Abstract
Neuropeptides have widespread effects on behavior, but how these molecules alter the activity of their target cells is poorly understood. We employed a new model system in Drosophila melanogaster to assess the electrophysiological and molecular effects of neuropeptides, recording in situ from larval motor neurons, which transgenically express a receptor of choice. We focused on two neuropeptides, pigment-dispersing factor (PDF) and small neuropeptide F (sNPF), which play important roles in sleep/rhythms and feeding/metabolism. PDF treatment depolarized motor neurons expressing the PDF receptor (PDFR), increasing excitability. sNPF treatment had the opposite effect, hyperpolarizing neurons expressing the sNPF receptor (sNPFR). Live optical imaging using a genetically encoded fluorescence resonance energy transfer (FRET)-based sensor for cyclic AMP (cAMP) showed that PDF induced a large increase in cAMP, whereas sNPF caused a small but significant decrease in cAMP. Coexpression of pertussis toxin or RNAi interference to disrupt the G-protein Gαo blocked the electrophysiological responses to sNPF, showing that sNPFR acts via Gαo signaling. Using a fluorescent sensor for intracellular calcium, we observed that sNPF-induced hyperpolarization blocked spontaneous waves of activity propagating along the ventral nerve cord, demonstrating that the electrical effects of sNPF can cause profound changes in natural network activity in the brain. This new model system provides a platform for mechanistic analysis of how neuropeptides can affect target cells at the electrical and molecular level, allowing for predictions of how they regulate brain circuits that control behaviors such as sleep and feeding.
Keywords: small neuropeptide F, pigment dispersing factor, cAMP, sleep
neuropeptides are the most numerous and diverse class of intercellular signaling molecules in animals (Clynen et al. 2010), and almost all influence target cells via an equally varied set of G protein-coupled receptors (GPCR) and downstream intracellular signaling pathways (Caers et al. 2012). Due to their number, low expression level, and signaling complexity, the functions and mechanisms of neuropeptides are poorly understood compared with classical small molecule neurotransmitters. The fruit fly Drosophila melanogaster provides an excellent system to study neuropeptide signaling mechanisms. Many genes have been identified that code for neuropeptides and their receptors in Drosophila (Hewes and Taghert 2001; Nassel and Winther 2010; Vanden Broeck 2001). Neuropeptidergic systems in fruit flies appear to be fundamentally similar to those in mammals (Nassel and Winther 2010; Taghert and Nitabach 2012), but in flies more powerful genetic tools (Venken et al. 2011) are available to manipulate the expression and function of specific receptors.
Here we have studied two neuropeptides, pigment-dispersing factor (PDF) and short neuropeptide F (sNPF). PDF is crucial for the regulation of daily rhythmicity (Chung et al. 2009; Lear et al. 2009; Renn et al. 1999; Wu et al. 2008; Yoshii et al. 2009) and promotes wakefulness and arousal (Chung et al. 2009; Parisky et al. 2008; Sheeba et al. 2008) Additionally, PDF may also function similarly to the mammalian neuropeptides vasoactive intestinal peptide (VIP) and orexin/hypocretin, which are expressed in small clusters of neurons that control circadian rhythms (Aton et al. 2005; Vosko et al. 2007) and promote wakefulness (Chemelli et al. 1999; de Lecea et al. 1998; Lin et al. 1999; Sakurai et al. 1998). PDF is believed to have a single receptor, PDFR, which is in the secretin family of GPCR and is related to mammalian receptors for CGRP, calcitonin, and VIP (Hewes and Taghert 2001; Hyun et al. 2005; Lear et al. 2005; Mertens et al. 2005).
The second neuropeptide examined in this study is sNPF, which promotes feeding and increases body size (Lee et al. 2004), stimulates insulin expression via the extracellular signal-regulated kinase (ERK) pathway (Kapan et al. 2012; Lee et al. 2008, 2009), and augments feeding-related odorant responses in a starvation-dependent manner (Root et al. 2011). A recent study from our laboratory also demonstrates that sNPF promotes sleep, partly through inhibition of the neurons that produce PDF (Shang et al. 2013). Like PDF, sNPF is thought to act through a single receptor (sNPFR), which shares homology with the vertebrate rhodopsin family neuropeptide Y (NPY) receptor Y2 (Garczynski et al. 2006; Mertens et al. 2002). Indeed, NPY signaling in mammals regulates behaviors similar to those controlled by sNPF in flies, including feeding and sleep/rhythms (Beck 2001; Dyzma et al. 2010; Larhammar and Salaneck 2004; Lawrence et al. 1999).
In this study, we set out to determine how PDF and sNPF act through their canonical receptors to influence the physiology of their target cells, using a combination of patch-clamp recordings and live fluorescent imaging of intracellular signaling molecules in central neurons.
METHODS
Fly lines.
Flies were raised on a 12:12-h light:dark cycle at 25°C. All experiments were done on third instar male and female larvae. The OK371-Gal4 driver line was used to drive expression of transgenes in larval motor neurons, either alone or with UAS-mCD8::GFP recombined onto the second chromosome. The UAS-sNPFR line was generated in the Yu laboratory (Lee et al. 2008). The UAS-PDFRmyc13 and UAS-PDFRm10 lines were obtained from Paul Taghert. The UAS-GCaMP3(10×)(aTTP40) line was developed by the Looger laboratory (Tian et al. 2009) and was obtained from the Janelia Farm Research Campus animal facility, and the UAS-Epac1-cAMPs lines (50AII and 55A) were obtained from Orie Shafer (Shafer et al. 2008). UAS-PTX.16 flies were obtained from Gregg Roman (Ferris et al. 2006). RNAi lines were obtained from the Bloomington Drosophila Stock Center (AC3: 28626, Gαo 47A: 34653, Gαi 65A: 35407 and 34924, Gαs 60A: 29576, Gαf 73B: 25930, Concertina: 31132, Gβ13F: 31134, Gβ5: 28310, Gβ76C: 28507, Gγ1: 25934 and 34372, and Gγ30A: 25932 and 34484) or from the Vienna Drosophila RNAi Center (Gαs 60A: 105485, AC3: 33217, and Nervy: 100273).
Neuropeptides.
PDF (H-NSELINSLLSLPKNMNDA-NH2) and sNPF-1 (H-AQRSPSLRLRF-NH2) were commercially synthesized (PolyPeptide). Peptides were stored as powder at room temperature and were then dissolved in DMSO at 20 mM. Aliquots were desiccated using a SpeedVac (Savant) and were stored at −20°C. Concentrations of peptides used for experiments were generally in the low micromolar range and were chosen based on previous studies (Mertens et al. 2002, 2005; Pírez et al. 2013; Root et al. 2011; Shafer et al. 2008) to reliably induce physiological effects.
Electrophysiology.
Third instar larvae were dissected and pinned in Sylgard (Dow Corning) in 0 mM Ca2+ modified A solution, containing the following (in mM): 118 NaCl, 2 NaOH, 2 KCl, 4 MgCl2, 22.3 sucrose, 5 trehalose, 5 HEPES pH 7.15, and 281 mosmol/kgH2O. In tests of PDF responses, recordings were carried out in situ, using the same external solution. For all other recordings, the brain was removed and pinned separately, and the preparation was shifted to a new external solution, adult hemolymph, containing the following (in mM): 108 NaCl, 5 KCl, 2 CaCl2, 8.2 MgCl2, 4 NaHCO3, 1 NaH2PO4, 5 trehalose, 10 sucrose, 5 HEPES pH 7.5, 265 mosmol/kgH2O (based on Wang et al. 2003). In all cases, protease XIV (Sigma-Aldrich) treatment (0.5–1% wt/vol) was used to dissolve the glial sheath, allowing access to the motor neurons. The photograph of a patch-clamp recording in progress was taken using a 4910 1/2'" CCD camera (Cohu), acquired using ATI's TV Wonder HD 600 USB HDTV tuner and VLC media player (VideoLAN). Patch electrodes were (in mm) 1.2 OD × 0.9 ID × 100 L (Friedrick & Dimmock) and were pulled and fire-polished to achieve a resistance of 3–7 MΩ. Pipettes were loaded with internal solution as per Choi et al. (2004), containing the following (in mM) 20 KCl, 0.1 CaCl2, 2 MgCl2, 1.1 EGTA, 120 K-gluconate, 10 HEPES pH 7.2, and 280 mosmol/kgH2O. Signals were acquired using the Axopatch 200B amplifier and Clampex (Molecular Devices). In our analysis, we did not distinguish between recordings from different cell subtypes within a cluster of motor neurons (Choi et al. 2004), because we did not qualitatively observe a difference in responses among subtypes. Current-clamp pulses to test excitability were 500 ms in duration and stepped in 20 pA increments from −20 to +140 pA, with a 10-s interpulse interval. Excitability was measured by averaging the spike rates across the +20 to +60 pA stimulation intensities, approximating the linear range of the response curve. PDF was bath applied at 10 μM for 20 min, after which resting membrane potential and assessment of excitability was carried out. sNPF was bath applied at 20 μM for 5 min, followed by measurements of resting membrane potential and excitability. Recovery was assessed after 10 min of washout. Forskolin (FSK; Sigma-Aldrich) was used at 10 μM.
