Abstract
Irritable bowel syndrome (IBS) is the most prevalent functional gastrointestinal disorder. It is a multifactorial disorder. Intestinal microbiota may cause the pathogenesis of IBS by contributing to abnormal gastrointestinal motility, low-grade inflammation, visceral hypersensitivity, communication in the gut-brain axis, and so on. Previous attempts to identify the intestinal microbiota composition in IBS patients have yielded inconsistent and occasionally contradictory results. This inconsistency may be due to the differences in the molecular techniques employed, the sample collection and handling methods, use of single samples that are not linked to fluctuating symptoms, or other factors such as patients’ diets and phenotypic characterizations. Despite these difficulties, previous studies found that the intestinal microbiota in some IBS patients was completely different from that in healthy controls, and there does appear to be a consistent theme of Firmicutes enrichment and reduced abundance of Bacteroides. Based on the differences in intestinal microbiota composition, many studies have addressed the roles of microbiota-targeted treatments, such as antibiotics and probiotics, in alleviating certain symptoms of IBS. This review summarizes the current knowledge of the associations between intestinal microbiota and IBS as well as the possible modes of action of intestinal microbiota in the pathogenesis of IBS. Improving the current level of understanding of host-microbiota interactions in IBS is important not only for determining the role of intestinal microbiota in IBS pathogenesis but also for therapeutic modulation of the microbiota.
Keywords: Irritable bowel syndrome, Intestinal microbiota, Dysbiosis, Antibiotics, Probiotics
Core tip: The intestinal microbiota is altered in some Irritable bowel syndrome (IBS) patients, and the symptoms of IBS can be alleviated by treatments that target the microbiota. Over the past several years, many studies have attempted to identify the intestinal microbiota composition in IBS patients and intestinal dysbiosis in IBS is characterized by Firmicutes enrichment and reduced abundance of Bacteroides. Based on the differences in intestinal microbiota composition, the roles of microbiota-targeted treatments, such as antibiotics and probiotics, were investigated in alleviating certain symptoms of IBS.
INTRODUCTION
Irritable bowel syndrome (IBS) is characterized by abdominal discomfort, bloating, and disturbed defecation in the absence of any identifiable abnormalities indicative of organic gastrointestinal disease[1]. IBS is the most commonly diagnosed gastrointestinal disorder, and it accounts for about 30% of all referrals to gastroenterologists[2]. In the general population worldwide, its prevalence has been reported to range from 5% to 25%[1,3-6]. IBS worsens patients’ quality of life significantly, and both patients and healthcare systems incur huge costs toward its treatment[6]. Several treatments and therapies help alleviate the symptoms of IBS; however, they do not cure this condition. Thus, the chronic nature of IBS and the challenge of controlling its symptoms can be frustrating for both patients and healthcare providers[1,2].
IBS is a multifactorial disorder, and its underlying pathophysiology is unclear[1]. Therapeutic strategies have traditionally focused on alterations in gastrointestinal motility and visceral hypersensitivity influenced heavily by stress[7]. However, some drugs that target gastrointestinal motility and visceral hypersensitivity, such as antidepressants, alosetron, and tegaserods, have only a narrow therapeutic window, limiting their clinical application, especially in mild cases of IBS[8]. Therefore, studying the pathophysiology of IBS is important, especially in light of the possibility of developing targeted therapies. More recent studies have focused on the role of altered intestinal microbiota[7,9,10].
Since prospective studies have demonstrated that 3%-36% of enteric infections lead to new, persistent IBS symptoms[10], the concept that gut microbes play an important role in the pathogenesis of IBS was confirmed. Recent studies have demonstrated an unimagined level of complexity in human intestinal microbiota, with thousands of phylotypes, 80% of which remain uncultured[11]. The introduction of culture-independent techniques for studying intestinal microbiota has increased our understanding of the role of intestinal microbiota in human diseases, and emerging studies have demonstrated changes in intestinal microbiota in patients with IBS[12-14]. The restoration of altered intestinal microbiota may be a new therapeutic option for treating IBS[15]. Previous randomized controlled trials (RCTs) have documented that the symptoms of IBS can be improved by treatments that target the microbiota, such as antibiotics and probiotics[7]. Herein, the evidence of associations between the intestinal microbiota composition and IBS is reviewed, and the possible roles of specific microbial groups in IBS management are discussed in light of the most recent findings.
HUMAN INTESTINAL MICROBIOTA
The human body is inhabited by a complex community of microbes that are collectively referred to as human microbiota. The human intestinal microbiota constitutes a complex and metabolically active ecosystem that is now well recognized for its impact on human health and disease[16]. It is estimated that the human microbiota number more than 1014 cells, which exceeds the number of human cells in our bodies[7]. The microbiota is taxonomically classified according to the traditional biological nomenclature (phylum-class-order-family-genus-species), and currently, more than 50 bacterial phyla have been described, of which 10 inhabit the colon and three bacterial phyla, Firmicutes, Bacteroidetes and Actinobacteria predominate[17]. Genotypic sequencing studies based on the 16S ribosomal RNA (16S rRNA)-encoding gene have been used for demonstrating that the human gastrointestinal tract can be populated by any of 1000-1150 different species[18]. Despite this diversity, a core of 18 species was found in all individuals, and 57 were found in 90% of individuals, indicating considerable dominance and inter-individual stability of these species across humans[18]. Faith et al[19] analyzed the fecal microbiota of 37 individuals and found that, on average, 60% of the bacterial strains present remained stable for up to 5 years; many were estimated to remain stable for decades.
Recent analyses of human-associated bacterial diversity have tried to categorize individuals into “enterotypes” based on the abundances of key bacterial genera in the intestinal microbiota[20]. Arumugam et al[21] reported that a set of 22 Sanger-sequenced European fecal metagenomes from Danish, French, Italian, and Spanish individuals was shown to fit into three distinct clusters (enterotypes), each characterized by variations in the numbers of Bacteroides (enterotype 1), Prevotella (enterotype 2), and Ruminococcus (enterotype 3). Recent meta-analysis including the 16S rRNA sequences and whole genome shotgun sequences from the Human Microbiome Project, Metagenomics of the Human Intestinal Tract consortium, and additional studies yielded only bimodal distributions of Bacteroides abundances in gut samples[20]. Enterotype identification depends not only on the structure of the data but also on the methods used for identifying clustering strength[20].
The diversity of intestinal microbiota within and among individuals is strongly influenced by factors such as age, diet, and diseases[9]. In a large cross-sectional study of an elderly population using pyrosequencing, the intestinal microbiota of the elderly subjects was found to be different from that of younger adults, with higher Bacteroides and Clostridia cluster IV, as well as some signature sequences that were present only in older people[22]. The impact of food intake on the microbiota is being explored. Habitual long-term diet has been shown to be strongly associated with enterotype, with protein/animal fat being associated with Bacteroides abundances and carbohydrate being associated with Prevotella abundances[23]. In a comparative study in children from urban Europe and rural Africa, rural African children showed significant enrichment in Bacteroidetes and depletion in Firmicutes, with a unique abundance of bacteria from the genus Prevotella and Xylanibacter, which are known to contain a set of bacterial genes for cellulose and xylan hydrolysis and were completely lacking in the urban European children[24]. In addition, obese individuals show an increase in Firmicutes and a decrease in Bacteroidetes, probably owing partly to differences in diets[25]. Furthermore, manipulation of dietary macronutrients in gnotobiotic mice was shown to account for the majority of the change in their microbiota[26]. Moreover, many dietary prebiotics including oligo-fructose[27], lactulose[28], lupin kernel[29], inulin-containing juices[30], and arabinoxylan-oligosaccharides[31] significantly alter human fecal microbiota.
