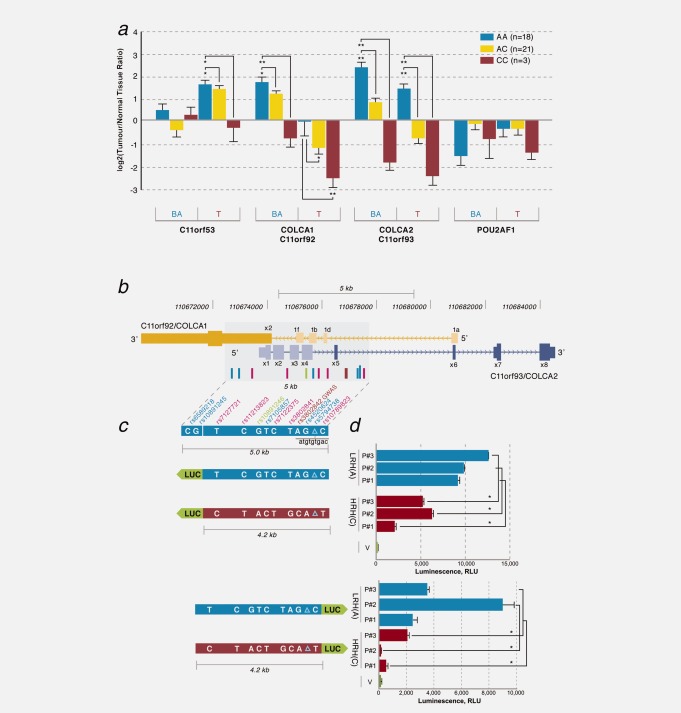
The region includes three uncharacterized protein-coding genes (C11orf53, C11orf92 and C11orf93). A nearby gene, POU2AF1 was also deemed a candidate as it is differentially expressed in lymphoma and leukemia.26,27 We characterized RNA abundances of these transcripts in tissues obtained during colon resections. We tested benign adjacent colonic tissues as well as colonic tumors from individuals homozygous for the lower risk (A) or higher risk (C) alleles of rs3802842, or heterozygous (A/C) (Fig. 2a). Lower risk alleles correlate with increased expression of C11orf92 and C11orf93 in benign adjacent colonic tissues as well as in tumors. We note increased expression of C11orf53 in tumor samples from individuals that are associated with the number of lower risk alleles, but no correlations are observed in benign adjacent colonic tissue. No association with POU2AF1 is detected.
Figure 2.
RNA expression, genomic organization and promoter analyses of COLCA1 and COLCA2. (a) Relative mRNA expression levels of 11q23 transcripts in benign adjacent (BA) and tumor (T) samples as a function of the rs3802842 genotype. Error bars indicate SEM. p values are derived from one-way ANOVA followed by Student-Newman-Keuls test. *p<0.01, **p<0.001. (b) Genomic organization of C11orf92/COLCA1 (gold) and C11orf93/COLCA2 (blue). (c) Luciferase (LUC)-based reporter constructs for COLCA1 (top) and COLCA2 (bottom). Lower risk (LRH) and higher risk (HRH) haplotypes are depicted as blue and red bars, respectively. (d) Luminescence results in HeLa cells. Values are expressed as mean ± SEM for n = 3. *p < 0.05. V is a promoter-less luciferase vector; p#1–3 denote patients 1–3.
C11orf92 and C11orf93 are arranged on opposite strands and share a regulatory region that contains genetic variants that are in high LD with rs3802842 (Fig. 2b). To investigate the cis-regulatory potential of the most common haplotypes associated with lower or higher risk, we cloned three independent triplicate DNA fragments of ∼4.2 kbp for each allele of rs3802842, as well as ten additional variants (9 SNPs and rs5794738, a 9 bp indel), into luciferase reporter vectors (Fig. 2c). When tested in both orientations, higher luciferase activity is observed for clones harboring the lower risk haplotype compared to those with the higher risk haplotype (Fig. 2d).
COLCA1 structure and expression features
C11orf92 (Colorectal Cancer Associated 1 or COLCA1) required further investigations of gene structure to complement annotations present in public databases (Supporting Information Figs. 1 and 2). COLCA1 has multiple alternative 5’ non-coding exons, and one constant exon that codes for a 124-amino acid protein. COLCA1 is a primate-specific gene (Supporting Information Fig. 3). COLCA1 does not show homology to other proteins in public databases. Its secondary structure is predicted to possess a signal peptide, a transmembrane domain (Supporting Information Fig. 4) and O-linked glycosylation.