Imaging.
Imaging experiments were performed using a naked brain preparation. Briefly, each brain was dissected in chilled 0 mM Ca2+ modified A solution (see above) and was transferred to an RC-26 chamber on a P1 platform (Warner Instruments) and pinned in Sylgard. The brain was then perfused with adult hemolymph (see above) by gravity feed at 3–4 ml/min. All experiments were performed using an Olympus BX51WI microscope, and optical signals were recorded using a back-illuminated CCD camera (Hamamatsu Orca C472-80-12AG). For calcium imaging, a ×60, 0.9-NA water immersion lens (Olympus LUMPlanFl) was used, and μManager software (Edelstein et al. 2010) was used to acquire images at 2 Hz using a GFP filter, with 50 ms exposure and 4× binning. Filters used for calcium imaging were as follows: excitation HQ470/×40, Dichroic Q495LP, emission HQ525/50 m (Chroma). For cAMP imaging, 45-ms exposure stimulated the sensor, and cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) emissions were collected simultaneously using a splitter (Photometrics). Either a ×60, 0.9-NA or a ×40, 0.8-NA water immersion lens (Olympus LUMPlanFl) was used, and images were acquired at 1 Hz with the software Volocity (PerkinElmer), with 4× binning. Filters used for cAMP imaging were as follows: excitation 86002v1 JP4 filter (436, Chroma), and emission D480/30 m and D535/40 m (Optical Insights). Off-line data analysis was performed using ImageJ (National Institutes of Health) and Matlab (Mathworks). To limit photobleaching, a 25% neutral density filter (Chroma) was used for all experiments, and in the cAMP experiments brains were preexposed to light for 5 min before beginning recording, a period that allowed for the baseline fluorescence signal to stabilize. Recordings lasted 240 s, with 30 s of baseline before beginning drug treatment, which lasted through the duration of the recording, unless otherwise indicated. Switching between solutions was achieved using a three-way valve solenoid (Cole-Parmer) under manual control. The fluorescence resonance energy transfer (FRET) signal (CFP/YFP ratio) for each time point was calculated and normalized to the ratio of the first time point before drug application. The relative cAMP changes were determined by plotting this normalized CFP/YFP ratio (%) over time. When possible, two to three cell bodies were used as regions of interest (ROIs) for each recording, and regions of interest values were averaged. cAMP data were normalized to the baseline slope to control for residual bleaching during the experiment. For calcium imaging, the change in fluorescence was calculated using the following formula, in which Fn is the fluorescence at time point n, and F0 is the fluorescence at time point 0: ΔF/F = (Fn − F0)/F0 × 100%.
Statistics.
Statistical analysis was carried out using JMP, Version 5 (SAS Institute) and SPSS, Version 19. For analysis of the effects of PDF and sNPF on resting membrane potential, excitability, and maximal firing rate in larval motor neurons, for each genotype tested, mixed model ANOVAs were used, with treatment (PDF or sNPF vs. vehicle) as the between-subjects factor and time point (before vs. after treatment) as the within-subject factor, followed by Bonferroni-corrected post hoc tests. Similar analysis was used for analysis of the effects of sNPF on resting membrane potential in larval DILP cells. For analysis of electrophysiological effects of FSK, paired t-tests were used, comparing before and after treatment. For analysis of effects of G-protein subunit RNAi on resting membrane potential changes due to sNPF treatment, mixed model ANOVAs were used, with genotype as the between-subjects factor and time point (before vs. after treatment) as the within-subject factor, followed by Bonferroni-corrected post hoc tests. A Fisher's exact test was used to determine if sNPF and vehicle treatments had a different likelihood of causing cessation of calcium waves. For analysis of cAMP imaging data, normalized cAMP levels averaged from 150–240 s were compared using a one-way ANOVA or t-test, as appropriate. In all figures, numbers in parentheses represent the number of animals in each condition, error bars represent ± SE, and *P < 0.05.
RESULTS
Larval motor neuron patch-clamp recordings constitute a novel system to study the electrical and molecular effects of neuropeptides in Drosophila neurons.
The ideal approach to study neuropeptide effects would be to record electrical responses in neurons that naturally express the appropriate receptor. In the case of PDF, antibody staining (Helfrich-Forster 1995; Hyun et al. 2005; Lear et al. 2005), PDFR-GAL4 lines (Im and Taghert 2010; Parisky et al. 2008), functional imaging (Duvall and Taghert 2012; Pírez et al. 2013; Shafer et al. 2008), and the use of membrane-tethered PDF peptide (Choi et al. 2009, 2012) has led to a consensus that, within the central brain, the PDFR is present in a subset of the PDF-expressing small ventrolateral neurons (the small LNvs, or sLNvs), ellipsoid body cells, and some cells within the dorsal lateral neuron (LNd) and DN1 clock cell clusters. sNPFR expression has not been mapped extensively, but its mRNA has been localized to several body tissues (Mertens et al. 2002), with the highest enrichment in the brain (http://www.flyatlas.org/; Chintapalli et al. 2007). In the larval brain, antibody staining has found expression of sNPFR in a handful of clusters including the DILP2-positive insulin-producing cells (Lee et al. 2008).
Even when target cells have been identified conclusively, most of those cells are small and buried deep within the brain, making patch-clamp recordings difficult. Performing recordings in heterologous systems, such as Xenopus laevis oocytes or cultured cells, in which an individual receptor is artificially expressed, has been a useful tool to probe neurotransmitter/receptor action (Mertens et al. 2002, 2005; Wagner et al. 2000). However, the utility of this type of recording is limited, because those cells are from different species and may not express components expected to be found in brain cells. Ectopic receptor expression in Drosophila has been used previously to show that cAMP responses to PDF could be induced in otherwise unresponsive large LNvs by GAL4-driven expression of PDFR in those cells (Shafer et al. 2008). However, the electrophysiological characterization of the peptide response was not robustly feasible in that cell type.
Therefore, we developed a novel model system to study the effects of neuropeptides on the electrophysiology of target neurons that has advantages over previous systems. Patch-clamp recordings were performed in third instar larvae motor neurons (Fig. 1) in which either PDFR or sNPFR were expressed transgenically using the GAL4/UAS system (Duffy 2002). Advantages of this model system are that 1) the cells we record from are in fact Drosophila cells, and 2) they are central neurons, which are known to be endogenous targets for neuropeptides and are capable of exhibiting robust active electrophysiological responses, unlike mammalian tissue culture cells or Xenopus oocytes. Because of these characteristics, we did not have to express additional signaling components, such as G proteins or ion channels, to see a response through the receptor of interest. Additionally, the large somata and superficial location of these neurons make them accessible for patch clamping, and they have been well-characterized electrophysiologically (Choi et al. 2004; Rohrbough and Broadie 2002; Ryglewski et al. 2012; Schaefer et al. 2010; Srinivasan et al. 2012a,b; Worrell and Levine 2008).
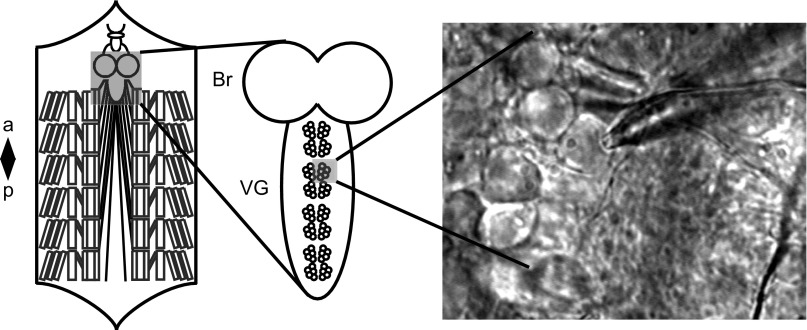
Fig. 1.
Larval motor neuron preparation for patch-clamp electrophysiology. Left: schematic of the larval preparation, showing the central nervous system in a gray box, with repeating bilateral sets of body wall muscles (a = anterior, p = posterior). Center: a zoom of the brain (Br) and ventral ganglion (VG) of the larval central nervous system, showing repeating bilateral clusters of motor neurons. Right: an ×800 photograph of a glass electrode making contact with one of the neurons within a fully dissected motor neuron cluster. Left and middle schematics adapted from Pulver and Griffith (2010).
PDF excites motor neurons expressing the PDFR.