Characterization of intestinal microbiota, however, has been limited to Western people. A recent study investigated the overall intestinal microbiota composition of 20 Koreans using pyrosequencing[32]. Microbial communities were dominated by five previously identified phyla: Actinobacteria, Firmicutes, Bacteroidetes, Fusobacteria, and Proteobacteria. Cluster analysis showed that the species composition of intestinal microbiota was host-specific and stable over the duration of the test period, but the relative abundance of each species varied among individuals. The results were compared with those of individuals from the United States, China, and Japan, and it was found that human intestinal microbiota differed among countries, but tended to vary less among individual Koreans. The gut microbial composition may be related to the internal and external characteristics of each country member, such as host genetics and dietary patterns[32].
INTESTINAL MICROBIOTA COMPOSITION OF IBS PATIENTS
Numerous diseases have been associated with alterations in the microbiota, which are referred to as dysbiosis, ranging from systemic disorders such as obesity and diabetes to gastrointestinal disorders such as IBS[9,33].The major physiological and immunological functions of the gut cannot be carried out in the absence of the intestinal microbiota[34,35]. The differences in the intestinal microbiota of IBS patients and those of healthy controls have been studied. A previous study that used cultures of fecal material obtained from patients with IBS reported decreased fecal Lactobacilli and Bifidobacteria, increased facultative bacteria dominated by Streptococci and Escherichia coli, as well as higher counts of anaerobic organisms such as Clostridium[36,37]. Traditional microbiology studies and microbial genome sequencing relied upon cultivated clonal cultures. Such culture-based assessment of fecal microbiota is cheap, widely available, and easy to use, but it grossly underestimates fecal populations because more than 80% of the bacteria in the human intestinal tract cannot be cultured using currently available methods[38].
A revolution in DNA sequencing technologies would be to define genetic material recovered directly from environmental samples. Metagenomics refers to culture-independent and sequencing-based studies of the collective set of genomes of mixed microbial communities (metagenomes) with the aim of exploring their compositional and functional characteristics[39]. In 1977, Woese et al[40] identified 16S rRNA, which is a component of the 30S small subunit of prokaryotic ribosomes, having relatively short gene sequences and highly conserved primer binding sites and containing hypervariable regions that can provide species-specific signature sequences useful for bacterial identification. Since then, the molecular profiling of bacterial communities via 16S rRNA-gene based approaches such as terminal restriction fragment length polymorphism, PCR temperature/denaturing gradient gel electrophoresis, and fluorescent in situ hybridization, has been performed[41]. In the last decade, Sanger sequencing was used for generating data in most microbial genomics and metagenomics sequencing projects; however, recent advances in molecular biology have resulted in the application of DNA microarrays and next-generation sequencing (NGS) technologies for studying complex intestinal microbiota. DNA microarrays comprising hundreds or thousands of DNA fragments arrayed on small glass slides were originally developed for gene expression profiling. These were subsequently applied to the study of different aspects of microbial ecology, including total microbial diversity and a range of biogeochemical functions[42]. Alternatively, NGS approaches, including pyrosequencing (introduced by 454 Life Sciences, Inc.) as well as other platforms such as Solexa (Illumina, Inc.) and SOLiD (ABI, Inc.), offer rapid and highly parallel sequencing of many DNA fragments from complex samples or transcriptomes[39]. Pyrosequencing is particularly suited to microbial ecology studies because of its relatively long read length compared with other NGS technologies platforms, and it has therefore been widely adopted by microbial ecology researchers; other platforms have also been recently adopted in this field[42]. Table 1 lists the advantages and disadvantages of the principal techniques used for characterizing intestinal microbiota.
Table 1.
Summary of molecular studies of intestinal microbiota in irritable bowel syndrome
| Ref. | Ethnicity | IBS patients, n | Mean age (range), yr | Male gender, n |
IBS subtype |
Controls, n | Sample | Method | Changes in intestinal microbiota composition in IBS | ||
| IBS-C | IBS-D | IBS-M | |||||||||
| Malinen et al[83] 2005 | Finland | 27 | 46.5 (20-65) | 7 | 9 | 12 | 6 | 22 (age, gender matching) | Feces | qPCR covering bacteria 300 bacterial species | IBS-D: ↓ lactobacillus spp. |
| IBS-C: ↑ veillonella spp. | |||||||||||
| Overall IBS: ↓ clostridium coccoides subgroup, Bifidobacterium catenulatum group | |||||||||||
| Mättö et al[46] 2005 | Finland | 26 | 46 (20-65) | 7 | 9 | 12 | 5 | 25 (age, gender matching) | Feces | Culture, PCR-DGGE | Temporal instability in the bacterial population↑ coliform bacteria |
| ↑ aerob:anaerob ratio | |||||||||||
| Maukonen et al[84] 2006 | Finland | 24 | 45 (24–64) | 5 | 6 | 7 | 3 | 16 | Feces | PCR-DGGE, Transcript analysis with the aid of affinity capture for Clostridial groups | Temporal instability in the bacterial population |
| IBS-C: ↓ clostridium coccoides-Eubacterium rectale group | |||||||||||
| Kassinen et al[43] 2007 | Finland | 24 | 47.3 (21–65) | 5 | 8 | 10 | 6 | 23 (age, gender matching) | Feces | GC-profiling + high-throughput 16S rRNA gene sequencing of 3753 clones | Coverage of the clone libraries of |
| IBS subtypes and control subjects differed | |||||||||||
| Kerckhoffs et al[85] 2009 | The Netherlands | 41 | 42 ± 2.12 | 12 | 11 | 11 | 16 | 26 | Feces, Duodenal mucosa | FISH, qPCR | ↓ bifidobacterium catenulatum |
| Krogius-Kurikka et al[86] 2009 | Finland | 10 | 46.5 | 4 | 0 | 10 | 0 | 22 | Feces | G + C (%G + C) -based profiling and fractioning combined with 16S rRNA gene clone library sequencing of 3267 clones | ↑ proteobacteria |
| ↑ firmicutes | |||||||||||
| ↓ actinobacteria | |||||||||||
| ↓ bacteroidetes | |||||||||||
| Lyra et al[87] 2009 | Finland | 20 | IBS-D: 43.6 (26-60), IBS-C: 48.6 (24-64), IBS-M: 50.8 (31-62) | 6 | 8 | 8 | 4 | 15 | Feces | qPCR | IBS-D: ↑ ruminococcus torques, ↓ clostridium thermosuccinogenes |
| IBS-C: ↑ ruminococcus bromii-like | |||||||||||
| IBS-M: ↓ ruminococcus torques, ↑ clostridium thermosuccinogenes | |||||||||||
| Tana et al[88] 2010 | Japan | 26 | 21.7 ± 2.0 | 13 | 11 | 8 | 7 | 26 (age, gender matching) | Feces | Culture, qPCR | ↑ veillonella spp. |
| ↑ lactobacillus spp. | |||||||||||
| Codling et al[89] 2010 | Ireland | 47 | 43.6 (24–66) | 0 | - | - | - | 33 | Feces, Colonic mucosa | PCR-DGGE | Significantly more variation in the gut microbiota of healthy volunteers than that of IBS patients |
| Ponnusamy et al[90] 2011 | South Korea | 11 | 47.5 (18-74) | 6 | - | -- | 8 | Feces | DGGE + qPCR of 16S rRNA genes | ↑ diversity of Bacteroidetes and Lactobacillus groups | |
| Rinttilä et al[91] 2011 | Finland | 96 | 47 (20-73) | 27 | 15 | 81 | 23 | Feces | qPCR | 17% of IBS samples (n = 15) tested positive for staphylococcus aureus | |
| Rajilić-Stojanović et al[45] 2011 | Finland | 62 | 49 (22−66) | 5 | 18 | 25 | 19 | 42 | Feces | Phylogenetic 16S rRNA microarray and qPCR | 2-fold ↑ firmicutes:Bacteroidetes ratio↓ bacteroidetes, ↑ dorea, ruminococcus, clostridium sppBifidobacterium faecalibacterium spp |
| Carroll et al[51] 2011 | United States | 16 | 35.6 (23–52) | 5 | 0 | 16 | 0 | 21 | Feces, Colonic biopsies | T-RFLP fingerprinting of 16S rRNA-PCR | ↓ microbial biodiversity in D-IBS fecal samples |
| Parkes et al[52] 2012 | United Kingdom | 53 | IBS-D: 36.2 (32.1–40.3), IBS-C: 32.4 (28.1–36.7) | 28 | 26 | 27 | 0 | 26 | Colonic mucosa | FISH, confocal microscopy | Expansion of mucosa-associated microbiota; mainly bacteroides and clostridia; association with IBS subgroups and symptoms |
| Jeffery et al[92] 2012 | Sweden | 37 | 37 ± 12 | 11 | 10 | 15 | 12 | 20 | Feces | Pyrosequencing 16SrRNA | Clustering of IBS patients: normal-like vs abnormal microbiota composition (increase of firmicutes-associated taxa and a depletion of bacteroidetes-related taxa) |
IBS: Irritable bowel syndrome; IBS-D: Diarrhea-predominant irritable bowel syndrome; IBS-C: Constipation-dominant irritable bowel syndrome; IBS-M: Alternating type or mixed irritable bowel syndrome; PCR-DGGE: PCR denaturing gradient gel electrophoresis; FISH: Fluorescent in situ hybridization; qPCR: Quantitative PCR; 16S rRNA: 16S ribosomal RNA.
