Abstract
Short nonprotein coding RNA molecules, known as microRNAs (miRNAs), are intracellular mediators of adaptive processes, including muscle hypertrophy, contractile force generation, and inflammation. During basal conditions and tissue injury, miRNAs are released into the bloodstream as “circulating” miRNAs (c-miRNAs). To date, the impact of extended-duration, submaximal aerobic exercise on plasma concentrations of c-miRNAs remains incompletely characterized. We hypothesized that specific c-miRNAs are differentially upregulated following prolonged aerobic exercise. To test this hypothesis, we measured concentrations of c-miRNAs enriched in muscle (miR-1, miR-133a, miR-499–5p), cardiac tissue (miR-208a), and the vascular endothelium (miR-126), as well as those important in inflammation (miR-146a) in healthy male marathon runners (N = 21) at rest, immediately after a marathon (42-km foot race), and 24 h after the race. In addition, we compared c-miRNA profiles to those of conventional protein biomarkers reflective of skeletal muscle damage, cardiac stress and necrosis, and systemic inflammation. Candidate c-miRNAs increased immediately after the marathon and declined to prerace levels or lower after 24 h of race completion. However, the magnitude of change for each c-miRNA differed, even when originating from the same tissue type. In contrast, traditional biomarkers increased after exercise but remained elevated 24 h postexercise. Thus c-miRNAs respond differentially to prolonged exercise, suggesting the existence of specific mechanisms of c-miRNA release and clearance not fully explained by generalized cellular injury. Furthermore, c-miRNA expression patterns differ in a temporal fashion from corollary conventional tissue-specific biomarkers, emphasizing the potential of c-miRNAs as unique, real-time markers of exercise-induced tissue adaptation.
Keywords: circulating microRNA, exercise physiology, cardiovascular biomarker, cardiorespiratory fitness biomarker, prolonged aerobic exercise
there are numerous physiological adaptations to endurance exercise training, including myocardial remodeling (7), skeletal muscle hypertrophy (31), and peripheral vascular angiogenesis (26). The mechanisms producing these organ-specific responses are initiated by exercise of sufficient intensity and/or duration to produce tissue stress/injury. Increases of circulating biomarkers of skeletal muscle damage [e.g., creatine phosphokinase (CPK)], cardiac muscle stress [e.g., NH2-terminal prohormone of brain natriuretic peptide (NT-proBNP)], cardiac muscle damage [e.g., cardiac-specific troponin, (cTn)], and systemic inflammation [e.g., high-sensitivity C-reactive protein (hsCRP)] have been well documented (2, 10, 15, 16, 24, 27, 29, 39, 43, 48, 49, 61, 66), but these conventional biomarkers provide limited insight into the adaptive processes stimulated by vigorous exercise.
Short and conserved nonprotein coding RNA molecules known as microRNAs (miRNAs) have been recently identified as essential intracellular mediators of processes inherent in exercise adaptation, such as inflammation (18), muscle hypertrophy, and contractile force generation (17, 65). miRNAs can alter cellular function by posttranscriptionally repressing translation and degrading messenger RNAs through binding the 3′ untranslated regions of messenger RNA molecules (50). miRNAs are secreted into the bloodstream at rest (38) and in response to tissue injury (35) and other pathological conditions (22, 28). Through several mechanisms, such as inclusion in phospholipid bilayer-encapsulated vesicles (63) and formation of RNA-binding protein complexes (25), these “circulating” miRNAs (c-miRNAs) are protected from degradation. Intracellular transfer of miRNAs to recipient tissues has been described by mechanisms including transport through gap junctions (32), as well as packaging into exosomes (59) and other microvesicle- or RNA-binding protein-associated transfer systems. Thus tissue-specific release appears to represent a key mechanism by which cellular functions are modulated in response to tissue stress (34, 42, 68), and such release may not be solely driven by generalized cellular injury, even during exercise. Furthermore, alterations in c-miRNAs appear to be rapidly upregulated during acute physiological stress and may, therefore, more accurately reflect “real-time” intracellular responses to tissue stress compared with conventional plasma-based markers (19, 44, 52).
Dynamic regulation of c-miRNAs during exercise has recently been documented, but our understanding of their roles in exercise response and adaptation is just emerging. Analysis of skeletal muscle biopsies in mice (4, 53) and humans (40) before and after exhaustive aerobic exercise has demonstrated upregulation of intramuscular miR-1 and miR-133, as well as other miRNAs (21). Exercise-induced alterations of miRNAs occur in tissue compartments beyond skeletal muscle, including inflammatory cells (45–47, 64) and cardiac cells (20, 57, 58), among others (36). Our laboratories previously characterized the dynamic regulation of specific plasma-based c-miRNAs after acute and relatively brief exhaustive exercise in healthy humans (6). Since then, other c-miRNAs have also been profiled after acute aerobic exercise (3), after acute resistance exercise (54), and after alterations in acute exercise modality (8). Furthermore, a recent study examined the extent of endothelial-specific miR-126 release under various modalities of acute and increasing time of exercise (62). However, c-miRNA expression after prolonged exercise remains incompletely defined, and there have been no direct comparisons among miRNAs originating from different tissue compartments and among miRNAs compared with more traditional protein markers of exercise-induced tissue perturbation. We hypothesized that specific plasma c-miRNAs are uniquely modulated following completion of prolonged, submaximal aerobic exercise (i.e., marathon run). These expression profiles may follow different kinetics of release and clearance than traditional plasma-based markers of exercise. To address this hypothesis, we quantified candidate c-miRNA species originating from muscle, cardiac tissue, and vascular endothelium, as well as those important in inflammation in healthy male marathon participants at rest, immediately following marathon completion, and 24 h after marathon completion. These profiles were compared with corollary conventional tissue-specific biomarkers of skeletal muscle damage (CPK), cardiac muscle injury and stress (cTn and NT-proBNP), and systemic inflammation (hsCRP). Finally, to determine the impact of exercise duration and intensity on c-miRNA expression, we also compared the levels of selected c-miRNAs among marathon runners to those observed among healthy male athletes during shorter-duration, exhaustive exercise.
MATERIALS AND METHODS
Ethical approval and marathon study participants.
Participants (N = 21) were healthy male marathon runners free of known cardiovascular or metabolic disease who participated in the 42-km Boston Athletic Association Marathon on April 18, 2011. Participants abstained from aspirin, acetaminophen, and nonsteroidal anti-inflammatory medications at 24 h before the initial study time point and through the 24-h postrace phlebotomy. Participants provided written, informed consent at the time of study enrollment. All protocol procedures complied with the Declaration of Helsinki and were approved by the institutional review board at Hartford Hospital (Hartford, CT).
General design of study for marathon exercise.
Participants were enrolled on the day before the marathon following a “tapering” period consisting of 1–2 wk of reduced exercise training. Exercise training histories for the 3 mo before the event and medical histories were assessed. Body mass, height, resting blood pressure, and heart rate (Welch Allen 52000 Vital Signs Monitor, Skaneateles Falls, NY) were measured at the time of enrollment. Conventional biomarker and c-miRNA profiles were assessed using a prospective, longitudinal, and repeated-measures study design. Specifically, venous blood was obtained under quiet resting conditions on the day before the marathon (PRE), immediately after completion of the marathon (FINISH), and the day following the marathon within 24 h of finishing time (POST-24).
General design of study for acute exhaustive exercise.
Eleven elite collegiate athletes [affiliated with the Harvard University Department of Athletics and who participated and were described in a previously published exercise study (6)] were included as part of this study for comparative purposes. As previously described (and reiterated here for clarity) (6), subjects over 18 yr of age who were recruited members of the men's competitive rowing program at Harvard were considered eligible for this study. Acute exhaustive cardiopulmonary exercise testing was performed during the morning hours. Participants abstained from any physical exercise for >24 h before testing and were tested after an overnight dietary fast. On the morning of testing, participants were permitted and encouraged to drink water (12–24 oz.) but were prohibited from drinking beverages with caloric or electrolyte content. All participants were required to abstain from nonsteroidal anti-inflammatory use (including ibuprofen, naproxyn, and aspirin) for at least 7 days before testing.