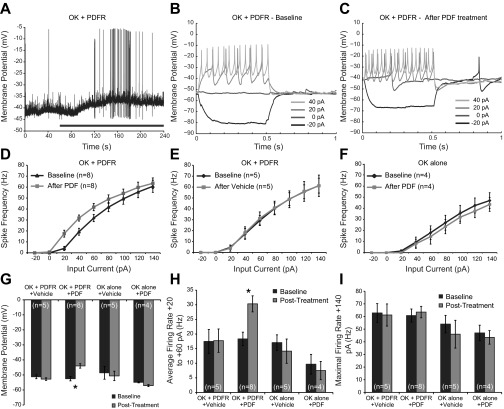
The PDFR (UAS-PDFRMYC13) was expressed under the control of the OK371-Gal4 driver (Mahr and Aberle 2006). Treatment with 10 μM PDF resulted in significant depolarization from baseline, typically within 1–2 min of treatment onset [F(1,11) = 14.916, P = 0.003], whereas vehicle treatment had no effect (Fig. 2, A and G). PDF treatment increased motor neuron excitability significantly compared with vehicle, as measured by the average firing rate in response to current injections from +20 to +60 pA [F(1,11) = 17.439, P = 0.002; Fig. 2, B–E, and H], although the maximum firing rate was not significantly affected by either PDF or vehicle treatment [F(1,11) = 2.088, P = 0.176; Fig. 2, D–E, and I]. Additionally, control larvae containing only the driver but not the UAS-PDFR transgene did not exhibit any depolarization [F(1,7) = 0.001, P = 0.983], change in excitability [F(1,7) = 0.035, P = 0.856], or alteration in maximal firing rate [F(1,7) = 0.782, P = 0.406] in response to PDF, showing that the effects of PDF were due to the transgenic expression of PDFR and not due to endogenous PDFR or another PDF-responsive receptor in motor neurons (Fig. 2, F–I).
Fig. 2.
Pigment-dispersing factor (PDF) causes excitation of larval motor neurons expressing the PDF receptor (PDFR). A: example recording shows a depolarization in resting membrane potential (RMP) induced by 10 μM PDF in flies expressing PDFRMYC13, resulting in increased basal spike rate. Black bar represents the period of treatment. OK, OK371-Gal4 driver. B: example current-clamp recording before treatment with PDF. Only the first 4 sweeps of the protocol are shown for clarity. C: current-clamp recording from the same cell after treatment with PDF. D: quantification of current-clamp data, plotting firing rate as a function of input current (F/I curve). PDF caused a significant leftward shift in the curve, indicating that the same input current caused a higher spike rate after PDF treatment. E: F/I curves comparing baseline with vehicle treatment in flies expressing PDFRMYC13. F: F/I curves comparing baseline with PDF treatment in flies that do not contain a PDFR transgene. G: summary of the effects of PDF or vehicle treatment on motor neuron RMP. Only the combination of transgenic PDFR expression and PDF treatment caused significant changes in RMP. H: summary of the effects of PDF or vehicle treatment on excitability, measured as the average firing rate across the three weakest positive input currents used (+20, +40, and +60 pA). I: summary of the effects of PDF or vehicle treatment on the maximal firing rate, measured at +140 pA.
PDF increases cAMP in larval motor neurons and muscles expressing the PDFR.
PDF treatment had previously been shown to increase cAMP levels in culture (Hyun et al. 2005; Mertens et al. 2005) and in small LNvs and other clock neurons that naturally express PDFR (Duvall and Taghert 2012; Shafer et al. 2008). Therefore, we performed experiments in which the OK371-Gal4 driver was used to express not only PDFR (UAS-PDFRM10) but also Epac-cAMP (Shafer et al. 2008), a transgene that uses FRET to report levels of the second messenger cAMP (Nikolaev et al. 2004; Nikolaev and Lohse 2006). PDF treatment produced a significant increase in cAMP from 150–240 s, when compared with vehicle treatment alone (P = 0.006; Fig. 3, A and C). This cAMP response was even larger than that induced by the potent adenylate cyclase activator FSK (Seamon and Daly 1986) at the same 10 μM concentration (data not shown). In fact, motor neurons that had been treated with PDF showed no further increase to subsequent FSK treatment, demonstrating that PDF caused a saturated cAMP response (data not shown). In motor neurons lacking transgenic PDFR expression (Fig. 3B), no effect of PDF was observed on cAMP levels when compared with the effects of vehicle treatment (P = 0.213). This indicated that exogenous PDFR expression was required in motor neurons for PDF to produce a cAMP response, although these cells were still able to respond to FSK (data not shown) and thus have functional cAMP signaling machinery. To determine if the same signaling pathway was utilized by the PDFR when expressed in a very different cell type, the MHC-Gal4 driver was used to express the PDFR in muscles, along with the Epac-cAMP sensor. As in motor neurons, PDF treatment caused a significantly larger cAMP response than vehicle in muscles (P = 0.0001; Fig. 3, D and F). The effect of PDF on cAMP levels was absent in muscles lacking transgenic PDFR expression (P = 0.569; Fig. 3E). These data demonstrate that the PDFR receptor can cause an increase in cAMP in two distinct cell types and that neither motor neurons nor muscles in third instar larvae are naturally responsive to PDF. Although PDF increased cAMP levels in both cell types, there were some qualitative differences between the responses in motor neurons and muscles. The magnitude of cAMP induction in muscles was considerably lower than in motor neurons, but the onset of the response was more rapid. This may be due to a combination of differences in the relative strength of the cell-specific drivers, the relative accessibility of the cells to peptide, or variations in cell membrane signaling.
Fig. 3.
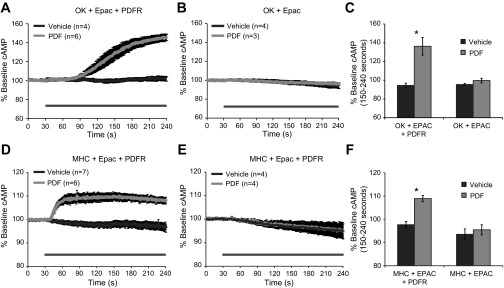
PDF increases cAMP levels in both motor neurons and muscle cells expressing the PDFR. A: PDF treatment caused a strong cAMP increase in motor neurons expressing PDFRM10. Time course of response is consistent with previous reports of PDF signaling in this type of preparation (Duvall and Taghert 2012; Shafer et al. 2008). B: PDF did not alter cAMP levels in motor neurons from flies lacking transgenic expression of PDFR. C: summary of motor neuron recordings, showing average cAMP levels from 150–240 s. D: PDF caused an increase in cAMP levels in body wall muscles of flies expressing PDFRM10 and exchange protein activated by cAMP (EPAC) under control of the muscle-specific MHC-Gal4 driver (MHC). E: PDF caused no change in levels of cAMP in muscles from flies lacking transgenic expression of PDFR. F: quantification of muscle recording data, showing average cAMP levels from 150–240 s. Black bars in A–F represent the period of treatment.
Direct stimulation of the cAMP pathway mimics the electrophysiological effects of PDF.
We next asked if stimulation of the cAMP pathway alone could recapitulate the electrophysiological effects of PDF. That is, is it likely that cAMP production causes the depolarization, or does depolarization result in increased cAMP production? To address this question, we performed patch-clamp recordings in third instar larval motor neurons and treated with FSK (10 μM). FSK produced an ∼10 mV depolarization of the resting membrane potential (Fig. 4, A and B; P = 0.0017), and increased the average firing rate in response to +20- to +60-pA current injections (P = 0.013; Fig. 4, B–D). This electrophysiological response was nearly identical to that produced by PDF and suggests that cAMP induction could be responsible for the electrophysiological effects of PDF.
Fig. 4.
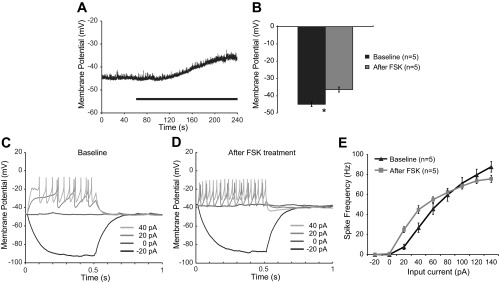
The adenylate cyclase activator forskolin (FSK) mimics the electrophysiological effects of PDF treatment on larval motor neurons. A: example recording shows a depolarization in RMP induced by 10 μM FSK. In this case, the action potential threshold was not quite reached during the depolarization, thus spontaneous spiking was not induced. Black bar represents the period of treatment. B: quantification of RMP responses to FSK. C: example current-clamp recording before treatment with PDF. Only the first 4 sweeps of the protocol are shown for clarity. D: current-clamp recording from the same cell after treatment with FSK. E: quantification of current-clamp data, plotting firing rate as a function of input current (F/I curve).
sNPF inhibits motor neurons expressing the sNPFR.