Studies using culture-independent molecular-based techniques revealed changes in the intestinal microbiota composition in IBS patients compared with those of healthy controls. Thus far, the results of studies on the intestinal microbiota of IBS patients are inconsistent and occasionally, contradictory (Table 1). This inconsistency in results may be ascribed to several reasons, including differences among the various molecular techniques employed, sample collection and handling methods, , as well as definitions of IBS and IBS subtypes[16]. Table 2 lists the advantages and disadvantages of the principal techniques used for characterizing intestinal microbiota. In studying human intestinal microbiota, classical approaches suffer from individual advantages and limitations[7,16]. NSG and phylogenic metagenomics update the bacterial community profiles of patients with IBS. The sample collection method can influence the intestinal microbiota composition. Namely, fecal samples show distal colonic luminal microbiota, whereas biopsy samples show only mucosa-attached microbiota. Although feces or fecal swabs are the most convenient samples, they do not accurately reflect the microbiota composition or activities in the proximal colon. Colon biopsies also do not represent the microbiota in its physiologic state because extensive colon preparation for cleaning intestinal contents removes some of the outer mucus layers and, in turn, the mucosa-attached microbes as well as their normal attachment sites[16]. In addition, different studies used different sample handing methods; some studies used frozen samples, whereas others used fresh samples. The use of single samples cannot be linked to fluctuating symptoms and probably to other factors such as diet and patients’ phenotypic characterization[7]. Although most studies used the Rome criteria for IBS, the proportions of the enrolled numbers of IBS subtypes differed among the studies. There is suggestive evidence of an association of intestinal microbiota in certain IBS subtypes. Kassinen et al[43] pooled fecal samples by an IBS subgroup diarrhea-predominant IBS (IBS-D), constipation-dominant irritable bowel syndrome (IBS-C), and IBS mixed type (IBS-M) and controls, extracted the bacterial DNA, and analyzed it using high-throughput 16S rRNA sequencing. Population analysis found significant differences between each IBS subgroup and controls[43].
Table 2.
Advantages and limitations of the principal techniques used in the characterization of the intestinal microbiota[16,39]
| Advantages | Limitations | |
| Culture | Cheap, easy to use | Limited estimate intestinal microbiota |
| PCR-T/DGGE | High sensitivity in detecting difference in bacterial populations, semi-quantitative | Does not identify bacteria unless bands on the gel are cut out and sequenced |
| FISH | Microbial in situ identification, high sensitivity, quantitative | Few species can be simultaneously detected, only known species are detected |
| T-RFLP | Low cost | Low biodiversity resolution, no species-level identification, not quantitative |
| Quantitative PCR | Can detect small number of bacteria and quantify them | Laborious |
| Phylogenetic microarray | High biodiversity resolution, quantitative | Only known species are detected |
| NGS phylogenetic analysis (e.g., pyrosequencing) | Enormous quantities of data at individual Species level | Very costly, need bioinformatics analysis |
16S rRNA: 16S ribosomal RNA; PCR-T/DGGE: PCR temperature/denaturing gradient gel electrophoresis; FISH: Fluorescent in situ hybridization; T-RFLP: Terminal restriction fragment length polymorphism; qPCR: Quantitative PCR; NGS: Next-generation sequencing.
It is difficult to determine whether alterations in microbiota are the primary events that lead to the development of IBS or merely the secondary effects of the syndrome. Despite these difficulties, previous studies found that the intestinal microbiota of some IBS patients was different from that of healthy controls, and there does appear to be a consistent theme of Firmicutes enrichment and reduced abundance of Bacteroides.
PATHOGENIC ROLE OF INTESTINAL DYSBIOSIS IN IBS
Intestinal microbiota can be divided into two distinct ecosystems: luminal bacteria, which are either dispersed in liquid feces or bound to food particles, and mucosa-associated bacteria, which are bound to a mucus layer adjacent to the intestinal epithelium[16]. Although microbial trafficking will occur between the two ecosystems with a distinct microenvironment, each ecosystem has the potential to play a different role in IBS symptomatology (Figure 1). Luminal microbiota constitutes the majority of the gastrointestinal tract microbiota and plays a crucial role in gut homeostasis. In IBS, luminal microbiota may play a key role in bloating and flatulence through carbohydrate fermentation and gas production. Bacterial fermentation of undigested carbohydrate leads to short-chain fatty acid production, with gaseous byproducts such as carbon dioxide, hydrogen, and methane. The metabolites and toxins of luminal microbiota can modulate the host immune system[44]. Rajilić-Stojanović et al[45] prepared a phylogenetic 16S rRNA microarray and performed qPCR using fecal samples from 62 IBS patients and 46 healthy adults. Adult patients with IBS had a two-fold greater ratio of Firmicutes to Bacteroidetes than controls, resulting from an approximately one-and-a-half-fold increase in the numbers of Dorea, Ruminococcus, and Clostridium spp. In addition, they observed a two-fold decrease in the number of Bacteroidetes and a one-and-a-half-fold decrease in the numbers of Bifidobacterium and Faecalibacterium spp[45]. Furthermore, the instability and temporal variation in the intestinal microbiota of IBS subjects was addressed, and a trend was noted wherein some Clostridium spp. increased and Eubacterium spp. decreased in IBS patients[46].
Figure 1.
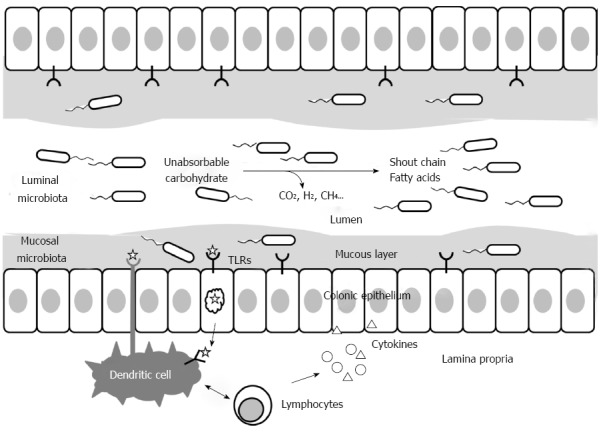
Luminal and mucosal intestinal microbiota and roles in gut homeostasis.