The exercise test consisted of uninterrupted, incremental cycling exercise using an upright cycle ergometer (CPE 2001, Medical Graphics, St. Paul, MN) equipped with an electronically braked ergometer (Warren Collins, Braintree, MA). Cycle ergometry was chosen as the exercise modality to limit the impact of training-induced (rowing) improvements in exercise economy. A tightly fitting mouthpiece and nose clips were in place during exercise to facilitate measurements of gas exchange and ventilation. Pulmonary gas exchange and minute ventilation were measured breath by breath with a commercially available metabolic cart (Medical Graphics CPX/D, St. Paul, MN). The pneumotachograph was calibrated with a 3-liter syringe at five different flow rates, and the zirconia cell O2 analyzer and single-beam CO2 analyzer were calibrated with room air and 5% CO2/12% O2 gas. Following a 3-min period of rest to facilitate ventilatory equilibration and a 1-min period of unloaded exercise (0 W/min), workload was increased by 25 W/min until volitional exhaustion. Subjects were considered to have reached their peak oxygen consumption (V̇o2 max) when the following criteria were met: 1) leveling off of oxygen consumption with increasing workload; 2) respiratory exchange ratio values >1.1; and 3) a heart rate of at least 90% of age-predicted maximum. V̇o2 max was defined as the highest 15-s average during the final minute of exercise. The ventilatory threshold was determined by the modified V-slope method (9). Blood pressures were determined by auscultation before and then at each minute interval during exercise and recovery (5 min). Standard 12-lead electrocardiography was performed at rest and at each minute during exercise and recovery. Continuous heart rate was recorded from lead V1 and continuously displayed on an oscilloscope.
c-miRNA profiles were assessed using a prospective, longitudinal, and repeated-measures study design for sampling before and after exercise. Specifically, venous blood plasma sampling was performed under quiet resting conditions before rowing training (PRE), immediately after completion of rowing training (FINISH), and 1 h following the acute exercise (POST-1H). Participants were excluded from the study if a history of anabolic steroid use was elicited. Written, informed consent was obtained from all participants, and ethical approval conformed to the standards of the Declaration of Helsinki. The Harvard University institutional review board and the Partners Human Research Committee approved the protocol before study initiation.
Ten milliliters of venous blood were collected in standard anticoagulant (EDTA)-treated vacutainer tubes at the three time points using a 20-gauge intravenous catheter placed into a hand or arm vein. Cellular elements were pelleted in each blood sample following blood draw via centrifugation at 2,000 g for 10 min. The supernatant plasma was then aliquoted and immediately frozen at −80°C to minimize freeze-thaw degradation.
Selection of traditional biomarkers and miRNAs.
Five conventional plasma-based proteins were selected for study based on their known associations with skeletal muscle damage, cardiac muscle stress and necrosis, and systemic inflammation. Selected proteins included: CPK, cardiac troponin I (con-cTn; Siemens Dimension Vista cTnI, Siemens Healthcare Diagnostics, Newark, NJ), high-sensitivity cTn (hs-cTn; Siemens Vista cTnI, Siemens Healthcare Diagnostics), NT-proBNP (Siemens Healthcare Diagnostics), and hsCRP (Siemens Healthcare Diagnostics). The limit of detection of the con-cTn assay was 15 ng/l, with a 10% coefficient of variation of 40 μg/l, and the limit of detection of hs-cTn assay was 0.5 ng/l, with a 10% coefficient of variation of 3 ng/l (5). Corollary c-miRNAs were selected for study based on their tissue-specific (endothelial, muscle, and cardiac muscle alone) and context-specific (systemic inflammation) expression patterns with previously documented physiological relevance to sustained exercise. Specifically, miR-1 and miR-133a were chosen as critical tissue-specific mediators of myocyte function via their repression of histone deacetylase-4, monocyte enhancer factor-2, and serum response factor (12, 13). Previous human muscle biopsy work has shown that both of these miRNAs are upregulated in skeletal muscle tissue by aerobic exercise (40). miR-499–5p was chosen as a skeletal- and cardiac-enriched miRNA, important in the determination of slow/fast myofiber gene programs (51). Plasma levels of c-miR-499–5p have been studied in the context of acute myocardial infarction (1). Furthermore, miR-499–5p has recently been defined to control muscle fitness and endurance via interfacing with specialized nuclear receptor signaling pathways (e.g., peroxisome proliferator-activated receptor-β/δ and estrogen-related receptor-γ) (23). miR-208a was selected as a known cardiomyocyte-specific regulator of cardiac function via transcriptional upregulation by its host gene α-myosin heavy chain (37). Recent data from mice studies suggest a role for c-miR-208a as a biomarker for myocardial injury (30). miR-146a was selected given its essential role in inflammatory signaling in multiple cell types, via its transcriptional upregulation by NF-κB and downstream downregulation of IL-1 receptor-associated kinase-1 and TNF receptor-activating factor 6 as direct targets (60). Thus we reasoned that alterations in c-miR-146a may reasonably reflect the overall inflammatory state in prolonged-duration aerobic exercise. miR-126 was selected as a highly abundant and endothelial-specific miRNA with previously described alterations in aerobic exercise in mice (57, 67) and importantly in humans after prolonged exercise (62). Finally, the brain-enriched miR-134 was chosen as an unrelated control to facilitate comparison with the above candidate c-miRNA species.
RNA extraction.
Although plasma-based c-miRNAs resist degradation even under a variety of harsh environmental conditions (38), the number of freeze-thawing cycles of plasma samples was kept to a minimum to further decrease the theoretical risk of c-miRNA degradation. Additionally, as described (6), all samples from a given individual were processed and analyzed in a single batch to reduce variability. The thawed plasma samples were centrifuged again (15,700 g, 10 min) to remove any remaining cellular contents. The plasma supernatant was then aliquoted into 150-μl volumes, which were stored at −80°C and kept for further analysis. Based on previous observations that endogenous miR-422b displays minimal expression in circulating plasma during various forms of volitional exercise (S. Y. Chan, unpublished observation, 2010), quantitative normalization of c-miRNA plasma levels was achieved by adding equivalent levels of exogenous miR-422b, as previously described (38). More specifically, 4 fmol of the chemically synthesized miRNA duplex mimic of miR-422b were added to 150 μl of each plasma sample (Life Technologies, Carlsbad, CA). Total RNA extraction was performed using a MicroRNA Extraction Kit (Benevbio, Mission Viejo, CA). Reverse transcription was performed to generate complementary DNA (cDNA), representing levels of mature c-miRNA molecules (MicroRNA Assay Kit, Life Technologies). The reliability of the c-miRNA extraction data was supported by additional extraction of the known quantities of the exogenous miR-422b, as previously performed (6).
Real-time PCR quantification of c-miRNA plasma levels.
Reverse transcription-quantitative “real-time” polymerase chain reaction was utilized to quantify levels of the candidate c-miRNAs. This method was chosen to minimize natural variability that can mistakenly be associated with false positive findings with the use of current high-throughput miRNA screening techniques. Because c-miRNAs typically exhibit lower concentration than intracellular miRNAs (41), taking into account such variability is particularly important, thus incentivizing the use of reverse transcription-quantitative real-time polymerase chain reaction.
Quantification of c-miRNA plasma levels was achieved via generation of cDNA from reverse transcription, thus representing levels of mature c-miRNA molecules (MicroRNA Assay Kit, Life Technologies). An Applied Biosystems 7900HT Fast Real Time PCR device was used to amplify cDNA using fluorescently labeled Taqman probe and primer sets. Taqman primer/probes for quantification were used as follows (Life Technologies): hsa-miR-1 [AB 4427975 (002222)], hsa-miR-133a [AB 4427975 (002246)], hsa-miR-499–5p [AB 4427975 (001352)], hsa-miR-126 [AB 4427975 (002228)], hsa-miR-134 [AB 4427975 (001186)], hsa-miR-146a [AB 4427975 (000468)], hsa-miR-208a [AB 4427975 (000511)], and hsa-miR-422b [AB 4427975 (000575)]. The raw Ct values produced, which represent the “real-time” cycle count at which miRNA probe fluorescence increases exponentially, were used to generate standard curves for each individual miRNA Taqman primer/probe set following dilutions of a synthetic oligonucleotide mimic for each mature miRNA (Life Technologies). On average, the level of detection for a miRNA by this method was approximately one to two copies in a given reaction, consistent with prior reports (55). This level of sensitivity allowed for detection of all candidate c-miRNAs before the marathon run, even in cases where some c-miRNAs were poorly expressed. To control for potential inconsistencies in c-miRNA extraction from plasma, as previously reported (55), copy numbers were then normalized using a ratio calculated from levels of a miR-422b reference control exogenously spiked into each given sample before extraction compared with an equivalent amount of miR-422b spiked in water (“gold standard”) before extraction. It is noteworthy that this normalization method is reliable because endogenous miR-422b exhibits relatively little expression compared with the exogenously added miR-422b control (6). Normalized miRNA copy number per microliter of plasma was then calculated based on amount of plasma sampled in the analysis. These normalized copy numbers were then used to calculate fold change by normalizing mean copy number measured at PRE to 1, to which each individual c-miRNA measurement was compared.