Protein variants for COLCA1 are rare, as confirmed by Sanger sequencing of an additional set of cases (Supporting Information Table 2). In silico predictions of functional correlates for alleles contained on the lower/higher risk haplotypes (Supporting Information Table 3) can be made, although their specific role in modifying cancer risk is unknown. One of the strongest CRC-associated variants at this locus is rs10891246 that is in LD with rs3802842 (r2 = 0.99). For COLCA1, rs10891246 coincides with a splice site resulting in a short and long version of exon 1, a non-coding exon (Supporting Information Fig. 5). In a survey of COLCA1 RNA in tissue panels representing the gastrointestinal tract and the immune system, we observe expression from the esophagus to the rectum (Supporting Information Fig. 6a), multiple immune organs (Supporting Information Fig. 6b) and other tissues such as prostate, testis and ovary (Supporting Information Fig. 7a). Western blot analyses provide preliminary evidence that COLCA1 has greater expression in benign adjacent tissues than in tumors obtained at the time of colon cancer or adenoma resections (Supporting Information Fig. 8). A single band is observed for COLCA1 in these tissues as well as lysates from peripheral blood, eosinophils, neutrophils and CD14+ monocytes, but not in lymphocytes. We observe greater protein expression in homozygotes having the lower risk A allele compared to homozygotes for the C allele (Supporting Information Fig. 8b).
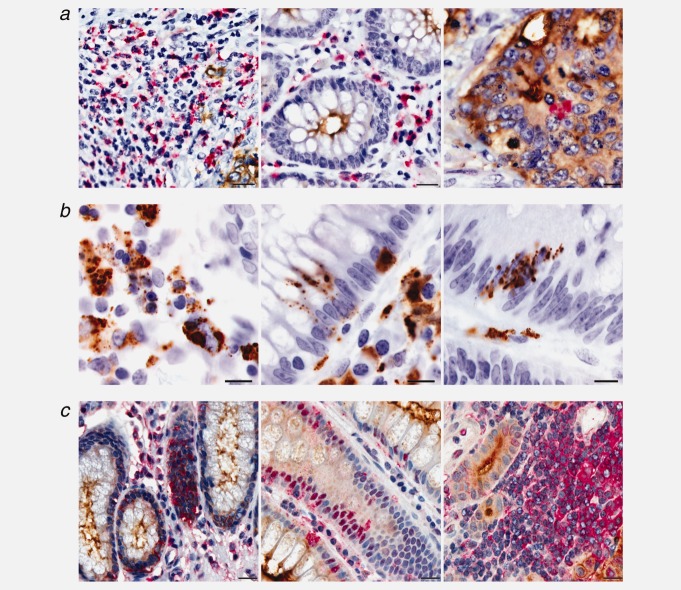
Immunohistochemistry with anti-COLCA1 in colon tissues reveals expression in the lamina propria, but not in normal epithelial cells or epithelium-derived neoplastic cells (Fig. 3a). At higher magnification (Fig. 3b), COLCA1 expression appears to be part of granular structures. In addition, extracellular COLCA1 is observed in normal adjacent tissue and in some cases the COLCA1 signal appears to infiltrate spaces between epithelial cells. Using cryosections of both benign adjacent colon and tumor tissues, triple immunofluorescence patterns show strong COLCA1 expression in eosinophils and moderate expression in mast cells, neutrophils, macrophages and dendritic cells (Fig. 4a). Within all COLCA1-positive immunofluorescent cells, COLCA1 signal is present in granular structures.
Figure 3.
COLCA1 and COLCA2 expression in paraffin-embedded colon biopsy samples. (a) Double immunohistochemical staining for COLCA1 (red; hematoxylin counterstain) and tumor-specific CEA (carcinoembryonic antigen; brown), scale bars, 20 µm. (b) 100× oil objective images (scale bars, 10 µm) of representative tissues immunostained with COLCA1 (brown; hematoxylin counterstain). (c) Double immunohistochemical staining for COLCA2 (red; hematoxylin counterstain) and CEA (brown), scale bars, 20 µm.
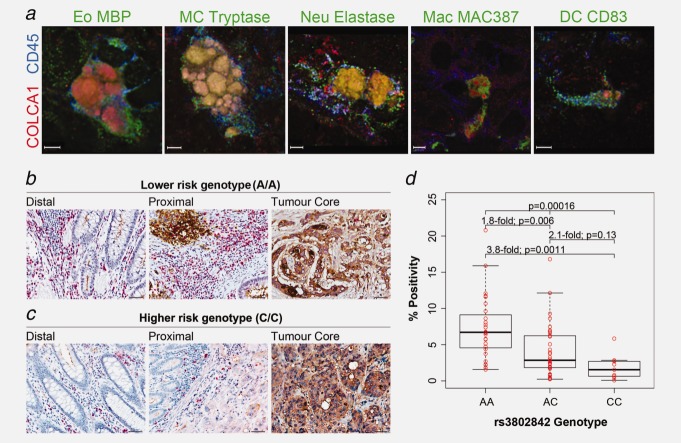
Figure 4.