We next used the same larval motor neuron system to ascertain the electrophysiological effects of the fly neuropeptide sNPF via its receptor, sNPFR. Drosophila expresses multiple sNPF peptides derived from the sNPF gene (CG13968) (Hewes and Taghert 2001; Nassel et al. 2008; reviewed in Nassel and Wegener 2011; Vanden Broeck 2001; Wegener et al. 2006), but in this study we focused on testing sNPF-1 (see methods for sequence), which oocyte and cell culture studies have suggested has high bioactivity (Feng et al. 2003; Mertens et al. 2002; Reale et al. 2004). The OK371-Gal4 driver was combined with the UAS-sNPFR transgene to express sNPFR in larval motor neurons. Perfusion of 20 μM sNPF caused a significant hyperpolarization in resting membrane potential, typically occurring within 1–2 min of treatment onset, whereas vehicle treatment had no effect [F(1,10) = 51.665, P = 0.00003; Fig. 5, A and G]. The hyperpolarization induced by sNPF was long lasting, only reversing partially after 10 min of washout (data not shown).
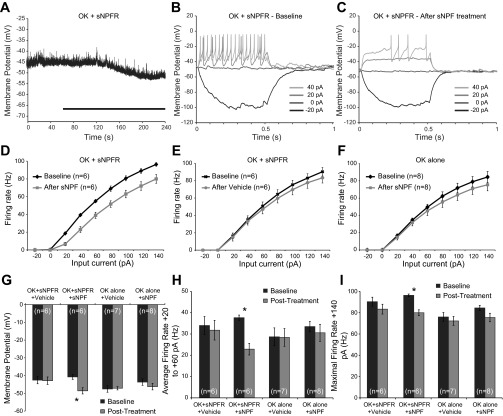
Fig. 5.
Small neuropeptide F (sNPF) inhibits larval motor neurons expressing the sNPF receptor (SNPFR). A: example recording shows a hyperpolarization in RMP induced by 20 μM sNPF in flies expressing sNPFR. Black bar represents the period of treatment. B: example current-clamp recording before treatment with sNPF. Only the first 4 sweeps of the protocol are shown for clarity. C: current-clamp recording from the same cell after treatment with sNPF. D: quantification of current-clamp data, plotting firing rate as a function of input current (F/I curve). sNPF caused a rightward shift in the curve, indicating that more current was required to elicit the same spike rate after sNPF treatment. E: F/I curves comparing baseline with vehicle treatment in flies expressing sNPFR. F: F/I curves comparing baseline with sNPF treatment in flies that do not contain a sNPFR transgene. G: summary of the effects of sNPF or vehicle treatment on motor neuron RMP. Only the combination of transgenic sNPFR expression and sNPF treatment caused a significant hyperpolarization in RMP. H: summary of the effects of sNPF or vehicle treatment on excitability, measured as the average firing rate across the three weakest positive input currents used (+20, +40, and +60 pA). I: summary of the effects of PDF or vehicle treatment on the maximal firing rate, measured at +140 pA.
sNPF-induced hyperpolarization was accompanied by a significant decrease in firing rate in response to current injections of +20 to +60 pA, relative to vehicle treatment [F(1,10) = 20.3, P = 0.001; Fig. 5, B-D, and H]. In this experiment, there was an overall decrease in maximal firing rate [F(1,10) = 17.133, P = 0.002] following treatment, but no significant interaction between treatments [F(1,10) = 2.741, P = 0.129; Fig. 5, D-F, and I]. This reflects a general trend that maximal firing capacity of the cells decreased over time.
sNPF treatment did not cause a significant hyperpolarization relative to vehicle in larvae expressing the OK371-Gal4 driver alone [F(1,13) = 2.516, P = 0.137]. sNPF also had no effect on firing rate in the absence of transgenic sNPFR expression [F(1,13) = 0.844, P = 0.375; Fig. 5, F and H]. There was again a significant overall effect of time on maximum firing rate [F(1,13) = 37.107, P = 0.00004], which was equivalent following sNPF or vehicle treatment (Fig. 5I). These findings demonstrate that the electrophysiological effects of sNPF are due to the transgenic expression of sNPFR and not due to endogenous sNPFR or another sNPF-responsive receptor in motor neurons.
sNPF inhibits larval DILP-expressing cells.
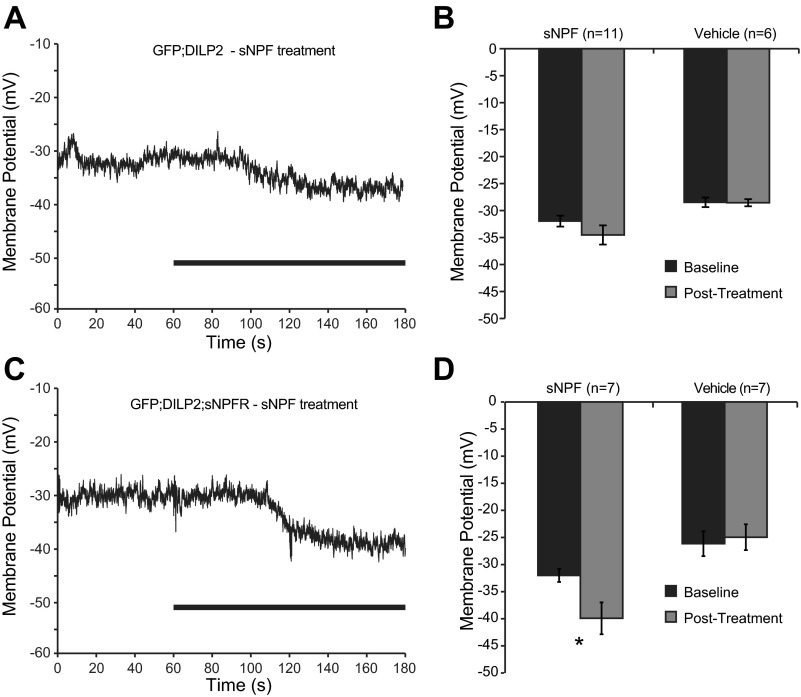
Our data demonstrate that motor neurons do not naturally have a significant electrical response to sNPF, suggesting that they do not express sNPFR endogenously. It is therefore possible that when expressed via transgene in this unnatural setting, sNPFR may couple to different downstream pathways than it would in a more natural setting. Therefore, we performed patch-clamp recordings in cells that express Drosophila insulin-like peptide DILP2 in the brains of third instar larvae. These cells had previously been suggested to express sNPFR (Lee et al. 2008). The electrical properties of these DILP-positive cells are quite different from those of motor neurons: their resting membrane potential is quite depolarized, between −25 and −40 mV, and they appear to be incapable of firing at high frequency; with increasing stimulation spikes become flat and wide or disappear completely. Therefore, measuring firing rates as a function of input current was not feasible in these cells. In larvae expressing only GFP under control of the DILP2-Gal4 driver, sNPF treatment induced small hyperpolarizations in a subset of recordings, but across all recordings the peptide had no significant effect [F(1,15) = 2.334, P = 0.147] on resting membrane potential (Fig. 6, A and B). However, in larvae additionally containing the UAS-sNPFR transgene, strong hyperpolarization responses were observed in response to sNPF treatment, whereas vehicle treatment caused no significant change [F(1,13) = 19.551, P = 0.001] in resting membrane potential (Fig. 6, C and D).
Fig. 6.
Electrical responses to sNPF in insulin-producing cells that naturally express sNPFR. A: example recording showing a small hyperpolarization in RMP induced by 20 μM sNPF in flies expressing GFP under control of the DILP2-Gal4 driver. Black bar represents the period of treatment. B: quantification of the effects of sNPF or vehicle treatment on DILP2 cell RMP. C: example recording showing a large hyperpolarization in RMP induced by 20 μM sNPF in flies expressing GFP and sNPFR under control of the DILP2-Gal4 driver. Black bar represents the period of treatment. D: quantification of the effects of sNPF or vehicle treatment on DILP2 + sNPFR cell RMP. sNPF caused a significant reduction in RMP.
Based on the previous report that DILP cells express sNPFR (Lee et al. 2008), it was surprising that DILP cells did not respond reliably to sNPF in the absence of sNPFR overexpression. This may have been because only subsets of DILP cells express the receptor, and our recordings were from some cells with sNPFR and some without, or because resting sNPFR levels were too low to obtain reliable responses. Interestingly, two recent studies have indicated that sNPFR is expressed in DILP cells during adulthood (Kapan et al. 2012) but not during larval stages (Carlsson et al. 2013). This differential developmental expression could explain the absence of a reliable electrophysiological effect of sNPF in larval DILP cells without exogenous sNPFR expression. Future electrophysiological studies in adult DILP cells could test the prediction that native sNPF responses might appear in that cell type during adulthood. In the Drosophila olfactory system, sNPFR has been shown to be upregulated in response to starvation (Root et al. 2011). Because DILP cells are also responsive to starvation, it would be interesting in future experiments to determine if natural sNPF responses in DILP cells are augmented by starvation or other stressors.
Electrophysiological effects of sNPF are mediated by Gαo signaling.