Statistical analysis.
Normality of data was assessed using the Shapiro-Wilk test. Raw data of c-miRNA profiles and traditional biomarkers were found to have a nonnormal distribution, while demographics and clinical characteristics were normally distributed. As a result, demographics and clinical characteristics are presented as means ± SE, and c-miRNA/traditional biomarker data are presented as median [interquartile range (IQR)]. All data are graphically presented as box and whisker plots in which lines within the boxes denote medians, boxes denote 25% and 75% IQRs, and whiskers represent maximum and minimum values. Medians and IQR (25% and 75% IQR) are also listed in Figs. 1–5. Multiple comparisons of c-miRNA values were made between the three points of venous blood collection via repeated-measures ANOVA, followed by a Student-Newman-Keuls post hoc test. Each plot is labeled with P values resulting from the post hoc test as well as the median (IQR). Values of P < 0.05 were considered significant. Correlations between changes in c-miRNAs and conventional biomarkers were assessed using the Pearson and Spearman techniques as appropriate for data distribution. Data analysis was performed using the Statisical Package for the Social Sciences (SPSS) software, version 16 (SPSS, Chicago, IL).
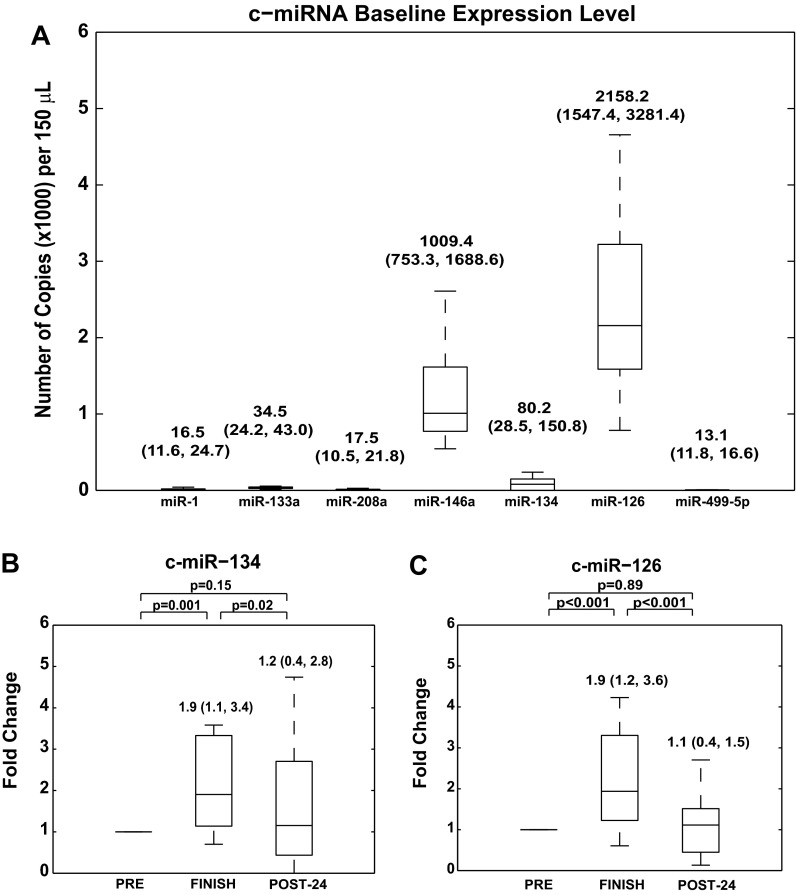
Fig. 1.
Baseline characteristics of candidate circulating microRNAs (c-miRNAs). A: under resting conditions before marathon run, c-miR-1, c-miR-133a, c-miR-208a, and c-miR-134 were nearly undetectable in plasma, whereas c-miR-146a and c-miR-126 were detectable at higher relative levels. Baseline steady-state c-miRNA levels displayed a relatively modest degree of variability among the athletes. Data are presented as box and whisker plots signifying copy numbers of c-miRNA expression levels (see materials and methods), where horizontal lines denote median, boxes denote 25% and 75% percentile confidence intervals, and error bars reflect maximum and minimum values. All plots are labeled with statistical medians (1st quartile, 3rd quartile). B: modest alterations in brain-enriched c-miR-134 in the context of prolonged aerobic exercise. c-miR-134 was poorly expressed at rest, but was modestly upregulated after the end of the run with a subsequent decrease at 24 h postmarathon. C: similar to the pattern of regulation for c-miR-134, modest alterations in the endothelial-specific c-miR-126 were observed after the end of the run, followed by a decrease at 24 h postmarathon. For each marathon athlete, baseline biomarker levels under resting condition are assigned a fold change of 1, to which measurements obtained during subsequent study time points (i.e., shortly after and 24 h after prolonged aerobic exercise) are compared. Data are represented as box and whisker plots signifying fold changes in c-miRNA levels and are labeled with statistical medians, interquartile range (IQR), and P values of comparisons between conditions. PRE, the day before the marathon; FINISH, immediately after completion of the marathon; POST-24, the day following the marathon within 24 h of finishing time.
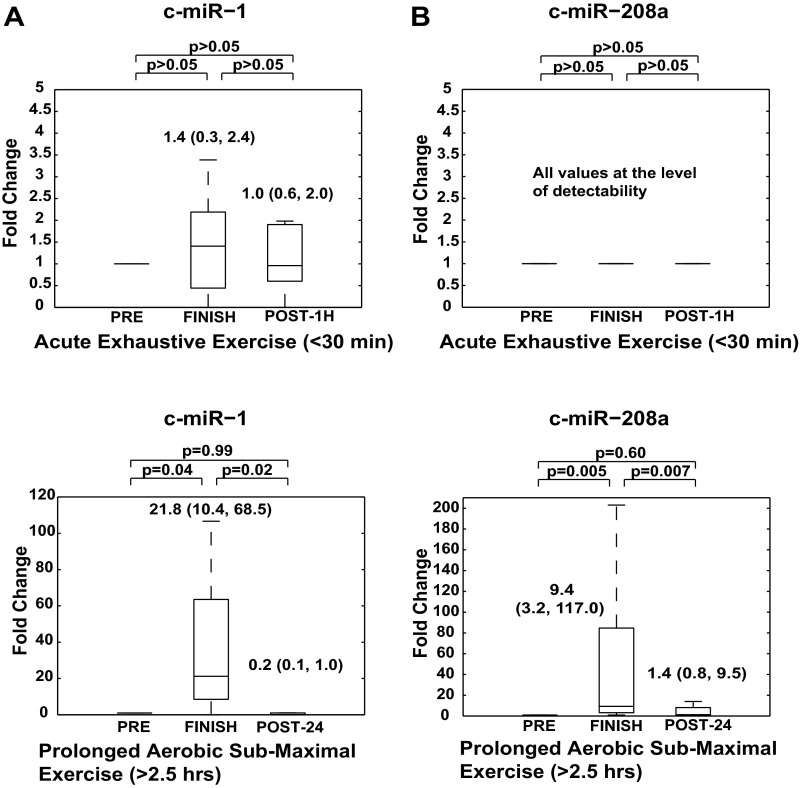
Fig. 5.