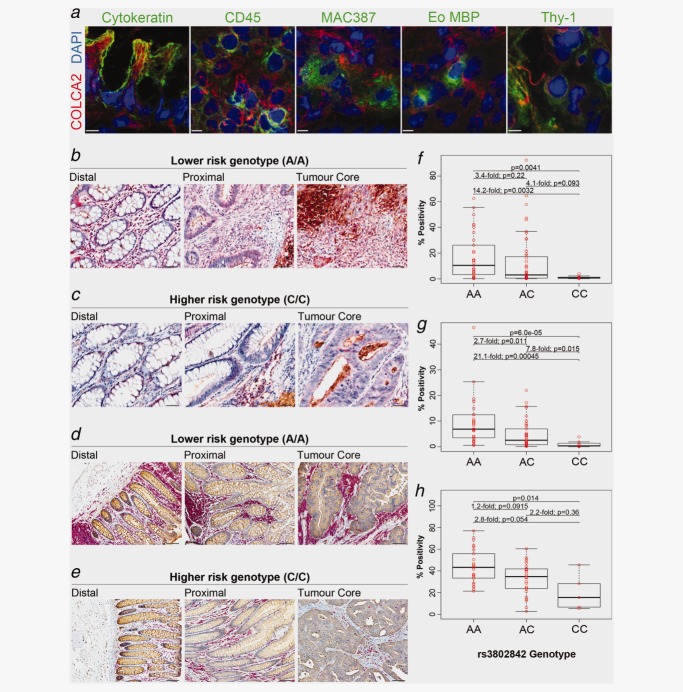
COLCA1 expression in immune cells in colon tissues and association with rs3802842. (a) Co-localization of COLCA1 with specific immune cell subsets markers was assessed on cryosections from colon tumor biopsies using three-dimensional deconvolution microscopy. High-power images of single immune cells were taken at sequential 0.1–0.3 µm z axis depth separation (scale bars, 3 µm). For all images, COLCA1 staining is shown in red and a pan-leukocyte marker (CD45) is shown in blue. Green signals represent different immune cell-specific markers: eosinophils (Eo) labeled with eosinophil major basic protein (MBP); mast cells (MC) labeled with mast cell tryptase; neutrophils (Neu) labeled with neutrophil elastase; macrophages (Mac) labeled with MAC387; dendritic cells (DC) labeled with CD83. (b, c) Double immunohistochemical staining for COLCA1, red and CEA, brown, in distal, proximal and tumor core of colon tissues from patients homozygous for the lower risk rs3802842 genotype (AA) (b), compared to the higher risk genotype (CC) (c) (scale bars, 50 µm). (d) Association analyses between genotypes at rs3802842 and COLCA1 expression in TMAs. Individual expression values and corresponding boxplots are illustrated. Thick horizontal lines represent median values.
To validate the correlations between rs3802842 and COLCA1 staining in the lamina propria surrounding tumor tissues (Figs. 4b and 4c), we employed a novel image analysis method (Supporting Information Fig. 9) to quantitate COLCA1 staining in tissue microarrays (TMAs). TMAs of colorectal cancers representing various stages from 96 patients were stained and scored. The correlation between the number of lower risk alleles of rs3802842 with increased staining for COLCA1 in the lamina propria was confirmed (Kruskal-Wallis test: p = 0.00016; Fig. 4d).
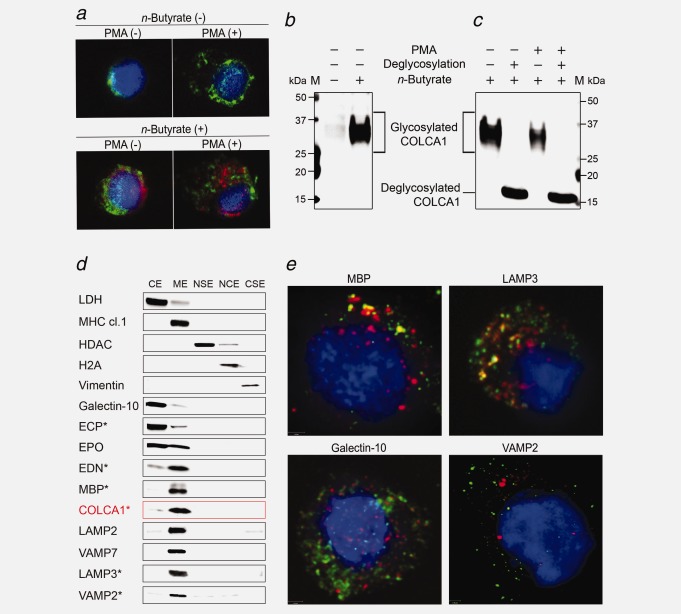
Although COLCA1 mRNA expression is mostly observed in cell lines of hematopoietic origin (lymphoid and myeloid) and in some cell lines derived from epithelial tumors including Caco-2 (colorectal adenocarcinoma) and Saos-2 (osteosarcoma) (Supporting Information Figs. 7b and 10), COLCA1 protein was not detected in HeLa cells, four colon cancer cell lines (including Caco-2) and a panel of lymphoid and myeloid cell lines (Supporting Information Figs. 11a and 11b). We hypothesized that only differentiated hematopoietic cells having the capacity to assemble granules would be able to express COLCA1. We first corroborated this in an in vitro model using human promyelocytic leukemia HL60 clone 15 cells differentiated into eosinophils using n-butyrate.28 Endogenous COLCA1 protein is observed when cells are stimulated to differentiate (Figs. 5a and 5b). In this model, COLCA1 is heavily glycosylated (Fig. 5c), a feature common to other granule-associated proteins.29 Several other in vitro models can be induced to express COLCA1 protein: (1) Cord blood mononuclear cells differentiated into CD45+CD34-CD117+CD11c-CD11b- mast cells (Supporting Information Fig. 11c) and CD11b+CD11c+ dendritic cells (Supporting Information Fig. 11d); (2) LAD2 mast cells30 transfected with GFP-fused COLCA1 cDNA (Supporting Information Fig. 12a) and (3) TLS-ERG transduced CD34+ TEX cells31 that are stimulated to differentiate into eosinophil-like cells using IL3, IL5 and GM-CSF (Supporting Information Fig. 12b).