Having shown that sNPF can act via sNPFR to hyperpolarize its target cells, we were interested to determine if particular G-protein subunits were required for this effect. Flies are thought to express six Gα, two Gβ, and three Gγ subunits (Katanayeva et al. 2010). We screened several RNAi constructs targeting specific G-protein subunits for effects on the electrical response to sNPF treatment in motor neurons expressing sNPFR. RNAi targeting Gαo 47A almost completely blocked the effect of sNPF, but sNPF still produced a significant hyperpolarization in flies in which other Gα subunits were knocked down [F(4,25) = 4.499, P = 0.007; post hoc tests found that sNPF treatment induced significant hyperpolarization in all groups but Gαo 47A RNAi; Fig. 8A]. RNAi targeting Gβ or Gγ subunits all showed equivalent hyperpolarization responses to sNPF [Gβ subunits: F(2,12) = 0.511, P = 0.612, Gγ subunits: F(3,15) = 0.1259, P = 0.9433; Fig. 7, A–C], suggesting either redundancy among these subunits or inefficient knock down.
Fig. 8.
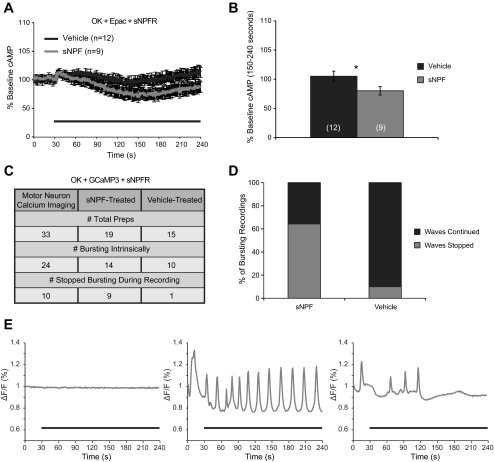
sNPF causes a slight reduction in cAMP and suppresses spontaneous calcium waves in motor neurons expressing the sNPFR. A: sNPF causes a slight but significant reduction in cAMP in larval motor neurons expressing the cAMP sensor Epac-cAMPs1 and sNPFR, measured by fluorescence resonance energy transfer (FRET) as in Fig. 3. Black bar represents the period of treatment. B: quantification of sNPF-induced reduction in cAMP from 150–240 s. For live imaging of calcium responses, the fluorescent intracellular calcium sensor GCaMP3 was expressed in motor neurons in conjunction with sNPFR. In many cases we observed spontaneous calcium waves passing up or down the ventral ganglion, representative of larval crawling rhythms. See Supplemental Movie S1 for an example, which corresponds to the trace shown in E, right. C: summary of the results of calcium imaging analysis. Based on the presence of the calcium waves mentioned above, recordings were categorized as either bursting or nonbursting preparations, and were then divided based on whether bursting was suppressed during treatment or if it continued throughout the recording. D: sNPF treatment halted calcium waves in a majority of bursting recordings, whereas calcium waves ceased in only 1 of 10 vehicle-treated preparations. E: examples of the 3 categories of recordings we observed. From left to right: nonbursting, bursting that continued through the recording, and bursting that ceased. Black bars represent the period of treatment. ΔF/F = (Fn − F0)/F0 × 100%, where Fn is the fluorescence at time point n, and F0 is the fluorescence at time point 0.
Fig. 7.
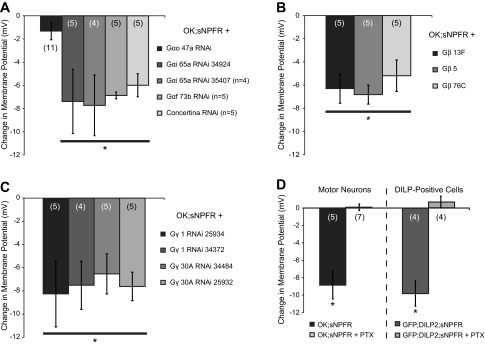
sNPF-induced hyperpolarization in motor neurons depends on signaling via the G-protein Gαo. Motor neuron recordings were performed in larvae expressing sNPFR alone or in conjunction with either an RNAi construct targeting a specific G-protein subunit or the Gαo inhibitor pertussis toxin (PTX). For clarity of presentation, the change in membrane potential induced by 20 μM sNPF treatment is shown for each group. A: RNAi targeting Gα subunits. Only Gαo knockdown resulted in a smaller sNPF response than in the other groups. B: RNAi targeting Gβ subunits. None of the tested RNAi significantly altered the response to sNPF. C: RNAi targeting Gγ subunits. Two different RNAi constructs were used for each Gγ subunit, but none significantly affected the response to sNPF. D: sNPF responses in motor neurons (left) or DILP neurons (right) expressing sNPFR alone or in conjunction with the Gαo inhibitor PTX. PTX completely blocked the effect of sNPF on membrane potential in both cell types.
Because RNAi constructs can sometimes have off-target effects (Jackson et al. 2003; Seinen et al. 2011), we validated this finding with an alternative approach. We used a transgene that expresses pertussis toxin (PTX), which in mammals blocks both Gαo and Gαi but in flies is a selective inhibitor of Gαo (reviewed in Ferris et al. 2006; Jiang and Bajpayee 2009). PTX coexpression with sNPFR in motor neurons completely blocked sNPF-induced hyperpolarization [F(1,10) = 40.676, P = 0.00008; Fig. 7D], confirming that Gαo is required for the effects of sNPF. We also found that coexpression of PTX with sNPFR blocked sNPF responses in DILP-positive cells [F(1,6) = 42.65, P = 0.00; Fig. 7D], indicating that Gαo is a common mechanism downstream of sNPFR in two different neuronal cell types.
sNPF reduces cAMP levels and suppresses rhythmic network activity in motor neurons expressing the sNPFR.
Signaling by Gαo can be mediated by a number of different output pathways. To determine if cAMP was one of them, we performed live imaging of motor neurons from third instar larvae expressing the OK371-Gal4 driver, UAS-sNPFR1, and the FRET-based cAMP sensor UAS-Epac-cAMPs. Treatment with sNPF caused a significant reduction in cAMP levels (P = 0.038; Fig. 8, A and B). While this does not rule out a role for Gαo in direct regulation of a leak current, it suggests that sNPF could act in part by blocking the effects of agents that activate cyclase.
The ability of sNPF to hyperpolarize neurons expressing sNPFR and reduce cAMP levels suggests that sNPFR mediates native inhibitory signaling and that it could have profound effects on a rhythmically active network. To test this idea, we took advantage of the fact that previous studies had observed spontaneous organized activity in the larval ventral ganglion that likely reflects output of the locomotion central pattern generator (Cattaert and Birman 2001; Fox et al. 2006). To visualize network activity, we performed live imaging in third instar larvae expressing the OK371-Gal4 driver, UAS-sNPFR, and the calcium-sensitive sensor UAS-GCaMP3 (Fig. 8B). Consistent with previous reports (Cattaert and Birman 2001; Fox et al. 2006), we observed spontaneous calcium waves in 24 of the 33 total brains from which we recorded (Fig. 8, D and E; Supplemental Movie S1; Supplemental Material for this article is available online at the J Neurophysiol website). None of the nine nonbursting preparations began bursting during the recording, and no effect of sNPF or vehicle treatment was observed on calcium levels in those animals. However, sNPF-treated brains were significantly more likely to cease bursting (9 of 14) during treatment than vehicle-treated brains (1 of 10; P = 0.0129, Fisher's exact test; Fig. 8, C and D). Examples of nonbursting, continuing bursting, and halted bursting recordings are shown in Fig. 8E. These data suggest that sNPF does not act via direct changes in calcium levels to affect physiology but that the hyperpolarization induced by sNPF is often strong enough that it can block spontaneous network activity. These results allow us to predict that endogenous sNPF signaling might be capable of acting as an inhibitory modulator in networks associated with the behaviors it regulates, including sleep (see Shang et al. 2013) and feeding (Lee et al. 2004, 2008).
DISCUSSION
Neuropeptides are typically expressed in small numbers of cells and influence a limited set of behaviors (reviewed in Nassel and Winther 2010). Neuropeptide systems are therefore appealing targets for the design of drugs that could selectively target particular behaviors, such as sleep, while limiting side effects. For comparison, most current drugs for sleep disorders modulate GABA signaling (Harrison 2007). Unfortunately, GABA is the major inhibitory transmitter in both vertebrates and invertebrates and is produced in an estimated 20% of the neurons in the mammalian brain (Hendry et al. 1987; Sahara et al. 2012). It is involved in practically all behaviors, making side effects due to drugs targeting GABA inevitable. Understanding the mechanisms by which neuropeptides influence activity is a critical step in manipulating their function.
This report establishes motor neuron patch-clamp recording as a viable model system to test the effects of Drosophila neuropeptides on central neuron targets. One benefit of this technique is the ability to record direct electrical responses to application of peptide, allowing for measurement of the time course of the response. The characterization of receptors in Drosophila neurons also allows fly genetic tools to be brought to bear to identify signaling mechanisms downstream of the peptide receptor of interest. In addition, those cell signaling components do not need to be heterologously expressed as they would be in Xenopus oocytes, for example. The current study focused on PDF and sNPF, but this approach is also promising for examination of other neuropeptides.