Meanwhile, the mucosal microbiota, although fewer in number, may influence the host via immune-microbial interactions[35]. Recently, mucosal microbiota has attracted increased research interest. Mucosal microbiota is bound to a mucus layer consisting of glycosylated polysaccharides and glycocalyx. The mucus layer contains binding sites for commensal and pathogenic bacteria that help minimize adherence to the intestinal epithelium below. The vast majority of the microbiota is trapped in a complex biofilm containing a diverse population, and only those bacteria that are able to penetrate the mucus and that possess suitable adhesion proteins can directly interface with the apical surface[47]. Luminal interaction occurs via pattern recognition receptors such as toll-like receptors (TLRs) and NOD2. TLRs are expressed on the apical and basolateral membranes of enterocytes and on the processes of dendritic cells that pass from the lamina propria into the lumen through tight enterocyte junctions. Differential expression of TLRs was observed in patients with IBS, with increased TLR-4 and TLR-5 expression and decreased TLR-7 and TLR-8 expression compared with controls[48]. In addition, bacteria can pass through the epithelial layer and are presented to dendritic cells. The pathogenicity of the bacteria determines whether the dendritic cells either auto-induce tolerance via the secretion of anti-inflammatory cytokines such as IL-10 and TGF-β or respond aggressively. Studies have also shown that bacteria such as Bifidobacteria and Lactobacilli stimulate IL-10 and TGF-β production by dendritic cells and inhibit the release of proinflammatory cytokines from dendritic cells[49]. A recent study revealed that some Bifidobacterium strains showed the highest production of IL-17 as well as poor secretion of interferon γ and tumor necrosis factor α, suggesting stimulation of the Th17 pathway[50]. The plasticity of Treg/Th17 populations and the commensal bacteria play a key role in mucosal tolerance and T cell reprogramming[50]. It is, therefore, readily apparent that a disturbance in the mucosal microbiota could lead to an upregulation of the immune system. However, recent studies that examined the mucosal microbiota of IBS patients reported different results. Carroll et al[51] performed microbial community composition analyses on fecal and mucosal samples from patients with IBS-D and healthy controls using terminal-restriction fragment length polymorphism fingerprinting of the bacterial 16S rRNA gene. There were compositional differences in the luminal- and mucosal-associated microbiota of IBS-D patients and those of healthy controls as well as diminished microbial biodiversity in the IBS-D fecal samples. There were no differences in the biodiversities of the mucosal samples of IBS-D patients and healthy controls[51]. In contrast, Parkes et al[52] performed an analysis of frozen rectal biopsies taken at colonoscopy and bacterial quantification by hybridizing frozen sections with bacterial-group-specific oligonucleotide probes. They found expansion of mucosa-associated microbiota in IBS patients, mainly Bacteroides and Clostridia, and association with IBS subgroups and symptoms. In addition, they found that the mucosal Bifidobacteria were lower in IBS-D patients than in controls, together with a negative correlation between mucosal Bifidobacteria and the number of days patients experienced pain or discomfort. However, the studies on the mucosal microbiota of IBS patients are limited because doing so requires endoscopic examination of subjects’ gastrointestinal tracts and carrying out biopsy, unlike the luminal microbiota, which can be readily examined in feces.
Intestinal microbiota may be involved in the pathogenesis of IBS by contributing to abnormal gastrointestinal motility, low-grade inflammation, visceral hypersensitivity, communication in the gut-brain axis, and so on. Lactobacillus paracasei NCC2461 significantly attenuated muscle dysfunction in a murine model of postinfective IBS[53]. The probiotic yeast Saccharomyces boulardii modulated the expression of neuronal markers in the submucous plexus of pigs[54]. There also seems to be an inflammatory component and dysregulation of pro- and anti-inflammatory cytokines in IBS patients[55]. Most interestingly, Bifidobacterium infantis (B. infantis) 35624 was shown to restore the balance of pro- and anti-inflammatory cytokines in patients[56]. Lactobacillus farciminis treatment prevented stress-induced hypersensitivity, increase in colonic paracellular permeability, and colonocyte myosin light chain phosphorylation in rats[57,58]. Modulation of the microbiota induces visceral hypersensitivity in mice, which is reduced by L. paracasei NCC 2461-secreted products[53]. Recently, Rousseaux et al[59] demonstrated that Lactobacillus acidophilus (L. acidophilus) contributes to the modulation and restoration of the normal perception of visceral pain through the NF-κB pathway and by inducing mu-opioid receptor 1 (MOR1) and cannabinoid receptor 2 (CB2) expression. Only the L. acidophilus NCFM strain was able to induce a significant in vitro expression of MOR1 and CB2 messenger in RNA and protein, respectively. To confirm these results in vivo, the researchers administered L. acidophilus NCFM orally to rats and mice at a clinically relevant concentration (109 CFU) and compared colonic samples from these rodents with those from untreated control rodents. MOR1 and CB2 expression was induced in 25%-60% of the intestinal epithelial cells from treated animals compared with only 0%-20% of those from the control group. In addition, visceral perception was assessed in rats using colorectal distension. Oral administration of the L. acidophilus NCFM strain for 15 d decreased normal visceral perception in the rats and increased their pain threshold by 20%. In further experiments of chronic colonic hypersensitivity on a rat model, treatment with L. acidophilus NCFM resulted in an analgesic effect similar to that of 1 mg morphine administered subcutaneously, thus increasing the colorectal distension threshold by 44% compared with that in untreated rats[59]. Transient perturbation of the microbiota with antimicrobials alters brain-derived neurotrophic factor expression, exploratory behavior, and colonization of germ-free mice, suggesting that the impact of the intestinal microbiota is not limited to the gut and the immune system[60].
SMALL INTESTINAL BACTERIAL OVERGROWTH AND ANTIBIOTICS
Since Pimentel et al[61] reported that 84% of IBS patients had small intestinal bacterial overgrowth (SIBO) and that patients with IBS were over 26 times more likely to harbor SIBO than controls, the potential role of SIBO in IBS pathogenesis has gained considerable research attention[62]. In addition, bacterial fermentation in IBS has been highlighted in recent studies on SIBO[16]. Bacterial overgrowth in stagnant sections of the small intestine leads to malabsorption, diarrhea, bloating, and pain, and it can be treated with antibiotics. However, a subsequent study on the SIBO-IBS link showed similar results, whereas other studies were unable to establish an association[62].
A SIBO diagnosis test includes jejuna aspirate and culture, 14C-xylose breath test, and hydrogen (H2) breath tests (HBT) using either glucose (GHBT) or lactulose (LHBT) as the substrate. Jejunal aspirate and culture is considered as the gold standard (> 105 CFU after 48 h of culture); however, it is invasive and time consuming. In contrast, HBT is noninvasive and cheap, but prone to error. Following the ingestion of glucose or lactulose, serial breath H2 measurements are performed. SIBO is defined by either a rise in H2 > 20 ppm in < 90 min or a “double peak” demonstrating distinct small intestinal and colonic bacterial populations[63]. Meta-analysis of 12 studies containing 1921 subjects meeting the Rome criteria for IBS revealed that the pooled prevalence of a positive LHBT or GHBT was 54% (95%CI: 32%-76%) and 31% (95%CI: 14%-50%), respectively, but showed marked statistical heterogeneity between study results[64]. In addition, the prevalence of a positive jejunal aspirate and culture was only 4% (95%CI: 2%-9%). These results suggested that it is premature to accept a firm etiologic link between SIBO and IBS. Moreover, despite a decade of investigation on the relationship between SIBO and IBS, it remains unclear whether SIBO causes IBS or is a bystander of something else altogether[62].
However, the idea of treating IBS patients with an antibiotic was developed as a consequence of the SIBO concept[65]. Neomycin therapy eradicated SIBO and reduced symptoms of IBS[61,66]. Considering the chronic, relapsing nature of IBS and the undesirability of long-term systemic antibiotic therapy, the efficacy of rifaximin, a nonabsorbable antibiotic, began to be explored in IBS[67]. In a RCT, rifaximin treatment for 10 d resulted in symptom improvement that lasted for up to 10 wk in some IBS patients who did not document bacterial overgrowth[68]. Subsequently, a double-blind, placebo-controlled trial phase III study reported that rifaximin treatment for 2 wk provided significant relief from IBS symptoms such as bloating, abdominal pain, and loose or watery stools[69]. A recent meta-analysis of 5 studies found rifaximin to be efficacious for global IBS symptom improvement (OR = 1.57, 95%CI: 1.22-2.01) and more likely to improve bloating (OR = 1.55, 95%CI: 1.23-1.96) compared with a placebo[70].