Figure 5.
Expression of COLCA1 in granules of HL60 clone 15 cells. (a) Cells were treated with n-butyrate (0.5 mM, lower panels) for 6 days, stimulated with phorbol 12-myristate 13-acetate (162 nM, right panels) for 4 hr, and immunostained for COLCA1 (red), eosinophil major basic protein (green) and DAPI nuclear stain (blue). (b) Western blot of COLCA1 in lysates from n-butyrate treated cells. The molecular weight of the immunoreactive signal is higher than predicted. (c) Western blot of glycosylated and deglycosylated COLCA1 in lysates from n-butyrate treated cells in the presence or absence of PMA. (d) Western blot of COLCA1 in subcellular fractions of cells: cytosolic (CE), membrane (ME), nuclear soluble (NSE), chromatin-bound (NCE) and cytoskeletal protein extracts (CSE). The asterisks indicate post-deglycosylation. (e) Co-localization of COLCA1-GFP with proteins associated with eosinophil granules. Confocal immunofluorescence images of COLCA1-GFP-transfected cells (green) immunostained with antibodies to eosinophil granule proteins (red) and counterstained with DAPI (blue).
To identify cellular compartment that contain COLCA1, we first employed a method that causes partial membrane permeabilization followed by differential centrifugation to separate cytosolic, nucleosolic, nuclear insoluble and membrane components of HL60 clone 15 cells treated with n-butyrate to produce COLCA1 (Fig. 5d). COLCA1 is absent in fractions specific to the cytosol, nucleus and cytoskeleton, but enriched in a fraction that contains a diversity of membrane proteins. We observe that COLCA1 co-sediments with membrane proteins associated with eosinophilic granules and secretory vesicles, including LAMP2, CD63/LAMP3, VAMP2 and VAMP7.32 Crystalloid-specific proteins (core: MBP; matrix: EDN) are mostly detected in the membrane extract, a pattern that could be explained by low permeabilization of crystalloid granule membranes. Galectin-10, a primary-granule specific protein is found in the cytosolic extract and ECP, a matrix protein of small granules and crystalloid granules is mostly found in the cytosolic extract.32 EPO, a matrix protein of lipid bodies and crystalloid granules32 is found in both the cytosolic and membrane extracts. The data are consistent with complete permeabilization of membranes for lipid bodies, primary granules and small granules.
Imaging studies using HL60 clone 15 cells transfected with GFP-COLCA1 and pre-treated with n-butyrate were conducted to identify eosinophil granule proteins that co-localize with COLCA1 (Fig. 5e, Supporting Information Fig. 13). Proteins that co-localize the most with COLCA1 are known to be associated with crystalloid granules, the most common type of granules in eosinophils; these include the membrane protein LAMP3, the core protein MBP and matrix proteins EPO and ECP.32 Galectin-10, which is associated with primary granules32 shows no co-localization with COLCA1. VAMP-2, a specific protein associated with a class of secretory vesicles also shows no co-localization.
COLCA2 structure and expression features
C11orf93 (Colorectal Cancer Associated 2 or COLCA2) has several orthologs in mammals (Supporting Information Fig. 14). The revised gene model for COLCA2 includes eight exons, with variable exons 1–4 added in various combinations to constant exons 5–8 to generate a minimum of five confirmed transcripts yielding protein isoforms ranging from 154 to 379 amino acids in length (Supporting Information Figs. 1, 15 and Supporting Information Table 4). Additional protein isoforms are predicted from Western blots. Immunoreactive bands ranging from 17 to 47 kDa that potentially represent eight COLCA2 protein isoforms are observed in different permutations in tested samples, including colonic tissues, peripheral blood and 17 cell lines (Supporting Information Fig. 16; some data not shown) representing multiple cell types. In silico predictions of functional correlates for COLCA2 variants (Supporting Information Table 3) can be made for rs10891246, such that isoforms having an alanine at this position correlate with the lower risk allele while the higher risk haplotype encodes for a threonine at this position. Another CRC-associated variant in LD with rs3802842 is rs7105857 (r2=0.91); this SNP disrupts a splice site resulting in a longer version of exon 4 and a longer protein isoform.