Muscle-specific and cardiac-specific miRNAs are elevated in circulating plasma after prolonged aerobic exercise, but not after acute exhaustive exercise. A, top: the plasma of 11 athletes (N = 11) was analyzed in conjunction with acute exhaustive exercise. At baseline resting conditions before acute exhaustive exercise, c-miR-1 was poorly expressed. At the end of exercise, c-miR-1 did not show any significant alterations in expression. Bottom: in 21 marathon athletes (N = 21) analyzed for this study, c-miR-1 levels were significantly increased at FINISH and returned to baseline by POST-24. B, top: c-miR-208a expression was at the level of detectability at baseline resting conditions and following acute exhaustive exercise (N = 11). Bottom: in marathon runners, c-miR-208a levels were significantly upregulated at FINISH and returned to baseline by POST-24 (N = 21). Data are presented as box and whisker plots signifying fold changes in c-miRNA levels, where horizontal lines denote median, boxes denote 25% and 75% percentile confidence intervals, and error bars reflect maximum and minimum values. Baseline biomarker levels under resting condition are assigned a fold change of 1, to which measurements obtained during subsequent study time points (i.e., FINISH and POST-24) are compared. All panels are labeled with statistical medians, IQR, and P values of comparisons between conditions.
RESULTS
Subject characteristics.
All 21 participants were Caucasian men with a mean age of 51.8 ± 1.4 yr, height of 176.5 ± 1.7 cm, and body mass of 74.2 ± 2.4 g/m2. Participants completed the marathon in an average of 233 ± 8 min. There were no significant changes in hemoglobin levels across the three study time points (PRE = 14.8 g/dl vs. FINISH = 14.9 g/dl vs. POST-24 = 13.8 g/dl, P = nonsignificant), suggesting minimal changes in intravascular blood volume throughout the study period.
Consistent plasma levels of candidate miRNAs under resting conditions.
The majority of candidate c-miRNAs (miR-1, miR-133a, miR-208a, miR-134, and miR-499–5p) displayed extremely low expression levels in plasma at PRE, as anticipated based on previous studies in healthy individuals (Fig. 1A) (38). In contrast, miR-146a and miR-126 were expressed at relatively higher levels at PRE, consistent with prior data of miR-146a from resting trained athletes (6). None of the candidate c-miRNAs displayed excessively wide ranges of variability at PRE, suggesting minimal technical inconsistencies in plasma processing or c-miRNA quantification. Hence, differences in c-miRNA expression between PRE and subsequent postmarathon time points likely reflect a specific physiological process.
Alterations of brain-enriched c-miR-134 and endothelial-specific c-miR-126 in response to prolonged exercise.
To delineate c-miRNA-dependent processes specific to muscle and/or inflammation during prolonged exercise, the brain-enriched miR-134 was chosen as an unrelated control. Similar to most of the other candidate c-miRNAs tested in this study, c-miR-134 was minimally expressed at PRE (Fig. 1A). In response to the marathon run, plasma levels were increased at FINISH [1.9 (1.1, 3.4), median (IQR) fold change vs. PRE, P = 0.001], but decreased toward baseline values at POST-24 [1.2 (0.4, 2.8) fold change vs. PRE, P = 0.15; Fig. 1B]. Importantly, such alterations were modest compared with alterations of specific muscle, cardiac, and inflammatory miRNAs described below (Figs. 2–4). Thus, these smaller alterations of a brain-enriched c-miR-134 provide a reference baseline upon which changes of other candidate c-miRNAs can be interpreted.
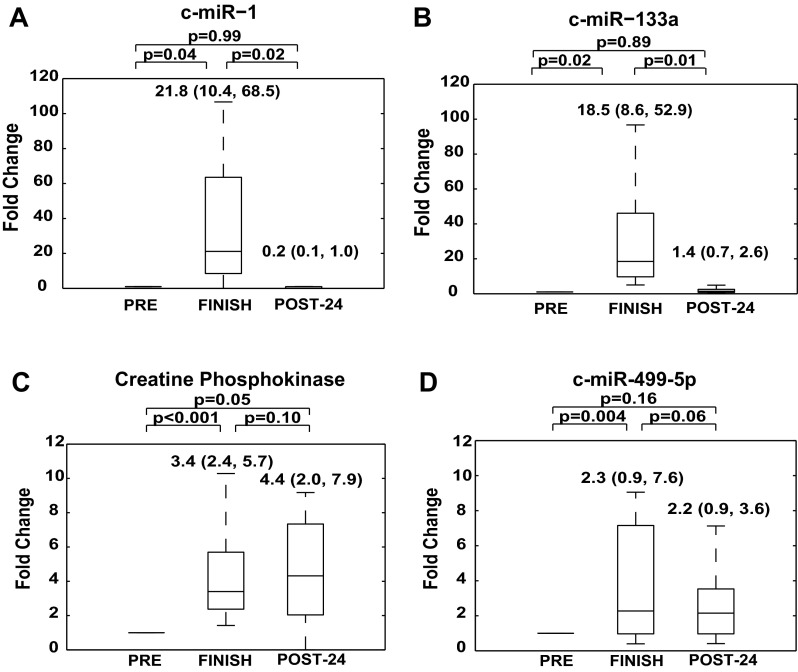
Fig. 2.
Distinct muscle-specific c-miRNAs are elevated after prolonged aerobic exercise and are downregulated 24 h postexercise. c-miR-1 (A) and c-miR-133a (B) were increased after prolonged aerobic exercise and returned very close to baseline by POST-24. C: in contrast, plasma CPK levels increased after exercise and remained elevated POST-24. D: c-miR-499–5p was increased after prolonged aerobic exercise and displayed a mixture of elevated and nonelevated measurements by POST-24. In contrast with c-miR-1 or c-miR-133a, the magnitude of increase of c-miR-499–5p was modest and was more similar to the profiles of c-miR-134 (Fig. 1B) or c-miR-126 (Fig. 1C). For each marathon athlete, baseline biomarker levels under resting condition are assigned a fold change of 1, to which measurements obtained during subsequent study time points (i.e., shortly after and 24 h after prolonged aerobic exercise) are compared. In all panels, data are represented as box and whisker plots signifying fold changes in c-miRNA levels, where horizontal lines denote medians, boxes denote 25% and 75% percentile confidence intervals, and error bars reflect maximum and minimum values. All panels are labeled with statistical medians, IQR, and P values of comparisons between conditions.
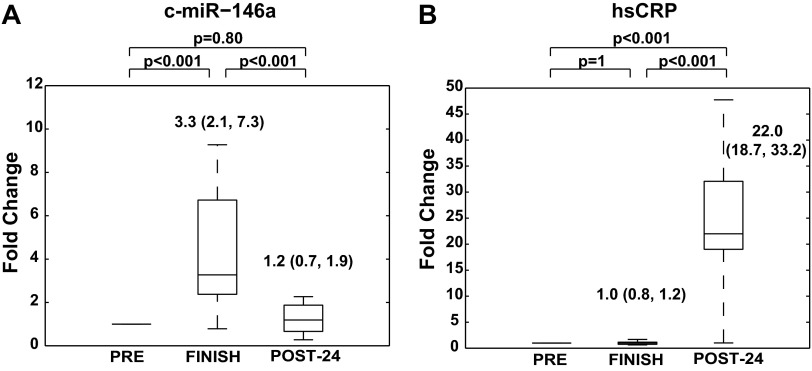
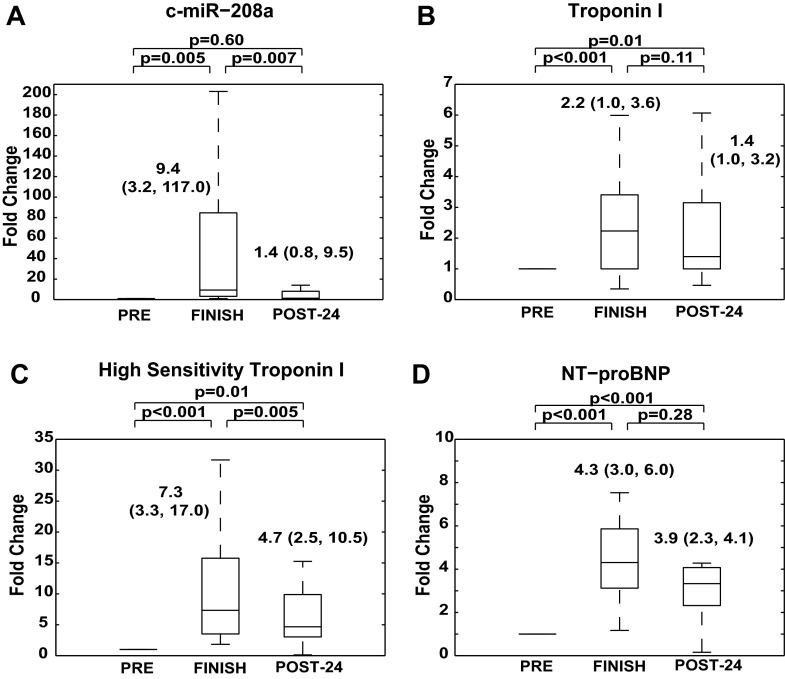
Fig. 4.