EVIDENCE OF THE ROLE OF POTENTIALLY PROBIOTIC BACTERIA IN IBS
An improved understanding of host-microbiota interactions in IBS is not only important for its pathogenesis but also for assessing the possible benefits of potential probiotic strains in IBS management. Probiotics are defined as live organisms that when ingested in adequate amounts yield a health benefit to the host[9]. Clinically acceptable probiotics should be species-specific; should be of human origin; should survive passage from the oral cavity through the gastric acid barrier, digestive enzymes, and bile acids; should travel down the small bowel into the colon; nidate; and should proliferate therein[54]. Probiotics offer protection against potential pathogens through enhancement of mucosal barrier function by secreting mucins; providing colonization resistance; producing bacteriocins; increasing production of secretory immunoglobulin A; producing a balanced T-helper cell response; and increasing production of IL-10 and TGF-β, both of which play a role in the development of immunologic tolerance to antigens. For example, a specific strain of B. infantis 35624 has been shown to prevent NF-κB and IL-8 activation as well as to inhibit the secretion of chemokine ligand 20 in response to Salmonella typhimurium, Clostridium difficile, and Mycobacterium paratuberculosis[71]. Current evidence suggests that probiotic effects are strain-specific[72].
Probiotics should be administered at an adequate dose, preferably greater than 10 billion CFU/g in adults; their viability and concentration should be maintained; and they should have a dependably measurable shelf life at the time of purchase and administration. When these criteria are fulfilled, randomized, placebo-controlled, double-blind trials should be performed on an appropriate population. Five systematic reviews with RCTs of adult IBS patients were published[73-77]. Most of the meta-analyses indicated a beneficial effect of probiotics on global symptoms, abdominal pain, and flatulence, whereas the influence on bloating was equivocal (Table 3). However, aggregation of the effects of different probiotics into a meta-analysis should be undertaken with caution. Different probiotics have different microbiological characteristics, which inevitably influence their efficacy. The most commonly studied probiotic species are Lactobacilli and Bifidobacteria. Products range in delivery systems (e.g., yogurts, fermented milk drinks, powders, and capsules) and dose (106-1010 CFU). Lactobacillus plantarum, B. infantis, and VSL 3 (Lactobacillus casei, L. plantarum, L. acidophilus, Lactobacillus delbrueckii, Bifidobacterium longum, Bifidobacterium breve, B. infantis, and Streptococcus salivarius) have demonstrated efficacy in patients with IBS[56,78,79].
Table 3.
Systemic reviews for randomized controlled trials of probiotics in irritable bowel syndrome
| Ref. | Selection criteria | n of identified studies | Results |
| McFarland et al[73] 2008 | RCTs in humans published as full articles or meeting abstracts in peer-reviewed journals | 20 RCTs | Global IBS symptoms: RR = 0.77 (95%CI: 0.62-0.94)/ abdominal pain: RR = 0.78 (95%CI: 0.69-0.88) |
| Brenner et al[76] 2009 | RCTs; adults with IBS defined by Manning or Rome II criteria; single or combination probiotic vs placebo; improvement in IBS symptoms and/or decrease in frequency of adverse events reported | 16 RCTs → 11 studies showed suboptimal study design | Bifidobacterium infantis 35624 has shown efficacy for improvement of IBS symptoms. Most RCTs about the utility of probiotics in IBS have not used an appropriate study design |
| Hoveyda et al[74] 2009 | RCTs compared the effects of any probiotic therapy with placebo in patients with IBS | 14 RCTs → 7 RCTs providing outcomes as dichotomous variable and 6 RCTs providing outcomes as continuous variable | Overall symptoms: dichtomous data - OR = 0.63 (95%CI: 0.45-0.83)/continuous data - mean ± SD, 0.23 (95%CI: 0.07-0.38) Trials varied in relation to the length of treatment (4-26 wk), dose, organisms and strengths of probiotics used |
| Moayyedi et al[75] 2010 | RCTs comparing the effect of probiotics with placebo or no treatment in adult patients with IBS (over the age of 16 yr) | 19 RCTs → 10 RCTs providing outcomes as a dichotomous variable | Probiotics appear to be efficacious in IBS (Probiotics were statistically significantly better than placebo, but there was statistically significant heterogeneity). The magnitude of benefit and the most effective species and strain are uncertain |
| Ortiz-Lucas et al[77] 2013 | RCTs comparing probiotics with placebo in treating IBS symptoms | 24 RCTs → 10 RCTs providing continuous data performed with continuous data summarized using mean ± SD and 95%CIs | Pain scores: improved by probiotics containing Bifidobacterium breve, Bifidobacterium longum, or Lactobacillus acidophilus species Distension scores: improved by probiotics containing B. breve, B. infantis, Lactobacillus casei, or Lactobacillus plantarum species Flatulence: improved by probiotics containing B. breve, B. infantis, L. casei, L. plantarum, B. longum, L. acidophilus, Lactobacillus bulgaricus, and Streptococcus salivarius ssp. thermophilus |
IBS: Irritable bowel syndrome; RCT: Randomized controlled trial.
Recently, we isolated have been isolated new strains, i.e., L. acidophilus-SDC 2012, 2013, from Korean infants’ feces[8]. In Korea, the prevalence of IBS is reported to be around 2.2%-6.6%[1], while that in Western countries is around 10%-20%[2]. Based on the relatively lower prevalence of IBS in Korea and previous reports on the efficacy of probiotics for treating IBS symptoms, we hypothesized that the newly isolated L. acidophilus-SDC 2012, 2013 may help control the symptoms of IBS patients. The result of our RCT showed that L. acidophilus-SDC 2012, 2013 were effective in alleviating IBS symptoms, irrespective of the bowel habit subtype[8]. Although Lactobacilli or Bifidobacteria have demonstrated efficacy in IBS patients, the benefits of one given species or organism have not been found to be better than that of other species or organisms. In an RCT of composite probiotics, Kim et al[80] reported that VSL3 reduced flatulence and retarded colonic transit without altering bowel function in patients with IBS and bloating.
Recent guidelines published by the British Dietetic Association have therefore made strain-specific recommendations considering the limited weak evidence for B. lactis DN 173010 in improving overall symptoms, abdominal pain, and urgency in constipation-predominant IBS and the limited weak evidence for VSL3 in reducingg flatulence in IBS patients[32]. People with IBS who choose to try probiotics should be advised to consume a given product for at least 4 wk while monitoring the effect. Probiotics should be consumed at the dose recommended by the manufacturer[75,76,81].
A number of RCTs have been performed for investigating the effectiveness of probiotics in IBS. However, most RCTs of probiotics had a suboptimal study design with inadequate blinding, trial length, sample size, and/or lack of intention-to-treat analysis. Despite these limitations, there is a possibility of greater efficacy of probiotics in patients whose IBS pathogenesis is known to be related to the intestinal microbiota. In addition, the probiotics include strains present in normal intestinal microbiota, and probiotic-associated adverse events are very rare. Thus, probiotics are good candidates for controlling the symptoms of IBS, especially when treatment safety is paramount in a nonlethal disorder such as IBS[82]. The evidence from clinical trials and systematic reviews are largely supportive of the use of specific probiotics strains in IBS[9].
CONCLUSION
Multiple recent studies have consistently proven that intestinal dysbiosis is associated with this IBS. An improved understanding of host-microbiota interactions in IBS is important not only for determining its pathogenesis but also for enabling therapeutic modulation of the microbiota. In addition, such evidence has encouraged investigations of the potential roles of antibiotics and probiotics in this disorder. Although the interactions of microbiota-targeted treatments with the host immune and visceral nervous systems are yet to be fully understood, they have the potential to play a key role in the management of IBS.