Western blots and immunofluorescence analyses for COLCA2 reveal protein expression in the cytoplasm of many cell types of epithelial (normal and tumoral), lymphoid, myeloid, mesenchymal (including fibroblasts) and other origins (Figs. 3c, 6a and Supporting Information methods). We also observe an increase in COLCA2 protein expression in colon tissues from individuals with the lower risk AA genotype (Figs. 6b and 6c). TMAs with this biomarker validate the association between the lower risk allele of rs3802842 with increased staining for COLCA2, and allow us to observe correlations in two distinct areas: (1) epithelium (p = 0.004; Fig. 6f); and (2) lamina propria (p = 6 × 10−5, Fig. 6g).
Figure 6.
COLCA2 expression and lymphocyte density in colon correlates with rs3802842. (a) Co-localization of COLCA2 with cell-specific markers was assessed on cryosections from colon tumor biopsies using three-dimensional deconvolution microscopy. High-power images of COLCA2-positive cells were taken at sequential 0.1–0.3 µm z axis depth separation (scale bars, 5 µm). For all images, COLCA2 staining is shown in red and DAPI is shown in blue. Green signals represent different cell-type specific markers: epithelial cells labeled for cytokeratin; leukocytes labeled with a pan-CD45 marker; macrophages labeled with MAC387; eosinophils (Eo) labeled with eosinophil major basic protein (MBP); fibroblasts labeled with Thy-1. (b, c) COLCA2 expression in colon tissues. Double staining for COLCA2, red and CEA, brown, in distal, proximal and tumor core of colon tissues from patients homozygous for the lower risk rs3802842 genotype (AA) in (b) compared to the higher risk genotype (CC) in (c) (scale bars, 50 µm). (d, e) Double immunohistochemical staining for lymphocytes (CD45 clones 2B11 and PD7/26), red and CEA, brown, in distal, proximal and tumor core of colon tissues from patients homozygous for the lower risk rs3802842 genotype (AA) in (d) compared to the higher risk genotype (CC) in (e) (scale bars, 50 µm). (f, g) Association analyses between genotypes at rs3802842 and COLCA2 in colon tumor epithelial cells (f) and in the lamina propria (g). (h) Association analyses between genotypes at rs3802842 and CD45 expression in the lamina propria. Individual expression values and corresponding boxplots are illustrated. Thick horizontal lines represent median values.
Lymphoid cell aggregates and lymphocyte density correlate with CRC-associated variants at 11q23
Histological review of tissue sections from patients having different genotypes at rs3802842 suggested that lymphoid cell aggregates are more common in colon biopsies from patients with the AA genotype. This observation was validated using combined monoclonal antibodies for CD45 (clones 2B11 and PD7/26) that are immunoreactive for lymphoid cells (and mast cells) but non-reactive to polymorphonucear leukocytes and nonhematopoietic tissues33 Figs. 6d and 6e). TMAs stained with these anti-lymphocyte antibodies show significant association at rs3802842 (p = 0.014; Fig. 6h).
Discussion
Several lines of evidence link COLCA1 and COLCA2 with CRC: (1) genetic association at 11q23; (2) correlation between risk alleles with COLCA1 and COLCA2 RNA levels in normal colon and CRC tissues; (3) correlation between risk alleles with COLCA1 and COLCA2 protein levels in CRC tissues; (4) presence in many mucosal immune cells of the colon implicated in tumor immunity.
In this study, COLCA1 and COLCA2 were screened for germline mutations in 40 individuals from pedigrees displaying autosomal dominant transmission, but no highly penetrant CRC mutation was discovered. An expanded analysis in sporadic cases revealed no plausible protein-coding variant in COLCA1 that can explain the association identified by GWAS. A common variant (rs10891246) in high LD (r2 = 0.97) with the GWAS variant (rs3802842) codes for an alanine to threonine polymorphism in COLCA2 that is present on two protein isoforms.
We observe that several variants in a ∼5kb haplotype containing rs3802842 correlate with expression levels of COLCA1 and COLCA2. Some of the variants in high LD with rs3802842 overlap with transcription factor binding sites: p53/rs6589218, CREB/rs10891245, E2F/rs11213823, FoxB1/rs4520624 and FoxB1/rs5794738 (a 9-bp deletion) (Supporting Information Table 3). It is interesting to note that this locus was reported to have a differential epigenomic profile at an enhancer overlapping rs3802842 between colon and non-colon derived tissues and cell lines,34 leading to the hypothesis that rs3802842 may affect enhancer function. We provided experimental evidence in support of differential transcriptional activity for the higher and lower risk haplotypes using luciferase reporter constructs.