Inflammation-specific c-miR-146a is elevated after prolonged aerobic exercise and downregulated 24 h postexercise. A: c-miR-146a increased significantly following prolonged aerobic exercise and nearly returned to baseline POST-24. B: high-sensitivity C-reactive protein (hsCRP), however, was not altered at the end of run, but substantially increased POST-24. A and B: for each marathon athlete, biomarker levels are presented in a similar manner to that of Fig. 1B. All panels are labeled with statistical medians, IQR, and P values of comparisons between conditions.
In the setting of prior reports identifying functions of miR-126, a well-established, highly abundant, and endothelial-specific miRNA, in aerobic exercise (57, 62, 67), we measured expression of c-miR-126. Consistent with previously published results by Uhlemann and colleagues (62), plasma levels were increased at FINISH [1.9 (1.2, 3.6) fold change, vs. PRE, P < 0.001, Fig. 1C]. Levels decreased toward baseline values at POST-24 [1.1 (0.4, 1.5) fold change vs. PRE, P = 0.89; Fig. 1C]. Importantly, these alterations in our cohort were modest and closely mirrored the degree of upregulation seen for the unrelated control miR-134 (Fig. 1B).
CPK and muscle-specific c-miRNAs.
Compared with PRE, CPK was significantly elevated at FINISH [3.4 (2.4, 5.7) fold change vs. PRE, P < 0.001] (Fig. 2C). Similarly, both c-miR-1 [21.8 (10.4, 68.5) fold change vs. PRE, P = 0.04] and c-miR-133a [18.5 (8.6, 52.9) fold change vs. PRE, P = 0.02] were increased significantly immediately after completion of the marathon run (Fig. 2, A and B, FINISH). PRE to FINISH changes in c-miR-1 were significantly correlated with corresponding changes in c-miR-133a (R = 0.57, P = 0.007). However, changes in neither c-miR-1 nor c-miR-133a correlated significantly with changes in CPK. Although relatively high variability in CPK levels was observed at POST-24, expression profiles of both c-miR-1 and miR-133a differed significantly from that of CPK at POST-24. Notably, CPK levels in general remained elevated at POST-24 [4.4 (2.0, 7.9) fold change vs. PRE, P = 0.05; Fig. 2C], with a statistically nonsignificant trend toward further upregulation at POST-24 compared with FINISH [4.4 (2.0, 7.9) fold change at POST-24 vs. 3.4 (2.4, 5.7) fold change at FINISH, P = 0.10; Fig. 2C]. In contrast, levels of c-miR-1 [0.2 (0.1, 1.0) fold change vs. PRE, P = 0.99] and c-miR-133a [1.4 (0.7, 2.6) fold change vs. PRE, P = 0.89] returned to baseline values at POST-24 (Fig. 2, A and B). Thus, unlike CPK, which began to rise immediately following exercise and remained elevated at POST-24, c-miR-1 and c-miR-133a demonstrated a rapid increase at FINISH and subsequent near-complete clearance by POST-24.
Interestingly, c-miR-499–5p [2.3 (0.9, 7.6) fold change vs. PRE, P = 0.004] was also increased significantly at FINISH, followed by a mixture of elevated and nonelevated measurements at POST-24 [2.2 (0.9, 3.6) fold change vs. PRE, P = 0.16; vs. FINISH, P = 0.06] (Fig. 2D). Considering that miR-1, miR-133a, and miR-499–5p are all enriched with nearly comparable expression in skeletal and cardiac muscle (56), such distinct profiles of c-miRNA expression during exercise support the existence of separate processes of c-miRNA release from muscle tissue that cannot be explained solely by tissue injury during prolonged aerobic exercise.
Cardiac troponin, NT-proBNP, and cardiac-specific c-miR-208a.
Conventional protein markers of cardiac stress/injury were increased at FINISH [con-cTn = 2.2 (1.0, 3.6) fold change vs. PRE, P < 0.001; hs-cTn = 7.3 (3.3, 17.0) fold change vs. PRE, P < 0.001; NT-proBNP = 4.3 (3.0, 6.0) fold change vs PRE, P < 0.001; Fig. 3, B–D] and remained elevated at 24-POST [con-cTn = 1.4 (1.0, 3.2) fold change vs. PRE, P = 0.01; hs-cTn = 4.7 (2.5, 10.5) fold change vs. PRE, P = 0.01; NT-proBNP = 3.9 (2.3, 4.1) fold change vs PRE, P < 0.001; Fig. 3, B–D]. Cardiac-specific c-miR-208a also displayed marked increase at FINISH [9.4 (3.2, 117.0) fold change vs. PRE, P = 0.005; Fig. 3A], but was substantially more downregulated at POST-24 [1.4 (0.8, 9.5) vs. PRE, P = 0.60] compared with the conventional cardiac biomarkers. Changes in c-miR-208a did not correlate with changes in either hs-cTn or NT-proBNP. Thus, as in the cases of c-miR-1 and c-miR-133a, the rapid regulation of c-miR-208a suggests its unique potential as a time-sensitive marker of cardiac muscle response to prolonged exercise.
Fig. 3.
Cardiac muscle-enriched c-miR-208a is elevated after prolonged aerobic exercise and is downregulated 24 h postexercise. A: c-miR-208a increased significantly after prolonged exercise and decreased nearly 50-fold by POST-24 to levels not significantly different from baseline. B: troponin I levels increased after prolonged aerobic exercise. However, despite falling significantly in the time period at FINISH and POST-24, troponin I levels remained significantly elevated POST-24 compared with prerun. C: ultrasensitive troponin I levels were significantly upregulated and increased after exercise but only subtly decreased POST-24. D: NH2-terminal prohormone of brain natriuretic peptide (NT-proBNP) levels were also significantly increased postexercise, but exhibited only a slight, nonstatistically significant decrease in the 24 h following completion of exercise. Thus levels at 24 h postexercise were still significantly higher than preexercise. In A–D, for each marathon athlete, biomarker levels are presented in a similar manner to that of Fig. 1B. All panels are labeled with statistical medians, IQR, and P values of comparisons between conditions.
hsCRP and inflammation-specific c-miR-146a.
The well-established inflammation biomarker hsCRP displayed no significant change in expression when measured at FINISH [1.0 (0.8, 1.2) vs. PRE, P = 1.0], but was significantly increased at POST-24 [22.0 (18.7, 33.2) vs. PRE, P < 0.001; Fig. 4B]. In contrast, c-miR-146a exhibited immediate increase at FINISH [3.3 (2.1, 7.3) fold change vs. PRE, P < 0.001], followed by complete normalization at POST-24 [1.2 (0.7, 1.9) vs. PRE, P = 0.8] (Fig. 4A). Furthermore, the inflammatory c-miR-146a increased to a greater magnitude than either c-miR-134 or c-miR-126 and cleared relatively quickly in response to marathon running. Thus, similar to miR-1, miR-133a, and miR-208a, the profile of c-miR-146a expression appears to rely, at least in part, on a different regulatory program than either c-miR-134 or c-miR-126 alone.