Despite its wide utility, this approach does have some limitations. For example, motor neurons may not express the same complement of downstream molecular components as cells that natively express the receptor. In addition, this method relies on GAL4/UAS-mediated expression of the receptor of interest, which may not mimic native expression levels. However, we have presented here several lines of evidence that motor neuron recordings are a useful model system, despite these potential concerns. First, our observed cAMP response to PDF closely mimics what has been seen in previous studies in clock cells (Duvall and Taghert 2012; Shafer et al. 2008), ellipsoid body cells (Pírez et al. 2013), in ureter muscles (Talsma et al. 2012), and in our own muscle recordings (Fig. 3). These combined findings demonstrate that very different cell types engage a similar signaling cascade induced by PDF, even when PDFR is overexpressed. In the case of sNPF a recent report from our group indicates that sNPF can act as an inhibitory modulator to regulate sleep (Shang et al. 2013). NPY, the mammalian homolog of sNPF, also typically has an inhibitory effect (van den Pol 2012). These results are consistent with our finding that sNPF induces hyperpolarization via sNPFR in motor neurons via Gαo. Additionally, exogenous expression of sNPFR within DILP cells led to a similar hyperpolarization response to sNPF (Fig. 6).
Of course, it is possible that particular cell types will have different electrical responses to a transmitter, even via the same molecular cascade, or initiate signaling through a different signaling system altogether. For example, in olfactory receptor neurons, sNPF appears to mediate enhancement of odor responses during starvation via activation of sNPFR autoreceptors, suggesting a stimulatory mode of action in those cells (Root et al. 2011). sNPF has also been found to act via Gαs/cAMP/PKA/CREB and to induce expression of the minibrain gene and promote feeding (Hong et al. 2012), and to couple positively to cyclase in a Drosophila cell line (Chen et al. 2013). PDF can have cell type-specific actions as well. Two recent studies (Duvall and Taghert 2012, 2013) found that PDF preferentially activated adenylate cyclase 3 (AC3) via Gαs in one subset of clock cells called sLNvs but acted through different adenylate cyclase subtypes in another cluster of clock neurons, the LNds, although PDF application increased cAMP levels in both cell populations. Future studies will be needed to determine the mechanisms by which neuropeptide receptors are coupled to cell-specific signaling mechanisms.
A handful of studies have pointed towards an excitatory role of PDF. The first found that PDF either increased firing in the terminal abdominal ganglion motor neuron nerve in the locust or changed the pattern of firing to increase bursting (Persson et al. 2001). However, their results suggested that this effect was likely through action on interneurons, not through direct effects on the motor neurons themselves. Because the interneurons could have been either excitatory or inhibitory, these data did not shed light on the direct actions of PDF. In Drosophila, PDF treatment was found to increase ureter circular muscle contractions, suggesting that PDF is excitatory in muscles expressing the PDFR (Talsma et al. 2012). Lastly, a recent study found that constitutive activation of PDFR caused a 5- to 10-mV depolarization in the resting membrane potential of circadian pacemaker neurons (Choi et al. 2012). However, it was unclear if this electrical alteration was due to acute effects of PDFR activation, compensatory changes due to chronic receptor stimulation, or developmental abnormalities.
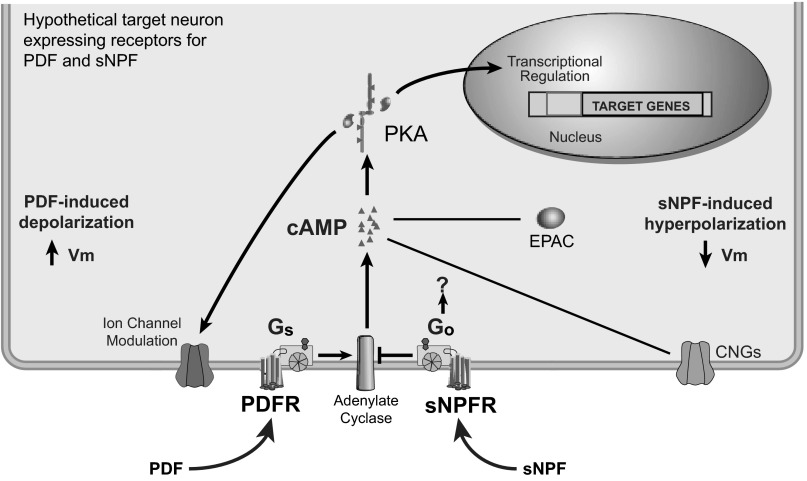
Using the motor neuron model system, we have been able to show for the first time that acute PDF exposure can be directly depolarizing, increasing excitability in cells expressing PDFR, the one identified receptor for PDF (see model in Fig. 9). This extends our understanding of how PDF influences target cells, predicting that PDF is capable of increasing excitability in its direct target cells, such as cells within the central clock (Duvall and Taghert 2012; Shafer et al. 2008) and the integrative locomotor control center, the ellipsoid body (Pírez et al. 2013). Activation of PDFR in motor neurons or in muscles also induced a strong cAMP response, confirming previous reports that PDFR signals via the cAMP pathway (Duvall and Taghert 2012; Hyun et al. 2005; Mertens et al. 2005; Shafer et al. 2008). Due to the functional and structural homology between PDFR and the mammalian receptor for VIP VPAC2 (Aton et al. 2005; Mertens et al. 2005; Pakhotin et al. 2006; Talsma et al. 2012; Vosko et al. 2007), our work will prove useful in understanding the similarities in brain circuits that control sleep in insects and humans.
Fig. 9.
Model diagram summarizing the identified signaling pathways downstream of the receptors for PDF and sNPF. Shown is a hypothetical target cell expressing receptors for both PDF and sNPF, with cell signaling events depicted as predicted by the current study. PDF increases production of cAMP via Gαs-mediated stimulation of adenylate cyclase. This induction couples to as-yet unidentified downstream effectors [likely PKA, EPAC, or cyclic nucleotide-gated channels (CNGs)] to alter membrane physiology, resulting in depolarizat ion and thus increased excitability. On the other hand, sNPF acts almost exclusively via Gαo, reducing cAMP levels through action on an as-yet-unidentified adenylate cyclase and resulting in membrane hyperpolarization and thus reduced excitability. It remains unclear which ion channels are modulated to produce the observed electrical effects or if either PDF or sNPF alter transcription to produce long-term changes in cellular function.
We have also shown for the first time that acute sNPF treatment can hyperpolarize Drosophila neurons expressing sNPFR, via a Gαo-dependent pathway (see model in Fig. 9). A previous study in Xenopus oocytes showed that sNPF could act via sNPFR to activate G-protein-coupled inward-rectifying potassium channels (GIRKs), although the targeted ion channels had to be coexpressed to observe any response (Reale et al. 2004). The authors found that GIRK activation was reduced by PTX preincubation, similar to our own findings. GIRK modulation by GPCR typically relies on Gβγ signaling, although the Gα subunit may be important for directing the specific downstream targets (reviewed in Betke et al. 2012). In this study, targeting Gβ or Gγ subunits by RNAi had no significant impact on the physiological effects of sNPF. It is possible that due to redundancy or promiscuity among the Gβ and Gγ subunits, multiple subunits would have to be reduced before an effect would be observed. However, our findings that inhibition of Gαo by two separate methods effectively blocked sNPF strongly suggest that Gαo is the crucial regulator of sNPF effects downstream of sNPFR.
Gαo has been shown to inhibit cAMP production, although it can also have cAMP-independent effects (reviewed in Jiang and Bajpayee 2009). In this study, we found that sNPFR activation reduced cAMP levels, but the effect was small, and it is certainly possible that Gαo targets additional downstream signaling mechanisms. Guo et al. (2011) found that hyperactivation of Gαo augmented sleep and this effect persisted in the adenylate cyclase mutant rutabaga or in the presence of inhibitors of PKA or CREB. The authors concluded that Gαo was acting in a cAMP-independent manner. However, the Drosophila genome contains several adenylate cyclase genes apart from rutabaga, and not all cAMP signaling is mediated by PKA/CREB. Thus future investigations will be necessary to determine conclusively which signaling pathways downstream of sNPFR/Gαo produce physiological inhibition.
The results described here help clarify the physiological effects of PDF and sNPF, neuropeptides that regulate processes such as odor sensitivity, feeding, metabolism, sleep, circadian rhythms, and locomotion. Because sNPF is expressed in at least two clusters of central brain neurons that control circadian rhythms (Johard et al. 2009), and because previous findings from our laboratory show that sNPF promotes sleep (Shang et al. 2013), sNPF may be an important signal within the circadian clock or as an output from the clock to regulate behavior. Our calcium imaging studies show that sNPF can hyperpolarize and thus silence rhythmic activity in neuronal networks. Future studies should build off of this work to determine if sNPF targets and inhibits wake-promoting neurons and if they utilize Gαo signaling to respond to sNPF. It will also be of interest to determine if sNPF acts via Gαo or through a different mechanism within brain circuits that control other behaviors such as feeding.