COLCA1 is a primate-specific gene whose recent evolution may be similar to many positively selected genes among mammals that are frequently involved in immunity and defense. COLCA1 is present in granular organelles in eosinophils, mast cells, neutrophils, macrophages and dendritic cells in the gastrointestinal microenvironment. We presented evidence that COLCA1 protein expression in cell line models is restricted to myeloid-derived cell lines that are differentiated into granule-producing cells. Cell fractionation and imaging co-localization studies are consistent with COLCA1 being a membrane protein of crystalloid granules in eosinophils, which contain the core protein MBP and matrix proteins (EPO, EDN and ECP).32
COLCA1 is expressed in many cells having a role in inflammation and cancer.35 The significance of eosinophils and mast cells in cancer remain controversial, particularly for mast cells which have been implicated in tumor promotion through angiogenesis, tissue remodeling and regulatory T-cell dysfunction.36,37 Mast cell depletion can lead to remission of polyps in mouse models.38 However, mast cells may also be damaging to tumor cells via cytokines and proteolytic enzyme secretion. In a breast cancer study, stromal mast cells correlate with favorable prognosis.39 The literature provides a stronger case for eosinophils having tumoricidal functions. Abundance of eosinophils in gastrointestinal cancers is a favorable prognostic factor.40 Eosinophils may induce apoptosis and kill tumor cells, via the release of ECP, EDN, TNF-α and granzyme.41 Eosinophil products can degrade necrotic materials from tumor cells through production of reactive oxygen species.42 Eosinophils, which are the highest expressors of COLCA1, contain granular structures that are known to harbor pre-formed proteins. Extracellular COLCA1 staining in colon tissues has a similar pattern that has been described for extracellular eosinophil-derived granules.43
COLCA2 is present in many cell types of epithelial, mesenchymal and hematopoietic origins and has orthologs in multiple mammals. Since COLCA2 expression is reduced in tumor cells from subjects with higher risk alleles, it is possible that COLCA2 has critical functions that suppress tumor formation in epithelial cells. Although not characterized as a cancer gene, we note the recent identification of two protein-coding mutations in 2 of 25 melanoma samples.44 Higher levels of COLCA2 in immune and other cells of the microenvironment may also provide protection against cancer cell growth.
The role of genetic variation linking the immune system with colon cancer has been recognized. Both juvenile polyposis and Lynch Syndromes are caused by germline mutations that in addition to promoting tumorigenesis within colon epithelial cells are associated with inflammatory cell infiltrates. Germline mutations in SMAD4 cause juvenile polyposis and Smad4 mutant mice have lymphocytic and eosinophilic infiltrates in colon tissues45 and provide evidence that the effect of genetic mutations in intestinal lymphocytes are important for maintaining intestinal homeostasis.46 Patients with colorectal tumors with microsatellite instability (MSI), including heritable forms that are part of Lynch Syndrome are also characterized by lymphocytic infiltrates, natural antitumor immune responses47 and improved prognosis compared with patients with non-MSI tumors.48 This is consistent with other reports linking tumor-infiltrating lymphocytes with improved survival in colon cancer,49 independent of MSI status.50
The characterization of novel proteins and inflammatory processes correlating with genotypes associated with colon cancer risk suggests that the locus may modulate a complex network involving immune and tumor cells. It is interesting to note that at this stage of our investigation, COLCA1 and COLCA2 functions do not interconnect with functions of other genes implicated in CRC. Further investigations are needed to determine the precise functions of COLCA1 and COLCA2 and histological changes correlating with genotypes at 11q23.
Acknowledgments
The authors thank Joe Paino, Teresa Selander, Gordana Kuruzar, Mona Reid, Darshana Daftary, Jeanette Joans, Sugy Kodeeswaran, Alexandra Kollara, Caroline Dunk, Vivek Sharma and Jun-Fen Ji for technical advice, support and sample management. The authors acknowledge the contribution of Alexandre Montpetit, François Bacot, Pierre Lepage and Joelle Fontaine of the McGill University and Génome Québec Innovation Centre, Montréal, Canada for genotyping the 10,640-bead iSelect array from Illumina and sequencing C11orf92. The authors also thank Arnold S. Kirshenbaum at NIAID, NIH for providing LAD2 cells.
Glossary
- CEA
carcinoembryonic antigen
- COLCA1
colorectal cancer associated 1
- COLCA2
colorectal cancer associated 2
- CRC
colorectal cancer
- EDN
eosinophil-derived neurotoxin
- ECP
eosinophilic cationic protein
- GWAS
genome-wide association study
- LD
linkage disequilibrium
- MSI
microsatellite instability
- OFCCR
Ontario Familial Colorectal Cancer Registry
- OR
Odds ratio
- Q–Q
quantile–quantile
- SNP
single nucleotide polymorphism
- TMA
tissue microarray
Supporting Information
Additional Supporting Information may be found in the online version of this article.