Footnotes
P- Reviewers: Ducrotte P, Krogsgaard LR, Lee YY S- Editor: Qi Y L- Editor: A E- Editor: Wang CH
References
- 1.Rhee PL. Definition and epidemiology of irritable bowel syndrome. Korean J Gastroenterol. 2006;47:94–100. [PubMed] [Google Scholar]
- 2.Drossman DA, Camilleri M, Mayer EA, Whitehead WE. AGA technical review on irritable bowel syndrome. Gastroenterology. 2002;123:2108–2131. doi: 10.1053/gast.2002.37095. [DOI] [PubMed] [Google Scholar]
- 3.Wilson S, Roberts L, Roalfe A, Bridge P, Singh S. Prevalence of irritable bowel syndrome: a community survey. Br J Gen Pract. 2004;54:495–502. [PMC free article] [PubMed] [Google Scholar]
- 4.Gwee KA, Wee S, Wong ML, Png DJ. The prevalence, symptom characteristics, and impact of irritable bowel syndrome in an asian urban community. Am J Gastroenterol. 2004;99:924–931. doi: 10.1111/j.1572-0241.2004.04161.x. [DOI] [PubMed] [Google Scholar]
- 5.Saito YA, Schoenfeld P, Locke GR. The epidemiology of irritable bowel syndrome in North America: a systematic review. Am J Gastroenterol. 2002;97:1910–1915. doi: 10.1111/j.1572-0241.2002.05913.x. [DOI] [PubMed] [Google Scholar]
- 6.Koloski NA, Talley NJ, Boyce PM. Epidemiology and health care seeking in the functional GI disorders: a population-based study. Am J Gastroenterol. 2002;97:2290–2299. doi: 10.1111/j.1572-0241.2002.05783.x. [DOI] [PubMed] [Google Scholar]
- 7.Simrén M, Barbara G, Flint HJ, Spiegel BM, Spiller RC, Vanner S, Verdu EF, Whorwell PJ, Zoetendal EG. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut. 2013;62:159–176. doi: 10.1136/gutjnl-2012-302167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinn DH, Song JH, Kim HJ, Lee JH, Son HJ, Chang DK, Kim YH, Kim JJ, Rhee JC, Rhee PL. Therapeutic effect of Lactobacillus acidophilus-SDC 2012, 2013 in patients with irritable bowel syndrome. Dig Dis Sci. 2008;53:2714–2718. doi: 10.1007/s10620-007-0196-4. [DOI] [PubMed] [Google Scholar]
- 9.Whelan K, Quigley EM. Probiotics in the management of irritable bowel syndrome and inflammatory bowel disease. Curr Opin Gastroenterol. 2013;29:184–189. doi: 10.1097/MOG.0b013e32835d7bba. [DOI] [PubMed] [Google Scholar]
- 10.Spiller R, Garsed K. Postinfectious irritable bowel syndrome. Gastroenterology. 2009;136:1979–1988. doi: 10.1053/j.gastro.2009.02.074. [DOI] [PubMed] [Google Scholar]
- 11.Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–1615. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]
- 12.Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141:1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chassard C, Dapoigny M, Scott KP, Crouzet L, Del’homme C, Marquet P, Martin JC, Pickering G, Ardid D, Eschalier A, et al. Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment Pharmacol Ther. 2012;35:828–838. doi: 10.1111/j.1365-2036.2012.05007.x. [DOI] [PubMed] [Google Scholar]
- 14.Carroll IM, Ringel-Kulka T, Siddle JP, Ringel Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:521–530, e248. doi: 10.1111/j.1365-2982.2012.01891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salonen A, de Vos WM, Palva A. Gastrointestinal microbiota in irritable bowel syndrome: present state and perspectives. Microbiology. 2010;156:3205–3215. doi: 10.1099/mic.0.043257-0. [DOI] [PubMed] [Google Scholar]
- 16.Parkes GC, Brostoff J, Whelan K, Sanderson JD. Gastrointestinal microbiota in irritable bowel syndrome: their role in its pathogenesis and treatment. Am J Gastroenterol. 2008;103:1557–1567. doi: 10.1111/j.1572-0241.2008.01869.x. [DOI] [PubMed] [Google Scholar]
- 17.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA. 2011;108 Suppl 1:4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis S, Burmeister S, Brazier J. Effect of the prebiotic oligofructose on relapse of Clostridium difficile-associated diarrhea: a randomized, controlled study. Clin Gastroenterol Hepatol. 2005;3:442–448. doi: 10.1016/s1542-3565(04)00677-9. [DOI] [PubMed] [Google Scholar]
- 28.Bouhnik Y, Neut C, Raskine L, Michel C, Riottot M, Andrieux C, Guillemot F, Dyard F, Flourié B. Prospective, randomized, parallel-group trial to evaluate the effects of lactulose and polyethylene glycol-4000 on colonic flora in chronic idiopathic constipation. Aliment Pharmacol Ther. 2004;19:889–899. doi: 10.1111/j.1365-2036.2004.01918.x. [DOI] [PubMed] [Google Scholar]
- 29.Smith SC, Choy R, Johnson SK, Hall RS, Wildeboer-Veloo AC, Welling GW. Lupin kernel fiber consumption modifies fecal microbiota in healthy men as determined by rRNA gene fluorescent in situ hybridization. Eur J Nutr. 2006;45:335–341. doi: 10.1007/s00394-006-0603-1. [DOI] [PubMed] [Google Scholar]
- 30.Ramnani P, Gaudier E, Bingham M, van Bruggen P, Tuohy KM, Gibson GR. Prebiotic effect of fruit and vegetable shots containing Jerusalem artichoke inulin: a human intervention study. Br J Nutr. 2010;104:233–240. doi: 10.1017/S000711451000036X. [DOI] [PubMed] [Google Scholar]
- 31.Cloetens L, Broekaert WF, Delaedt Y, Ollevier F, Courtin CM, Delcour JA, Rutgeerts P, Verbeke K. Tolerance of arabinoxylan-oligosaccharides and their prebiotic activity in healthy subjects: a randomised, placebo-controlled cross-over study. Br J Nutr. 2010;103:703–713. doi: 10.1017/S0007114509992248. [DOI] [PubMed] [Google Scholar]
- 32.Nam YD, Jung MJ, Roh SW, Kim MS, Bae JW. Comparative analysis of Korean human gut microbiota by barcoded pyrosequencing. PLoS One. 2011;6:e22109. doi: 10.1371/journal.pone.0022109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shanahan F. The colonic microbiota in health and disease. Curr Opin Gastroenterol. 2013;29:49–54. doi: 10.1097/MOG.0b013e32835a3493. [DOI] [PubMed] [Google Scholar]
- 34.Sommer F, Bäckhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 35.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balsari A, Ceccarelli A, Dubini F, Fesce E, Poli G. The fecal microbial population in the irritable bowel syndrome. Microbiologica. 1982;5:185–194. [PubMed] [Google Scholar]
- 37.Wyatt GM, Bayliss CE, Lakey AF, Bradley HK, Hunter JO, Jones VA. The faecal flora of two patients with food-related irritable bowel syndrome during challenge with symptom-provoking foods. J Med Microbiol. 1988;26:295–299. doi: 10.1099/00222615-26-4-295. [DOI] [PubMed] [Google Scholar]
- 38.Hayashi H, Sakamoto M, Benno Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol. 2002;46:535–548. doi: 10.1111/j.1348-0421.2002.tb02731.x. [DOI] [PubMed] [Google Scholar]
- 39.Maccaferri S, Biagi E, Brigidi P. Metagenomics: key to human gut microbiota. Dig Dis. 2011;29:525–530. doi: 10.1159/000332966. [DOI] [PubMed] [Google Scholar]
- 40.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nocker A, Burr M, Camper AK. Genotypic microbial community profiling: a critical technical review. Microb Ecol. 2007;54:276–289. doi: 10.1007/s00248-006-9199-5. [DOI] [PubMed] [Google Scholar]