GRANTS
Financial support was provided by National Institute of Mental Health Grants F32-MH-090711 (to C. G. Vecsey) and R01-MH-067284 (to L. C. Griffith).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: C.G.V. and L.C.G. conception and design of research; C.G.V. and N.P. performed experiments; C.G.V. and N.P. analyzed data; C.G.V., N.P., and L.C.G. interpreted results of experiments; C.G.V. prepared figures; C.G.V. drafted manuscript; C.G.V., N.P., and L.C.G. edited and revised manuscript; C.G.V., N.P., and L.C.G. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Gregg Roman, Orie Shafer, Paul Taghert, and the Janelia Farm Research Campus for generously providing fly lines for these experiments; Gina Turrigiano, Sandrine Lefort, and Anne Joseph for the use of their CCD camera and expertise in setting it up; Stefan Pulver for providing larval motor neuron recording schematics; and Nathan Donelson for comments on drafts of this manuscript.
Current address for C. G. Vecsey: Biology Department, Swarthmore College, 500 College Avenue, Swarthmore, PA, 19081.
Current address for N. Pírez: Fundación Instituto Leloir and IIBBA-CONICET, Av. Patricias Argentinas 435 (1405-BWE), Buenos Aires, Argentina.
REFERENCES
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci 8: 476–483, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B. KO's and organisation of peptidergic feeding behavior mechanisms. Neurosci Biobehav Rev 25: 143–158, 2001 [DOI] [PubMed] [Google Scholar]
- Betke KM, Wells CA, Hamm HE. GPCR mediated regulation of synaptic transmission. Progr Neurobiol 96: 304–321, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caers J, Verlinden H, Zels S, Vandersmissen HP, Vuerinckx K, Schoofs L. More than two decades of research on insect neuropeptide GPCRs: an overview. Front Endocrinol (Lausanne) 3: 151, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson MA, Enell LE, Nassel DR. Distribution of short neuropeptide F and its receptor in neuronal circuits related to feeding in larval Drosophila. Cell Tissue Res 353: 511–523, 2013 [DOI] [PubMed] [Google Scholar]
- Cattaert D, Birman S. Blockade of the central generator of locomotor rhythm by noncompetitive NMDA receptor antagonists in Drosophila larvae. J Neurobiol 48: 58–73, 2001 [DOI] [PubMed] [Google Scholar]
- Chemelli RM, Willie JT, Sinton CM, Elmquist JK, Scammell T, Lee C, Richardson JA, Williams SC, Xiong Y, Kisanuki Y, Fitch TE, Nakazato M, Hammer RE, Saper CB, Yanagisawa M. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell 98: 437–451, 1999 [DOI] [PubMed] [Google Scholar]
- Chen W, Shi W, Li L, Zheng Z, Li T, Bai W, Zhao Z. Regulation of sleep by the short neuropeptide F (sNPF) in Drosophila melanogaster. Insect Biochem Mol Biol 43: 809–819, 2013 [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet 39: 715–720, 2007 [DOI] [PubMed] [Google Scholar]
- Choi C, Cao G, Tanenhaus AK, McCarthy EV, Jung M, Schleyer W, Shang Y, Rosbash M, Yin JC, Nitabach MN. Autoreceptor control of peptide/neurotransmitter corelease from PDF neurons determines allocation of circadian activity in drosophila. Cell Rep 2: 332–344, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Fortin JP, McCarthy E, Oksman L, Kopin AS, Nitabach MN. Cellular dissection of circadian peptide signals with genetically encoded membrane-tethered ligands. Curr Biol 19: 1167–1175, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JC, Park D, Griffith LC. Electrophysiological and morphological characterization of identified motor neurons in the Drosophila third instar larva central nervous system. J Neurol 91: 2353–2365, 2004 [DOI] [PubMed] [Google Scholar]
- Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R. The GABA(A) receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol 19: 386–390, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clynen E, Reumer A, Baggerman G, Mertens I, Schoofs L. Neuropeptide biology in Drosophila. Adv Exp Med Biol 692: 192–210, 2010 [DOI] [PubMed] [Google Scholar]
- de Lecea L, Kilduff TS, Peyron C, Gao X, Foye PE, Danielson PE, Fukuhara C, Battenberg EL, Gautvik VT, Bartlett FS, Frankel WN, 2nd, van den Pol AN, Bloom FE, Gautvik KM, Sutcliffe JG. The hypocretins: hypothalamus-specific peptides with neuroexcitatory activity. Proc Natl Acad Sci USA 95: 322–327, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JB. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis 34: 1–15, 2002 [DOI] [PubMed] [Google Scholar]
- Duvall LB, Taghert PH. The circadian neuropeptide PDF signals preferentially through a specific adenylate cyclase isoform AC3 in M pacemakers of Drosophila. PLoS Biol 10: e1001337, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall LB, Taghert PH. E and M circadian pacemaker neurons use different PDF receptor signalosome components in Drosophila. J Biol Rhythms 28: 239–248, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyzma M, Boudjeltia KZ, Faraut B, Kerkhofs M. Neuropeptide Y and sleep. Sleep Med Rev 14: 161–165, 2010 [DOI] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using microManager. Curr Protoc Mol Biol 14: Unit 14.20, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Reale V, Chatwin H, Kennedy K, Venard R, Ericsson C, Yu K, Evans PD, Hall LM. Functional characterization of a neuropeptide F-like receptor from Drosophila melanogaster. Eur J Neurosci 18: 227–238, 2003 [DOI] [PubMed] [Google Scholar]
- Ferris J, Ge H, Liu L, Roman G. G(o) signaling is required for Drosophila associative learning. Nat Neurosci 9: 1036–1040, 2006 [DOI] [PubMed] [Google Scholar]
- Fox LE, Soll DR, Wu CF. Coordination and modulation of locomotion pattern generators in Drosophila larvae: effects of altered biogenic amine levels by the tyramine beta hydroxlyase mutation. J Neurosci 26: 1486–1498, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garczynski SF, Brown MR, Crim JW. Structural studies of Drosophila short neuropeptide F: occurrence and receptor binding activity. Peptides 27: 575–582, 2006 [DOI] [PubMed] [Google Scholar]
- Guo F, Yi W, Zhou M, Guo A. Go signaling in mushroom bodies regulates sleep in Drosophila. Sleep 34: 273–281, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison NL. Mechanisms of sleep induction by GABA(A) receptor agonists. J Clin Psychiatry 68, Suppl 5: 6–12, 2007 [PubMed] [Google Scholar]
- Helfrich-Forster C. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc Natl Acad Sci USA 92: 612–616, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry SH, Schwark HD, Jones EG, Yan J. Numbers and proportions of GABA-immunoreactive neurons in different areas of monkey cerebral cortex. J Neurosci 7: 1503–1519, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewes RS, Taghert PH. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res 11: 1126–1142, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Lee KS, Kwak SJ, Kim AK, Bai H, Jung MS, Kwon OY, Song WJ, Tatar M, Yu K. Minibrain/Dyrk1a regulates food intake through the Sir2-FOXO-sNPF/NPY pathway in Drosophila and mammals. PLoS Genetics 8: e1002857, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun S, Lee Y, Hong ST, Bang S, Paik D, Kang J, Shin J, Lee J, Jeon K, Hwang S, Bae E, Kim J. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron 48: 267–278, 2005 [DOI] [PubMed] [Google Scholar]
- Im SH, Taghert PH. PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J Comp Neurol 518: 1925–1945, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotech 21: 635–637, 2003 [DOI] [PubMed] [Google Scholar]
- Jiang M, Bajpayee NS. Molecular mechanisms of go signaling. Neurosignals 17: 23–41, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johard HA, Yoishii T, Dircksen H, Cusumano P, Rouyer F, Helfrich-Forster C, Nassel DR. Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J Comp Neurol 516: 59–73, 2009 [DOI] [PubMed] [Google Scholar]
- Kapan N, Lushchak OV, Luo J, Nassel DR. Identified peptidergic neurons in the Drosophila brain regulate insulin-producing cells, stress responses and metabolism by coexpressed short neuropeptide F and corazonin. Cell Mol Life Sci 2012. July 25 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katanayeva N, Kopein D, Portmann R, Hess D, Katanaev VL. Competing activities of heterotrimeric G proteins in Drosophila wing maturation. PLoS One 5: e12331, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larhammar D, Salaneck E. Molecular evolution of NPY receptor subtypes. Neuropeptides 38: 141–151, 2004 [DOI] [PubMed] [Google Scholar]
- Lawrence CB, Turnbull AV, Rothwell NJ. Hypothalamic control of feeding. Curr Opin Neurobiol 9: 778–783, 1999 [DOI] [PubMed] [Google Scholar]
- Lear BC, Merrill CE, Lin JM, Schroeder A, Zhang L, Allada R. A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron 48: 221–227, 2005 [DOI] [PubMed] [Google Scholar]