References
- Chung CC, Chanock SJ. Current status of genome-wide association studies in cancer. Hum Genet. 2011;130:59–78. doi: 10.1007/s00439-011-1030-9. [DOI] [PubMed] [Google Scholar]
- Dunlop MG, Dobbins SE, Farrington SM, et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat Genet. 2012;44:770–76. doi: 10.1038/ng.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui R, Okada Y, Jang SG, et al. Common variant in 6q26-q27 is associated with distal colon cancer in an Asian population. Gut. 2011;60:799–05. doi: 10.1136/gut.2010.215947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnersley B, Migliorini G, Broderick P, et al. The TERT variant rs2736100 is associated with colorectal cancer risk. Br J Cancer. 2012;107:1001–08. doi: 10.1038/bjc.2012.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia WH, Zhang B, Matsuo K, et al. Genome-wide association analyses in east Asians identify new susceptibility loci for colorectal cancer. Nat Genet. 2013;45:191–96. doi: 10.1038/ng.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters U, Jiao S, Schumacher FR, et al. Identification of genetic susceptibility loci for colorectal tumors in a genome-wide meta-analysis. Gastroenterology. 2013;144:799–07. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick P, Carvajal-Carmona L, Pittman AM, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–17. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- Pomerantz MM, Ahmadiyeh N, Jia L, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–84. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuupanen SM, Turunen M, Lehtonen R, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–90. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- Pittman M, Twiss P, Broderick P, et al. The CDH1-160C>A polymorphism is a risk factor for colorectal cancer. Int J Cancer. 2009;125:1622–25. doi: 10.1002/ijc.24542. [DOI] [PubMed] [Google Scholar]
- Pittman AM, Naranjo S, Jalava SE, et al. Allelic variation at the 8q23.3 colorectal cancer risk locus functions as a cis-acting regulator of EIF3H. PLoS Genet. 2010;6:pii:e1001126. doi: 10.1371/journal.pgen.1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Dudley JT, Kimberly R, et al. Systematic functional regulatory assessment of disease-associated variants. Proc Natl Acad Sci U S A. 2013;110:9607–12. doi: 10.1073/pnas.1219099110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehringer G, Liu G, Pintilie M, et al. Association of the 15q25 and 5p15 lung cancer susceptibility regions with gene expression in lung tumor tissue. Cancer Epidemiol Biomarkers Prev. 2012;21:1097–04. doi: 10.1158/1055-9965.EPI-11-1123-T. [DOI] [PubMed] [Google Scholar]
- Fu YP, Kohaar I, Rothman N, et al. Common genetic variants in the PSCA gene influence gene expression and bladder cancer risk. Proc Natl Acad Sci U S A. 2012;109:4974–79. doi: 10.1073/pnas.1202189109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowper-Sal-lari R, Zhang X, Wright JB, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet. 2012;44:1191–98. doi: 10.1038/ng.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelly DA, Ronald J, Akey JM. Inherited variation in gene expression. Annu Rev Genomics Hum Genet. 2009;10:313–32. doi: 10.1146/annurev-genom-082908-150121. [DOI] [PubMed] [Google Scholar]
- Tenesa SM, Farrington SM, Prendergast JG, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–37. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman M, Webb E, Carvajal-Carmona L, et al. Refinement of the basis and impact of common 11q23.1 variation to the risk of developing colorectal cancer. Hum Mol Genet. 2008;17:3720–27. doi: 10.1093/hmg/ddn267. [DOI] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middeldorp A, Jagmohan-Changur S, van Eijk R, et al. Enrichment of low penetrance susceptibility loci in a Dutch familial colorectal cancer cohort. Cancer Epidemiol Biomarkers Prev. 2009;18:3062–67. doi: 10.1158/1055-9965.EPI-09-0601. [DOI] [PubMed] [Google Scholar]
- Xiong F, Wu C, Bi X, et al. Risk of genome-wide association study-identified genetic variants for colorectal cancer in a Chinese population. Cancer Epidemiol Biomarkers Prev. 2010;19:1855–61. doi: 10.1158/1055-9965.EPI-10-0210. [DOI] [PubMed] [Google Scholar]
- He J, Wilkens LR, Stram DO, et al. Generalizability and epidemiologic characterization of eleven colorectal cancer GWAS hits in multiple populations. Cancer Epidemiol Biomarkers Prev. 2011;20:70–81. doi: 10.1158/1055-9965.EPI-10-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Zhong R, Lou J, et al. Replication study in Chinese population and meta-analysis supports association of the 11q23 locus with colorectal cancer. PLoS One. 2012;7:e45461. doi: 10.1371/journal.pone.0045461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanke W, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- Wong KM, Hudson TJ, McPherson JD. Unraveling the genetics of cancer: genome sequencing and beyond. Annu. Rev. Genomics Hum Genet. 2011;12:407–30. doi: 10.1146/annurev-genom-082509-141532. [DOI] [PubMed] [Google Scholar]
- Auer RL, Starczynski J, McElwaine S, et al. Identification of a potential role for POU2AF1 and BTG4 in the deletion of 11q23 in chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2005;43:1–10. doi: 10.1002/gcc.20159. [DOI] [PubMed] [Google Scholar]