- 42.Roh SW, Abell GC, Kim KH, Nam YD, Bae JW. Comparing microarrays and next-generation sequencing technologies for microbial ecology research. Trends Biotechnol. 2010;28:291–299. doi: 10.1016/j.tibtech.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Kassinen A, Krogius-Kurikka L, Mäkivuokko H, Rinttilä T, Paulin L, Corander J, Malinen E, Apajalahti J, Palva A. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133:24–33. doi: 10.1053/j.gastro.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 44.Deplancke B, Gaskins HR. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. Am J Clin Nutr. 2001;73:1131S–1141S. doi: 10.1093/ajcn/73.6.1131S. [DOI] [PubMed] [Google Scholar]
- 45.Rajilić-Stojanović M, Biagi E, Heilig HG, Kajander K, Kekkonen RA, Tims S, de Vos WM. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 46.Mättö J, Maunuksela L, Kajander K, Palva A, Korpela R, Kassinen A, Saarela M. Composition and temporal stability of gastrointestinal microbiota in irritable bowel syndrome--a longitudinal study in IBS and control subjects. FEMS Immunol Med Microbiol. 2005;43:213–222. doi: 10.1016/j.femsim.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 47.Swidsinski A, Sydora BC, Doerffel Y, Loening-Baucke V, Vaneechoutte M, Lupicki M, Scholze J, Lochs H, Dieleman LA. Viscosity gradient within the mucus layer determines the mucosal barrier function and the spatial organization of the intestinal microbiota. Inflamm Bowel Dis. 2007;13:963–970. doi: 10.1002/ibd.20163. [DOI] [PubMed] [Google Scholar]
- 48.Brint EK, MacSharry J, Fanning A, Shanahan F, Quigley EM. Differential expression of toll-like receptors in patients with irritable bowel syndrome. Am J Gastroenterol. 2011;106:329–336. doi: 10.1038/ajg.2010.438. [DOI] [PubMed] [Google Scholar]
- 49.Hart AL, Lammers K, Brigidi P, Vitali B, Rizzello F, Gionchetti P, Campieri M, Kamm MA, Knight SC, Stagg AJ. Modulation of human dendritic cell phenotype and function by probiotic bacteria. Gut. 2004;53:1602–1609. doi: 10.1136/gut.2003.037325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.López P, González-Rodríguez I, Gueimonde M, Margolles A, Suárez A. Immune response to Bifidobacterium bifidum strains support Treg/Th17 plasticity. PLoS One. 2011;6:e24776. doi: 10.1371/journal.pone.0024776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carroll IM, Ringel-Kulka T, Keku TO, Chang YH, Packey CD, Sartor RB, Ringel Y. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2011;301:G799–G807. doi: 10.1152/ajpgi.00154.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parkes GC, Rayment NB, Hudspith BN, Petrovska L, Lomer MC, Brostoff J, Whelan K, Sanderson JD. Distinct microbial populations exist in the mucosa-associated microbiota of sub-groups of irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:31–39. doi: 10.1111/j.1365-2982.2011.01803.x. [DOI] [PubMed] [Google Scholar]
- 53.Verdú EF, Bercik P, Verma-Gandhu M, Huang XX, Blennerhassett P, Jackson W, Mao Y, Wang L, Rochat F, Collins SM. Specific probiotic therapy attenuates antibiotic induced visceral hypersensitivity in mice. Gut. 2006;55:182–190. doi: 10.1136/gut.2005.066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamm K, Hoppe S, Breves G, Schröder B, Schemann M. Effects of the probiotic yeast Saccharomyces boulardii on the neurochemistry of myenteric neurones in pig jejunum. Neurogastroenterol Motil. 2004;16:53–60. doi: 10.1046/j.1365-2982.2003.00458.x. [DOI] [PubMed] [Google Scholar]
- 55.Collins SM. Dysregulation of peripheral cytokine production in irritable bowel syndrome. Am J Gastroenterol. 2005;100:2517–2518. doi: 10.1111/j.1572-0241.2005.00246.x. [DOI] [PubMed] [Google Scholar]
- 56.O’Mahony L, McCarthy J, Kelly P, Hurley G, Luo F, Chen K, O’Sullivan GC, Kiely B, Collins JK, Shanahan F, et al. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology. 2005;128:541–551. doi: 10.1053/j.gastro.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 57.Lamine F, Fioramonti J, Bueno L, Nepveu F, Cauquil E, Lobysheva I, Eutamène H, Théodorou V. Nitric oxide released by Lactobacillus farciminis improves TNBS-induced colitis in rats. Scand J Gastroenterol. 2004;39:37–45. doi: 10.1080/00365520310007152. [DOI] [PubMed] [Google Scholar]
- 58.Ait-Belgnaoui A, Han W, Lamine F, Eutamene H, Fioramonti J, Bueno L, Theodorou V. Lactobacillus farciminis treatment suppresses stress induced visceral hypersensitivity: a possible action through interaction with epithelial cell cytoskeleton contraction. Gut. 2006;55:1090–1094. doi: 10.1136/gut.2005.084194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rousseaux C, Thuru X, Gelot A, Barnich N, Neut C, Dubuquoy L, Dubuquoy C, Merour E, Geboes K, Chamaillard M, et al. Lactobacillus acidophilus modulates intestinal pain and induces opioid and cannabinoid receptors. Nat Med. 2007;13:35–37. doi: 10.1038/nm1521. [DOI] [PubMed] [Google Scholar]
- 60.Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, Deng Y, Blennerhassett P, Macri J, McCoy KD, et al. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141:599–609, 609.e1-3. doi: 10.1053/j.gastro.2011.04.052. [DOI] [PubMed] [Google Scholar]
- 61.Pimentel M, Chow EJ, Lin HC. Eradication of small intestinal bacterial overgrowth reduces symptoms of irritable bowel syndrome. Am J Gastroenterol. 2000;95:3503–3506. doi: 10.1111/j.1572-0241.2000.03368.x. [DOI] [PubMed] [Google Scholar]
- 62.Spiegel BM. Questioning the bacterial overgrowth hypothesis of irritable bowel syndrome: an epidemiologic and evolutionary perspective. Clin Gastroenterol Hepatol. 2011;9:461–49; quiz e59. doi: 10.1016/j.cgh.2011.02.030. [DOI] [PubMed] [Google Scholar]
- 63.Simrén M, Stotzer PO. Use and abuse of hydrogen breath tests. Gut. 2006;55:297–303. doi: 10.1136/gut.2005.075127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ford AC, Spiegel BM, Talley NJ, Moayyedi P. Small intestinal bacterial overgrowth in irritable bowel syndrome: systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2009;7:1279–1286. doi: 10.1016/j.cgh.2009.06.031. [DOI] [PubMed] [Google Scholar]
- 65.Quigley EM. Antibiotics for irritable bowel syndrome: hitting the target, but what is it? Gastroenterology. 2011;141:391–393. doi: 10.1053/j.gastro.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 66.Pimentel M, Chow EJ, Lin HC. Normalization of lactulose breath testing correlates with symptom improvement in irritable bowel syndrome. a double-blind, randomized, placebo-controlled study. Am J Gastroenterol. 2003;98:412–419. doi: 10.1111/j.1572-0241.2003.07234.x. [DOI] [PubMed] [Google Scholar]
- 67.Esposito I, de Leone A, Di Gregorio G, Giaquinto S, de Magistris L, Ferrieri A, Riegler G. Breath test for differential diagnosis between small intestinal bacterial overgrowth and irritable bowel disease: an observation on non-absorbable antibiotics. World J Gastroenterol. 2007;13:6016–6021. doi: 10.3748/wjg.v13.45.6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pimentel M, Park S, Mirocha J, Kane SV, Kong Y. The effect of a nonabsorbed oral antibiotic (rifaximin) on the symptoms of the irritable bowel syndrome: a randomized trial. Ann Intern Med. 2006;145:557–563. doi: 10.7326/0003-4819-145-8-200610170-00004. [DOI] [PubMed] [Google Scholar]