DISCUSSION
This study was designed to determine how specific c-miRNAs are modulated by prolonged, submaximal aerobic exercise. To do so, we compared the profiles of c-miRNAs originating from various tissue sources or inflammation-specific contexts: muscle (miR-1, miR-133a, miR-499–5p), cardiac tissue (miR-208a), and the vascular endothelium (miR-126), as well as those prominent in the inflammatory response (miR-146a), before and after a 42-km marathon run. To determine further the uniqueness of c-miRNA regulation in this context, we next compared c-miRNA profiles to those of conventional biomarkers of skeletal muscle damage (CPK), cardiac muscle injury (con-cTn and hs-cTn), cardiac tissue stress (NT-proBNP), and generalized systemic inflammation (hsCRP). Our key findings are summarized as follows. First, although all candidate c-miRNAs were increased at FINISH, only specific ones originating from skeletal muscle (miR-1, miR-133a), cardiac muscle (miR-208a), and inflammatory processes (miR-146a) were robustly upregulated. Others, including the brain-enriched miR-134, the endothelial-specific miR-126, and the muscle-specific miR-499–5p were substantially less affected by the marathon run, thereby supporting the notion of distinct mechanisms of c-miRNA response to exercise rather than a general class effect originating simply from nonspecific tissue damage. Second, the magnitude of tissue-specific increases of c-miRNAs was substantially greater than that observed with the conventional biomarkers. This finding may suggest higher a priori intracellular c-miRNA concentrations or perhaps specific stress-induced upregulation of c-miRNA production and release compared with conventional biomarkers. Finally, as anticipated from prior studies (2, 10, 15, 16, 24, 27, 29, 39, 43, 48, 49, 61, 66), marathon completion led to significant skeletal muscle damage, cardiac muscle stress/injury, and systemic inflammation, as reflected by statistically significant increases in circulating CPK, NT-proBNP, con-cTn, hs-cTn, and hsCRP, respectively. In contrast, we found that the temporal sequence of response of certain key c-miRNAs was characterized by immediate elevation followed by rapid normalization and differed from the response of conventional biomarkers, which demonstrated relatively delayed peak expression (hsCRP and CPK) or persistent upregulation 24 h after exercise. These markedly different temporal patterns of c-miRNA compared with their tissue-associated conventional biomarkers emphasize a potential role for c-miRNAs as unique, real-time markers of exercise physiology.
Organ-specific adaptations to exercise, such as skeletal muscle hypertrophy, mitochondrial biogenesis, cardiomyocyte hypertrophy, and vascular angiogenesis, underlie human performance and are linked with the salubrious benefits associated with routine physical exercise. Adaptation requires exposure to a stimulus of sufficient intensity and duration to produce cellular activation, often to the level of stress and/or injury, followed by compensatory repair and recovery. While conventional biomarkers may be useful markers of tissue stress and or injury, they provide limited information about the cellular mechanisms of injury and recovery. Biomarkers like CPK and cTn, although important molecules for cellular energy storage and contractile function, respectively, have limited roles as direct mediators of cellular response to external stimuli. In contrast, miRNAs are increasingly recognized as inducible and direct regulators of numerous cellular functions and, therefore, play key roles in the cellular response to environmental stimuli. Accordingly, our results demonstrate that numerous tissue-specific miRNAs are released during sustained aerobic exercise and may reflect the acute responses to such physiological stress.
The potential roles of c-miRNAs as biomarkers of exercise provide a foundation for important future work. Follow-up studies correlating c-miRNA expression with the rate and magnitude of tissue (cardiac or muscle) breakdown, an individual's aerobic fitness, or prior exercise training will be informative. It is plausible that c-miRNAs may be useful to direct individual training and performance regimens, consistent with prior studies that suggest certain c-miRNAs reflect aerobic fitness (6, 11). Furthermore, the rapid normalization of c-miRNAs following exercise indicates that these molecules may be useful indexes of short-term tissue recovery. Accordingly, characterization of c-miRNA levels following repetitive bouts of exercise is needed. In addition, if c-miRNAs released from tissue indeed have as-of-yet undiscovered regulatory functions, future studies may be designed to explore the correlation of c-miRNA levels following a single bout of exercise in the presence or absence of repeated exercise training with the degree of physiological improvement (e.g., increase in V̇o2 max or muscle strength). Such endeavors could provide insight into the role of c-miRNAs as not only useful biomarkers, but also regulators, of the body's response to exercise. Finally, it is notable that the cardiac-specific c-miR-208a, upregulated during prolonged exercise, has also been proposed as a marker of underlying heart disease (30). Moreover, recent data suggest that alterations of miRNA expression with aerobic exercise may influence the development of atherosclerosis (67). It will be important to determine whether unique profiles of c-miR-208a and perhaps other c-miRNAs correlate with varying degrees of cardiovascular risk with exercise, thus aiding in the clinical identification and prevention of adverse exercise-induced events in putative at-risk populations.
Beyond their potential utility as physiological/pathological markers of exercise, our findings also set the stage for deeper inquiry into the dynamic biology of c-miRNAs in response to aerobic exertion. In contrast to their robust increase after marathon running (Figs. 2 and 3), muscle-specific miR-1 and cardiac-specific miR-208a revealed negligible alterations after acute exhaustive exercise [<30 min of graded, effort-limited upright cycling exercise designed to measure V̇o2 max] in 11 elite collegiate athletes (Fig. 5). Thus either the duration of exercise or the type of aerobic exercise (running vs. upright cycle ergometry) appears to be a key determinant of cardiac- and skeletal muscle-specific c-miRNA regulation. However, a difference in age is also notable in the present study comparing the more senior marathon runners and the younger crew athletes, which may have contributed to these distinct expression profiles. Given that microscopic cellular damage is common during sustained aerobic exercise (14), we suspect that some nonspecific release of miRNA contributed to the observed increases of c-miRNAs after prolonged, but not acute, exercise. It is, however, notable that, in marathon runners, the magnitude of fold change of specific c-miRNAs, such as miR-1, miR-133a, miR-208a, and miR-146a, greatly exceeded the magnitudes observed with c-miR-134 and c-miR-126, as well as other passively released biomarkers, such as CPK (Fig. 1, B and C). The differences of such changes were especially apparent when considering the distinct profiles of various c-miRNAs all expressed from the same skeletal and cardiac muscle compartment: a robust upregulation of miR-1 and miR-133a vs. a much more modest alteration in miR-499–5p (Fig. 2). Thus, as emphasized by differences in c-miRNA profiles stratified by specific miRNA identity, by tissue compartment of origin, and by temporal sequence of upregulation, we suspect that specialized c-miRNA secretory mechanisms are uniquely activated during exercise and not merely the result of generalized tissue breakdown. Our data also indicate that candidate c-miRNAs undergo significant downregulation 24 h following prolonged aerobic exercise. Although the mechanism of such clearance is unclear, increased handling by kidney or liver is possible for eventual excretion through urine or feces. Alternatively, rapid enzymatic degradation of exercise-induced c-miRNA could occur. Finally, as supported by recent findings in other contexts, such as the study of secretory mechanisms, intracellular transport, and genetic exchange (32, 34, 59, 63), transport of c-miRNA to distant recipient tissue to influence exercise adaptation is an intriguing possibility that should be interrogated in the future.
There are an increasing number of studies examining the potential role of c-miRNAs as mediators of physiological adaptation. It is noteworthy that many prior reports have been complicated biological and methodological limitations, including the following: 1) uncontrolled interindividual baseline miRNA variability; 2) confounding effects of hemolysis or cellular contamination on c-miRNA levels (41); and 3) an inability to identify the tissue sources of origin for more ubiquitously expressed c-miRNAs. The present study was designed to circumvent these issues. First, we utilized a repeated-measures study design to permit each participant's resting sample to serve as his own control, thereby facilitating accurate determination of c-miRNA secretion patterns, despite any interindividual baseline variability. Second, most of the candidate c-miRNAs selected for this study are tissue specific and not expressed in blood, thereby minimizing the potential impact of hemolysis. Third, we selected candidate c-miRNAs directly relevant to the key biological responses involved in exercise-induced tissue injury and adaptation. As a result of these technical advantages, many of the common confounding factors that plague the study of plasma-based c-miRNAs were avoided, thus providing greater confidence in the accuracy of the results.
There are, however, several study limitations. First, this study included a relatively small number of participants (N = 21). Thus, to facilitate analysis of c-miRNA profiles, we used a longitudinal repeated-measures study design. Second, to maximize the accuracy of this initial “proof-of-concept” study, quantitative c-miRNA analysis was restricted to a subset of carefully selected c-miRNAs with anticipated biological relevance rather than the use of a more comprehensive, but perhaps less reliable, high-throughput c-miRNA screening technique. Recently, a cohort of additional miRNAs has been identified that is dynamically regulated in cardiac tissue during adaptation to exercise training, including miR-27a/b, miR-143 (20), and the miR-29 family (58). Certainly, to define more completely the spectrum of exercise-induced c-miRNAs, a more complete characterization of these molecules and others in a larger number of human subjects is warranted. Finally, the study cohort was restricted to recreational marathon participants. Future study is required to determine whether these results are applicable to elite athletes or exercise-naive subjects.