- Herbeck R, Teodorescu Brînzeu D, Giubelan M, et al. B-cell transcription factors Pax-5, Oct-2, BOB.1, Bcl-6, and MUM1 are useful markers for the diagnosis of nodular lymphocyte predominant Hodgkin lymphoma. Rom J Morphol Embryol. 2011;52:69–74. [PubMed] [Google Scholar]
- Ahmed N, Williams JF, Weidemann MJ. The human promyelocytic HL60 cell line: a model of myeloid cell differentiation using dimethylsulphoxide, phorbol ester and butyrate. Biochem Int. 1991;23:591–02. [PubMed] [Google Scholar]
- Tiffany HL, Li F, Rosenberg HF. Hyperglycosylation of eosinophil ribonucleases in a promyelocytic leukemia cell line and in differentiated peripheral blood progenitor cells. J Leuk Biol. 1995;58:49–54. doi: 10.1002/jlb.58.1.49. [DOI] [PubMed] [Google Scholar]
- Kirshenbaum AS, Akin C, Wu Y, et al. Characterization of novel stem cell factor responsive human mast cell lines LAD 1 and 2 established from a patient with mast cell sarcoma/leukemia; activation following aggregation of FcepsilonRI or FcgammaRI. Leuk Res. 2003;27:677–82. doi: 10.1016/s0145-2126(02)00343-0. [DOI] [PubMed] [Google Scholar]
- Warner JK, Wang JC, Takenaka K, et al. Direct evidence for cooperating genetic events in the leukemic transformation of normal human hematopoietic cells. Leukemia. 2005;19:1794–05. doi: 10.1038/sj.leu.2403917. [DOI] [PubMed] [Google Scholar]
- Paige L, Moqbel R. Chapter 7—eosinophil signal transduction. In: Lee JJ, Rosenberg LF, editors. Eosinophils in health and disease. 1. Boston: Academic Press; 2013. pp. 167–27. [Google Scholar]
- Kurtin PJ, Pinkus GS. Leukocyte common antigen—a diagnostic discriminant between hematopoietic and nonhematopoietic neoplasms in paraffin sections using monoclonal antibodies: correlation with immunologic studies and ultrastructural localization. Hum Pathol. 1985;16:353–65. doi: 10.1016/s0046-8177(85)80229-x. [DOI] [PubMed] [Google Scholar]
- Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science. 2012;336:736–39. doi: 10.1126/science.1217277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesely MD, Kershaw MH, Schreiber RD, et al. Natural innate and adaptive immunity to cancer. Annu Rev Immunol. 2011;29:235–71. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- Maltby S, Khazaie K, McNagny KM. Mast cells in tumor growth: |angiogenesis, tissue remodelling and immune-modulation. Biochim Biophys Acta. 2009;1796:19–26. doi: 10.1016/j.bbcan.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatner NR, Bonertz A, Beckhove P, et al. In colorectal cancer mast cells contribute to systemic regulatory T-cell dysfunction. Proc Natl Acad Sci USA. 2010;107:6430–35. doi: 10.1073/pnas.0913683107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounaris E, Erdman SE, Restaino C, et al. Mast cells are an essential hematopoietic component for polyp development. Proc Natl Acad Sci USA. 2007;104:19977–82. doi: 10.1073/pnas.0704620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajput B, Turbin DA, Cheang MC, et al. Stromal mast cells in invasive breast cancer are a marker of favourable prognosis: a study of 4,444 cases. Breast Cancer Res Treat. 2008;107:249–57. doi: 10.1007/s10549-007-9546-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotfi R, Lee J, Lotze MT. Eosinophilic granulocytes and damage-associated molecular pattern molecules (DAMPs): role in the inflammatory response within tumors. J Immunother. 2007;30:16–28. doi: 10.1097/01.cji.0000211324.53396.f6. [DOI] [PubMed] [Google Scholar]
- Legrand F, Driss V, Delbeke M, et al. Human eosinophils exert TNF-α and granzyme A-mediated tumoricidal activity toward colon carcinoma cells. J Immunol. 2010;185:7443–51. doi: 10.4049/jimmunol.1000446. [DOI] [PubMed] [Google Scholar]
- Lotfi R, Herzog GI, DeMarco RA, et al. Eosinophils oxidize damage-associated molecular pattern molecules derived from stressed cells. J Immunol. 2009;183:5023–31. doi: 10.4049/jimmunol.0900504. [DOI] [PubMed] [Google Scholar]
- Neves JS, Perez SA, Spencer LA, et al. Eosinophil granules function extracellularly as receptor-mediated secretory organelles. Proc Natl Acad Sci USA. 2008;105:18478–83. doi: 10.1073/pnas.0804547105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–06. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaku K, Miyoshi H, Matsunaga A, et al. Gastric and duodenal polyps in Smad4 (Dpc4) knockout mice. Cancer Res. 1999;59:6113–6117. [PubMed] [Google Scholar]
- Kim BG, Li C, Qiao W, et al. Smad4 signalling in T cells is required for suppression of gastrointestinal cancer. Nature. 2006;441:1015–19. doi: 10.1038/nature04846. [DOI] [PubMed] [Google Scholar]
- de Miranda NF, Goudkade D, Jordanova ES, et al. Infiltration of Lynch colorectal cancers by activated immune cells associates with early staging of the primary tumor and absence of lymph node metastases. Clin Cancer Res. 2012;18:1237–45. doi: 10.1158/1078-0432.CCR-11-1997. [DOI] [PubMed] [Google Scholar]
- Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–87. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon J, Costes A, Sanchez-Cabo F, et al. Type, |density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–64. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- Ogino S, Nosho K, Irahara N, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15:6412–20. doi: 10.1158/1078-0432.CCR-09-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.