- 69.Pimentel M, Lembo A, Chey WD, Zakko S, Ringel Y, Yu J, Mareya SM, Shaw AL, Bortey E, Forbes WP. Rifaximin therapy for patients with irritable bowel syndrome without constipation. N Engl J Med. 2011;364:22–32. doi: 10.1056/NEJMoa1004409. [DOI] [PubMed] [Google Scholar]
- 70.Menees SB, Maneerattannaporn M, Kim HM, Chey WD. The efficacy and safety of rifaximin for the irritable bowel syndrome: a systematic review and meta-analysis. Am J Gastroenterol. 2012;107:28–35; quiz 36. doi: 10.1038/ajg.2011.355. [DOI] [PubMed] [Google Scholar]
- 71.Sibartie S, O’Hara AM, Ryan J, Fanning A, O’Mahony J, O’Neill S, Sheil B, O’Mahony L, Shanahan F. Modulation of pathogen-induced CCL20 secretion from HT-29 human intestinal epithelial cells by commensal bacteria. BMC Immunol. 2009;10:54. doi: 10.1186/1471-2172-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kekkonen RA, Lummela N, Karjalainen H, Latvala S, Tynkkynen S, Jarvenpaa S, Kautiainen H, Julkunen I, Vapaatalo H, Korpela R. Probiotic intervention has strain-specific anti-inflammatory effects in healthy adults. World J Gastroenterol. 2008;14:2029–2036. doi: 10.3748/wjg.14.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McFarland LV, Dublin S. Meta-analysis of probiotics for the treatment of irritable bowel syndrome. World J Gastroenterol. 2008;14:2650–2661. doi: 10.3748/wjg.14.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoveyda N, Heneghan C, Mahtani KR, Perera R, Roberts N, Glasziou P. A systematic review and meta-analysis: probiotics in the treatment of irritable bowel syndrome. BMC Gastroenterol. 2009;9:15. doi: 10.1186/1471-230X-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moayyedi P, Ford AC, Talley NJ, Cremonini F, Foxx-Orenstein AE, Brandt LJ, Quigley EM. The efficacy of probiotics in the treatment of irritable bowel syndrome: a systematic review. Gut. 2010;59:325–332. doi: 10.1136/gut.2008.167270. [DOI] [PubMed] [Google Scholar]
- 76.Brenner DM, Moeller MJ, Chey WD, Schoenfeld PS. The utility of probiotics in the treatment of irritable bowel syndrome: a systematic review. Am J Gastroenterol. 2009;104:1033–1049; quiz 1050. doi: 10.1038/ajg.2009.25. [DOI] [PubMed] [Google Scholar]
- 77.Ortiz-Lucas M, Tobías A, Saz P, Sebastián JJ. Effect of probiotic species on irritable bowel syndrome symptoms: A bring up to date meta-analysis. Rev Esp Enferm Dig. 2013;105:19–36. doi: 10.4321/s1130-01082013000100005. [DOI] [PubMed] [Google Scholar]
- 78.Niedzielin K, Kordecki H, Birkenfeld B. A controlled, double-blind, randomized study on the efficacy of Lactobacillus plantarum 299V in patients with irritable bowel syndrome. Eur J Gastroenterol Hepatol. 2001;13:1143–1147. doi: 10.1097/00042737-200110000-00004. [DOI] [PubMed] [Google Scholar]
- 79.Kim HJ, Camilleri M, McKinzie S, Lempke MB, Burton DD, Thomforde GM, Zinsmeister AR. A randomized controlled trial of a probiotic, VSL#3, on gut transit and symptoms in diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther. 2003;17:895–904. doi: 10.1046/j.1365-2036.2003.01543.x. [DOI] [PubMed] [Google Scholar]
- 80.Kim HJ, Vazquez Roque MI, Camilleri M, Stephens D, Burton DD, Baxter K, Thomforde G, Zinsmeister AR. A randomized controlled trial of a probiotic combination VSL# 3 and placebo in irritable bowel syndrome with bloating. Neurogastroenterol Motil. 2005;17:687–696. doi: 10.1111/j.1365-2982.2005.00695.x. [DOI] [PubMed] [Google Scholar]
- 81.Kekkonen RA, Kajasto E, Miettinen M, Veckman V, Korpela R, Julkunen I. Probiotic Leuconostoc mesenteroides ssp. cremoris and Streptococcus thermophilus induce IL-12 and IFN-gamma production. World J Gastroenterol. 2008;14:1192–1203. doi: 10.3748/wjg.14.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi SC, Kim BJ, Rhee PL, Chang DK, Son HJ, Kim JJ, Rhee JC, Kim SI, Han YS, Sim KH, et al. Probiotic Fermented Milk Containing Dietary Fiber Has Additive Effects in IBS with Constipation Compared to Plain Probiotic Fermented Milk. Gut Liver. 2011;5:22–28. doi: 10.5009/gnl.2011.5.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Malinen E, Rinttilä T, Kajander K, Mättö J, Kassinen A, Krogius L, Saarela M, Korpela R, Palva A. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373–382. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- 84.Maukonen J, Satokari R, Mättö J, Söderlund H, Mattila-Sandholm T, Saarela M. Prevalence and temporal stability of selected clostridial groups in irritable bowel syndrome in relation to predominant faecal bacteria. J Med Microbiol. 2006;55:625–633. doi: 10.1099/jmm.0.46134-0. [DOI] [PubMed] [Google Scholar]
- 85.Kerckhoffs AP, Samsom M, van der Rest ME, de Vogel J, Knol J, Ben-Amor K, Akkermans LM. Lower Bifidobacteria counts in both duodenal mucosa-associated and fecal microbiota in irritable bowel syndrome patients. World J Gastroenterol. 2009;15:2887–2892. doi: 10.3748/wjg.15.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krogius-Kurikka L, Lyra A, Malinen E, Aarnikunnas J, Tuimala J, Paulin L, Mäkivuokko H, Kajander K, Palva A. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009;9:95. doi: 10.1186/1471-230X-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lyra A, Rinttilä T, Nikkilä J, Krogius-Kurikka L, Kajander K, Malinen E, Mättö J, Mäkelä L, Palva A. Diarrhoea-predominant irritable bowel syndrome distinguishable by 16S rRNA gene phylotype quantification. World J Gastroenterol. 2009;15:5936–5945. doi: 10.3748/wjg.15.5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tana C, Umesaki Y, Imaoka A, Handa T, Kanazawa M, Fukudo S. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol Motil. 2010;22:512–519, e114-e115. doi: 10.1111/j.1365-2982.2009.01427.x. [DOI] [PubMed] [Google Scholar]
- 89.Codling C, O’Mahony L, Shanahan F, Quigley EM, Marchesi JR. A molecular analysis of fecal and mucosal bacterial communities in irritable bowel syndrome. Dig Dis Sci. 2010;55:392–397. doi: 10.1007/s10620-009-0934-x. [DOI] [PubMed] [Google Scholar]
- 90.Ponnusamy K, Choi JN, Kim J, Lee SY, Lee CH. Microbial community and metabolomic comparison of irritable bowel syndrome faeces. J Med Microbiol. 2011;60:817–827. doi: 10.1099/jmm.0.028126-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rinttilä T, Lyra A, Krogius-Kurikka L, Palva A. Real-time PCR analysis of enteric pathogens from fecal samples of irritable bowel syndrome subjects. Gut Pathog. 2011;3:6. doi: 10.1186/1757-4749-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jeffery IB, O’Toole PW, Öhman L, Claesson MJ, Deane J, Quigley EM, Simrén M. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut. 2012;61:997–1006. doi: 10.1136/gutjnl-2011-301501. [DOI] [PubMed] [Google Scholar]