- Lear BC, Zhang L, Allada R. The neuropeptide PDF acts directly on evening pacemaker neurons to regulate multiple features of circadian behavior. PLoS Biol 7: e1000154, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Hong SH, Kim AK, Ju SK, Kwon OY, Yu K. Processed short neuropeptide F peptides regulate growth through the ERK-insulin pathway in Drosophila melanogaster. FEBS Lett 583: 2573–2577, 2009 [DOI] [PubMed] [Google Scholar]
- Lee KS, Kwon OY, Lee JH, Kwon K, Min KJ, Jung SA, Kim AK, You KH, Tatar M, Yu K. Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat Cell Biol 10: 468–475, 2008 [DOI] [PubMed] [Google Scholar]
- Lee KS, You KH, Choo JK, Han YM, Yu K. Drosophila short neuropeptide F regulates food intake and body size. J Biol Chem 279: 50781–50789, 2004 [DOI] [PubMed] [Google Scholar]
- Lin L, Faraco J, Li R, Kadotani H, Rogers W, Lin X, Qiu X, de Jong PJ, Nishino S, Mignot E. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell 98: 365–376, 1999 [DOI] [PubMed] [Google Scholar]
- Mahr A, Aberle H. The expression pattern of the Drosophila vesicular glutamate transporter: a marker protein for motoneurons and glutamatergic centers in the brain. Gene Expr 6: 299–309, 2006 [DOI] [PubMed] [Google Scholar]
- Mertens I, Meeusen T, Huybrechts R, De Loof A, Schoofs L. Characterization of the short neuropeptide F receptor from Drosophila melanogaster. Biochem Biophys Res Commun 297: 1140–1148, 2002 [DOI] [PubMed] [Google Scholar]
- Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron 48: 213–219, 2005 [DOI] [PubMed] [Google Scholar]
- Nassel DR, Enell LE, Santos JG, Wegener C, Johard HA. A large population of diverse neurons in the Drosophila central nervous system expresses short neuropeptide F, suggesting multiple distributed peptide functions. BMC Neurosci 9: 90, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassel DR, Wegener C. A comparative review of short and long neuropeptide F signaling in invertebrates: Any similarities to vertebrate neuropeptide Y signaling? Peptides 32: 1335–1355, 2011 [DOI] [PubMed] [Google Scholar]
- Nassel DR, Winther AM. Drosophila neuropeptides in regulation of physiology and behavior. Progr Neurobiol 92: 42–104, 2010 [DOI] [PubMed] [Google Scholar]
- Nikolaev VO, Bunemann M, Hein L, Hannawacker A, Lohse MJ. Novel single chain cAMP sensors for receptor-induced signal propagation. J Biol Chem 279: 37215–37218, 2004 [DOI] [PubMed] [Google Scholar]
- Nikolaev VO, Lohse MJ. Monitoring of cAMP synthesis and degradation in living cells. Physiology (Bethesda, Md) 21: 86–92, 2006 [DOI] [PubMed] [Google Scholar]
- Pakhotin P, Harmar AJ, Verkhratsky A, Piggins H. VIP receptors control excitability of suprachiasmatic nuclei neurones. Pflügers Arch 452: 7–15, 2006 [DOI] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M, Griffith LC. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60: 672–682, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson MG, Eklund MB, Dircksen H, Muren JE, Nassel DR. Pigment-dispersing factor in the locust abdominal ganglia may have roles as circulating neurohormone and central neuromodulator. J Neurobiol 48: 19–41, 2001 [DOI] [PubMed] [Google Scholar]
- Pírez N, Christmann BL, Griffith LC. Daily rhythms in locomotor circuits in Drosophila involve pigment-dispersing factor (PDF). J Neurophysiol 110: 700–708, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver SR, Griffith LC. Spike integration and cellular memory in a rhythmic network from Na+/K+ pump current dynamics. Nat Neurosci 13: 53–59, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reale V, Chatwin HM, Evans PD. The activation of G-protein gated inwardly rectifying K+ channels by a cloned Drosophila melanogaster neuropeptide F-like receptor. Eur J Neurosci 19: 570–576, 2004 [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99: 791–802, 1999 [DOI] [PubMed] [Google Scholar]
- Rohrbough J, Broadie K. Electrophysiological analysis of synaptic transmission in central neurons of Drosophila larvae. J Neurol 88: 847–860, 2002 [DOI] [PubMed] [Google Scholar]
- Root CM, Ko KI, Jafari A, Wang JW. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell 145: 133–144, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryglewski S, Lance K, Levine RB, Duch C. Ca(v)2 channels mediate low and high voltage-activated calcium currents in Drosophila motoneurons. J Physiol 590: 809–825, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara S, Yanagawa Y, O'Leary DD, Stevens CF. The fraction of cortical GABAergic neurons is constant from near the start of cortical neurogenesis to adulthood. J Neurosci 32: 4755–4761, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Amemiya A, Ishii M, Matsuzaki I, Chemelli RM, Tanaka H, Williams SC, Richarson JA, Kozlowski GP, Wilson S, Arch JR, Buckingham RE, Haynes AC, Carr SA, Annan RS, McNulty DE, Liu WS, Terrett JA, Elshourbagy NA, Bergsma DJ, Yanagisawa M. Orexins and orexin receptors: a family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell 92: 1 page following 696, 1998 [DOI] [PubMed] [Google Scholar]
- Schaefer JE, Worrell JW, Levine RB. Role of intrinsic properties in Drosophila motoneuron recruitment during fictive crawling. J Neurophysiol 104: 1257–1266, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seamon KB, Daly JW. Forskolin: its biological and chemical properties. Adv Cyclic Nucleotide Protein Phosphorylation Res 20: 1–150, 1986 [PubMed] [Google Scholar]
- Seinen E, Burgerhof JG, Jansen RC, Sibon OC. RNAi-induced off-target effects in Drosophila melanogaster: frequencies and solutions. Brief Funct Genomics 10: 206–214, 2011 [DOI] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron 58: 223–237, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Donelson NC, Vecsey CG, Guo F, Rosbash M, Griffith LC. Short neuropeptide F is a sleep-promoting inhibitory modulator. Neuron 80: 171–183, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, Holmes TC. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol 18: 1537–1545, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Lance K, Levine RB. Contribution of EAG to excitability and potassium currents in Drosophila larval motoneurons. J Neurophysiol 107: 2660–2671, 2012a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Lance K, Levine RB. Segmental differences in firing properties and potassium currents in Drosophila larval motoneurons. J Neurophysiol 107: 1356–1365, 2012b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert PH, Nitabach MN. Peptide neuromodulation in invertebrate model systems. Neuron 76: 82–97, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talsma AD, Christov CP, Terriente-Felix A, Linneweber GA, Perea D, Wayland M, Shafer OT, Miguel-Aliaga I. Remote control of renal physiology by the intestinal neuropeptide pigment-dispersing factor in Drosophila. Proc Natl Acad Sci USA 109: 12177–12182, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, Bargmann CI, Jayaraman V, Svoboda K, Looger LL. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods 6: 875–881, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Pol AN. Neuropeptide transmission in brain circuits. Neuron 76: 98–115, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Broeck J. Neuropeptides and their precursors in the fruitfly, Drosophila melanogaster. Peptides 22: 241–254, 2001 [DOI] [PubMed] [Google Scholar]
- Venken KJ, Simpson JH, Bellen HJ. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron 72: 202–230, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosko AM, Schroeder A, Loh DH, Colwell CS. Vasoactive intestinal peptide and the mammalian circadian system. Gen Comp Endocrinol 152: 165–175, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner CA, Friedrich B, Setiawan I, Lang F, Broer S. The use of Xenopus laevis oocytes for the functional characterization of heterologously expressed membrane proteins. Cell Physiol Biochem 10: 1–12, 2000 [DOI] [PubMed] [Google Scholar]
- Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell 112: 271–282, 2003 [DOI] [PubMed] [Google Scholar]
- Wegener C, Reinl T, Jansch L, Predel R. Direct mass spectrometric peptide profiling and fragmentation of larval peptide hormone release sites in Drosophila melanogaster reveals tagma-specific peptide expression and differential processing. J Neurochem 96: 1362–1374, 2006 [DOI] [PubMed] [Google Scholar]
- Worrell JW, Levine RB. Characterization of voltage-dependent Ca2+ currents in identified Drosophila motoneurons in situ. J Neurophysiol 100: 8 68–878, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Cao G, Nitabach MN. Electrical silencing of PDF neurons advances the phase of non-PDF clock neurons in Drosophila. J Biol Rhythms 23: 117–128, 2008 [DOI] [PubMed] [Google Scholar]
- Yoshii T, Wulbeck C, Sehadova H, Veleri S, Bichler D, Stanewsky R, Helfrich-Forster C. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila's clock. J Neurosci 29: 2597–2610, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]