In conclusion, we report unique alterations in c-miRNAs after prolonged aerobic exercise. Such alterations differ depending on c-miRNA identity, tissue of origin, or physiological context (i.e., inflammation). These c-miRNA profiles also substantially differ compared with the expression patterns seen with traditional protein markers. Thus these findings provide an essential framework for future studies aimed at interrogating both the potential use of c-miRNAs as real-time markers of exercise and the study of the direct biological function of these molecules.
GRANTS
This work was supported by the American Heart Association (A. L. Baggish), the National Heart, Lung, and Blood Institute (HL-096834), the McArthur-Radovsky, Lerner, Harris, and Watkins Funds, and the Pulmonary Hypertension Association (S. Y. Chan). The Gilead Research Scholars Fund also supported this work.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: A.L.B., B.A.T., P.D.T., C.T., and S.Y.C. conception and design of research; A.L.B., J.P., P.-K.M., S.I., M.T., A.H., and S.Y.C. analyzed data; A.L.B., J.P., P.-K.M., S.I., M.T., A.H., and S.Y.C. interpreted results of experiments; A.L.B. and S.Y.C. prepared figures; A.L.B., P.-K.M., and S.Y.C. drafted manuscript; A.L.B., P.-K.M., B.A.T., P.D.T., C.T., and S.Y.C. edited and revised manuscript; A.L.B., J.P., P.-K.M., S.I., B.A.T., P.D.T., C.T., P.D., S.D., M.T., A.H., and S.Y.C. approved final version of manuscript; J.P., P.K.M., and S.Y.C. performed experiments.
ACKNOWLEDGMENTS
We thank S. Tribuna for expert administrative assistance.
REFERENCES
- 1.Adachi T, Nakanishi M, Otsuka Y, Nishimura K, Hirokawa G, Goto Y, Nonogi H, Iwai N. Plasma microRNA 499 as a biomarker of acute myocardial infarction. Clin Chem 56: 1183–1185, 2010 [DOI] [PubMed] [Google Scholar]
- 2.Alardín AL, Varon J, Marik PE. Bench-to-bedside review: rhabdomyolysis–an overview for clinicians. Crit Care 9: 158–169, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoi W, Ichikawa H, Mune K, Tanimura Y, Mizushima K, Naito Y, Yoshikawa T. Muscle-enriched microRNA miR-486 decreases in circulation in response to exercise in young men. Front Physiol 4: 80, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoi W, Naito Y, Mizushima K, Takanami Y, Kawai Y, Ichikawa H, Yoshikawa T. The microRNA miR-696 regulates PGC-1α in mouse skeletal muscle in response to physical activity. Am J Physiol Endocrinol Metab 298: E799–E806, 2010 [DOI] [PubMed] [Google Scholar]
- 5.Apple FS, Collinson PO; IFCC Task Force on Clinical Applications of Cardiac Biomarkers Analytical characteristics of high-sensitivity cardiac troponin assays. Clin Chem 58: 54–61, 2012 [DOI] [PubMed] [Google Scholar]
- 6.Baggish AL, Hale A, Weiner RB, Lewis GD, Systrom D, Wang F, Wang TJ, Chan SY. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J Physiol 589: 3983–3994, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baggish AL, Wang F, Weiner RB, Elinoff JM, Tournoux F, Boland A, Picard MH, Hutter AM, Jr, Wood MJ. Training-specific changes in cardiac structure and function: a prospective and longitudinal assessment of competitive athletes. J Appl Physiol 104: 1121–1128, 2008 [DOI] [PubMed] [Google Scholar]
- 8.Banzet S, Chennaoui M, Girard O, Racinais S, Drogou C, Chalabi H, Koulmann N. Changes in circulating microRNAs levels with exercise modality. J Appl Physiol 115: 1237–1244, 2013 [DOI] [PubMed] [Google Scholar]
- 9.Beaver WL, Wasserman K, Whipp BJ. A new method for detecting anaerobic threshold by gas exchange. J Appl Physiol 60: 2020–2027, 1986 [DOI] [PubMed] [Google Scholar]
- 10.Bhalla V, Willis S, Maisel AS. B-type natriuretic peptide: the level and the drug–partners in the diagnosis of congestive heart failure. Congest Heart Fail 10: 3–27, 2004 [DOI] [PubMed] [Google Scholar]
- 11.Bye A, Røsjø H, Aspenes ST, Condorelli G, Omland T, Wisløff U. Circulating microRNAs and aerobic fitness–the HUNT-Study. PLos One 8: e57496, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Hoydal M, Autore C, Russo MA, Dorn GW, 2nd, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nat Med 13: 613–618, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38: 228–233, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarkson PM. Eccentric exercise and muscle damage. Int J Sports Med 18: S314–S317, 1997 [DOI] [PubMed] [Google Scholar]
- 15.Clyne B, Olshaker JS. The C-reactive protein. J Emerg Med 17: 1019–1025, 1999 [DOI] [PubMed] [Google Scholar]
- 16.Dahlqvist JR, Voss LG, Lauridsen T, Krag TO, Vissing J. A pilot study of muscle plasma protein changes following exercise. Muscle Nerve 49: 261–266, 2014 [DOI] [PubMed] [Google Scholar]
- 17.Davidsen PK, Gallagher IJ, Hartman JW, Tarnopolsky MA, Dela F, Helge JW, Timmons JA, Phillips SM. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. J Appl Physiol 110: 309–317, 2011 [DOI] [PubMed] [Google Scholar]
- 18.Davidson-Moncada J, Papavasiliou FN, Tam W. MicroRNAs of the immune system: roles in inflammation and cancer. Ann N Y Acad Sci 1183: 183–194, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devaux Y, Vausort M, Goretti E, Nazarov PV, Azuaje F, Gilson G, Corsten MF, Schroen B, Lair ML, Heymans S, Wagner DR. Use of circulating microRNAs to diagnose acute myocardial infarction. Clin Chem 58: 559–567, 2012 [DOI] [PubMed] [Google Scholar]
- 20.Fernandes T, Hashimoto NY, Magalhães FC, Fernandes FB, Casarini DE, Carmona AK, Krieger JE, Phillips MI, Oliveira EM. Aerobic exercise training-induced left ventricular hypertrophy involves regulatory microRNAs, decreased angiotensin-converting enzyme-angiotensin II, and synergistic regulation of angiotensin-converting enzyme 2-angiotensin (1–7). Hypertension 58: 182–189, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandes T, Magalhães FC, Roque FR, Phillips MI, Oliveira EM. Exercise training prevents the microvascular rarefaction in hypertension balancing angiogenic and apoptotic factors: role of microRNAs-16, -21, and -126. Hypertension 59: 513–520, 2012 [DOI] [PubMed] [Google Scholar]
- 22.Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Roxe T, Muller-Ardogan M, Bonauer A, Zeiher AM, Dimmeler S. Circulating microRNAs in patients with coronary artery disease. Circ Res 107: 677–684, 2010 [DOI] [PubMed] [Google Scholar]
- 23.Gan Z, Rumsey J, Hazen BC, Lai L, Leone TC, Vega RB, Xie H, Conley KE, Auwerx J, Smith SR, Olson EN, Kralli A, Kelly DP. Nuclear receptor/microRNA circuitry links muscle fiber type to energy metabolism. J Clin Invest 123: 2564–2575, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Greef J, Funk M, Vermaak WJ, Perumal NS, Libhaber CD, Vangu MD. NT-proBNP and the diagnosis of exercise-induced myocardial ischaemia. Cardiovasc J Afr 19: 264–267, 2008 [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta SK, Bang C, Thum T. Circulating microRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ Cardiovasc Genet 3: 484–488, 2010 [DOI] [PubMed] [Google Scholar]
- 26.Gute D, Fraga C, Laughlin MH, Amann JF. Regional changes in capillary supply in skeletal muscle of high-intensity endurance-trained rats. J Appl Physiol 81: 619–626, 1996 [DOI] [PubMed] [Google Scholar]
- 27.Hekimsoy Z, Oktem IK. Serum creatine kinase levels in overt and subclinical hypothyroidism. Endocr Res 31: 171–175, 2005 [DOI] [PubMed] [Google Scholar]
- 28.Heneghan HM, Miller N, Kerin MJ. MiRNAs as biomarkers and therapeutic targets in cancer. Curr Opin Pharmacol 10: 543–550, 2010 [DOI] [PubMed] [Google Scholar]
- 29.Huffman KM, Samsa GP, Slentz CA, Duscha BD, Johnson JL, Bales CW, Tanner CJ, Houmard JA, Kraus WE. Response of high-sensitivity C-reactive protein to exercise training in an at-risk population. Am Heart J 152: 793–800, 2006 [DOI] [PubMed] [Google Scholar]
- 30.Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem 55: 1944–1949, 2009 [DOI] [PubMed] [Google Scholar]
- 31.Kiessling KH, Pilstrom L, Karlsson J, Piehl K. Mitochondrial volume in skeletal muscle from young and old physically untrained and trained healthy men and from alcoholics. Clin Sci 4: 547–554, 1973 [DOI] [PubMed] [Google Scholar]
- 32.Kizana E, Cingolani E, Marban E. Non-cell-autonomous effects of vector-expressed regulatory RNAs in mammalian heart cells. Gene Ther 16: 1163–1168, 2009 [DOI] [PubMed] [Google Scholar]
- 33.König D, Neubauer O, Nics L, Kern N, Berg A, Bisse E, Wagner KH. Biomarkers of exercise-induced myocardial stress in relation to inflammatory and oxidative stress. Exerc Immunol Rev 13: 15–36, 2007 [PubMed] [Google Scholar]
- 34.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem 285: 17442–17452, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laterza OF, Lim L, Garrett-Engele PW, Vlasakova K, Muniappa N, Tanaka WK, Johnson JM, Sina JF, Fare TL, Sistare FD, Glaab WE. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem 55: 1977–1983, 2009 [DOI] [PubMed] [Google Scholar]
- 36.Liu G, Detloff MR, Miller KN, Santi L, Houlé JD. Exercise modulates microRNAs that affect the PTEN/mTOR pathway in rats after spinal cord injury. Exp Neurol 233: 447–456, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malizia AP, Wang DZ. MicroRNAs in cardiomyocyte development. Wiley Interdiscip Rev Syst Biol Med 3: 183–190, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 105: 10513–10518, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neumayr G, Gaenzer H, Pfister R, Sturm W, Schwarzacher SP, Eibl G, Mitterbauer G, Hoertnagl H. Plasma levels of cardiac troponin I after prolonged strenuous endurance exercise. Am J Cardiol 87: 369–371, A10, 2001 [DOI] [PubMed] [Google Scholar]
- 40.Nielsen S, Scheele C, Yfanti C, Akerstrom T, Nielsen AR, Pedersen BK, Laye M. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle. J Physiol 588: 4029–4037, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parikh VN, Chan SY. Analysis of microRNA niches: techniques to measure extracellular microRNA and intracellular microRNA in situ. Methods Mol Biol 1024: 157–172, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A 107: 6328–6333, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pepys MB, Hirschfield GM. C-reactive protein: a critical update. J Clin Invest 111: 1805–1812, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pleister A, Selemon H, Elton SM, Elton TS. Circulating miRNAs: novel biomarkers of acute coronary syndrome? Biomark Med 7: 287–305, 2013 [DOI] [PubMed] [Google Scholar]
- 45.Radom-Aizik S, Zaldivar F, Haddad F, Cooper DM. Impact of brief exercise on peripheral blood NK cell gene and microRNA expression in young adults. J Appl Physiol 114: 628–636, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radom-Aizik S, Zaldivar F, Jr, Leu SY, Adams GR, Oliver S, Cooper DM. Effects of exercise on microRNA expression in young males peripheral blood mononuclear cells. Clin Transl Sci 5: 32–38, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Radom-Aizik S, Zaldivar F, Jr, Oliver S, Galassetti P, Cooper DM. Evidence for microRNA involvement in exercise-associated neutrophil gene expression changes. J Appl Physiol 109: 252–261, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rauramaa R, Halonen P, Väisänen SB, Lakka TA, Schmidt-Trucksäss A, Berg A, Penttila IM, Rankinen T, Bouchard C. Effects of aerobic physical exercise on inflammation and atherosclerosis in men: the DNASCO study a six-year randomized, controlled trial. Ann Intern Med 140: 1007–1014, 2004 [DOI] [PubMed] [Google Scholar]
- 49.Rifai N, Douglas PS, O'Toole M, Rimm E, Ginsburg GS. Cardiac troponin T and I, echocardiographic [correction of electrocardiographic] wall motion analyses, and ejection fractions in athletes participating in the Hawaii Ironman Triathlon. Am J Cardiol 83: 1085–1089, 1999 [DOI] [PubMed] [Google Scholar]
- 50.van Rooij E, Olson E. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest 117: 2369–2376, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Rooji E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell 17: 662–673, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rotkrua P, Shimada S, Mogushi K, Akiyama Y, Tanaka H, Yuasa Y. Circulating microRNAs as biomarkers for early detection of diffuse-type gastric cancer using a mouse model. Br J Cancer 108: 932–940, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Safdar A, Abadi A, Akhtar M, Hettinga BP, Tarnopolsky MA. miRNA in the regulation of skeletal muscle adaptation to acute endurance exercise in C57Bl/6J male mice. PLos One 4: e5610, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawada S, Kon M, Wada S, Ushida T, Suzuki K, Akimoto T. Profiling of circulating microRNAs after a bout of acute resistance exercise in humans. PLos One 8: e70823, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, Chen C. Real-time PCR quantification of precursor and mature microRNA. Methods 44: 31–38, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shieh JT, Huang Y, Gilmore J, Srivastava D. Elevated miR-499 levels blunt the cardiac stress response. PLos One 6: e19841, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.da Silva ND, Jr, Fernandes T, Soci UP, Monteiro AW, Phillips MI, Oliveira EMDE. Swimming training in rats increases cardiac MicroRNA-126 expression and angiogenesis. Med Sci Sports Exerc 44: 1453–1462, 2012 [DOI] [PubMed] [Google Scholar]
- 58.Soci UP, Fernandes T, Hashimoto NY, Mota GF, Amadeu MA, Rosa KT, Irigoyen MC, Phillips MI, Oliveira EM. MicroRNAs 29 are involved in the improvement of ventricular compliance promoted by aerobic exercise training in rats. Physiol Genomics 43: 665–673, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stoorvogel W. Functional transfer of microRNA by exosomes. Blood 119: 646–648, 2012 [DOI] [PubMed] [Google Scholar]
- 60.Taganov K, Boldin M, Chang K, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 103: 12481–12486, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thygesen K, Mair J, Giannitsis E, Mueller C, Lindahl B, Blankenberg S, Huber K, Plebani M, Biasucci LM, Tubaro M, Collinson P, Venge P, Hasin Y, Galvani M, Koenig W, Hamm C, Alpert JS, Katus H, Jaffe AS. How to use high-sensitivity cardiac troponins in acute cardiac care. Eur Heart J 33: 2252–2257, 2012 [DOI] [PubMed] [Google Scholar]
- 62.Uhlemann M, Möbius-Winkler S, Fikenzer S, Adam J, Redlich M, Möhlenkamp S, Hilberg T, Schuler GC, Adams V. Circulating microRNA-126 increases after different forms of endurance exercise in healthy adults. Eur J Prev Cardiol. In press [DOI] [PubMed] [Google Scholar]
- 63.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9: 654–659, 2007 [DOI] [PubMed] [Google Scholar]
- 64.Wessner B, Gryadunov-Masutti L, Tschan H, Bachl N, Roth E. Is there a role for microRNAs in exercise immunology? A synopsis of current literature and future developments. Exerc Immunol Rev 16: 22–39, 2010 [PubMed] [Google Scholar]
- 65.Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol 21: 461–469, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woods JA, Vieira VJ, Keylock KT. Exercise, inflammation, and innate immunity. Neurol Clin 24: 585–599, 2006 [DOI] [PubMed] [Google Scholar]
- 67.Wu XD, Zeng K, Liu WL, Gao YG, Gong CS, Zhang CX, Chen YQ. Effect of aerobic exercise on miRNA-TLR4 signaling in atherosclerosis. Int J Sports Med. In press [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, Sun Q, Wang K, Ba Y, Wang Q, Wang D, Yang J, Liu P, Xu T, Yan Q, Zhang J, Zen K, Zhang CY. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell 39: 133–144, 2010 [DOI] [PubMed] [Google Scholar]
- 69.Zhao JL, Rao DS, O'Connell RM, Garcia-Flores Y, Baltimore D. MicroRNA-146a acts as a guardian of the quality and longevity of hematopoietic stem cells in mice. Elife 2: e00537, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]