Abstract
AIM: To investigate if the presence of relevant genetic polymorphisms has effect on the effectual clearance of bacteria by monocytes and granulocytes in patients with Crohn’s disease (CD).
METHODS: In this study, we assessed the differential responses in phagocytosis by measuring the phagocytic activity and the percentage of active phagocytic monocytes and granulocytes in inflammatory bowel disease patients as well as healthy controls. As both autophagy related like 1 (ATG16L1) and immunity-related guanosine triphosphatase gene are autophagy genes associated with CD and more recently nucleotide-binding ligomerization domain-containing protein 2 (NOD2) has been identified as a potent inducer of autophagy we genotyped the patients for these variants and correlated this to the phagocytic reaction. The genotyping was done with restriction fragment length polymorphisms analysis and the phagocytosis was determined with the pHrodo™ Escherichia coli Bioparticles Phagocytosis kit for flowcytometry.
RESULTS: In this study, we demonstrate that analysis of the monocyte and granulocyte populations of patients with CD and ulcerative colitis showed a comparable phagocytic activity (ratio of mean fluorescence intensity) between the patient groups and the healthy controls. CD patients show a significantly higher phagocytic capacity (ratio mean percentage of phagocytic cells) compared to healthy controls (51.91% ± 2.85% vs 37.67% ± 7.06%, P = 0.05). The extend of disease was not of influence. However, variants of ATG16L1 (WT: 2.03 ± 0.19 vs homozygoot variant: 4.38 ± 0.37, P < 0.009) as well as NOD2 (C-ins) (heterozygous variant: 42.08 ± 2.94 vs homozygous variant: 75.58 ± 4.34 (P = 0.05) are associated with the phagocytic activity in patients with CD.
CONCLUSION: Monocytes of CD patients show enhanced phagocytosis associated with the presence of ATG16L1 and NOD2 variants. This could be part of the pathophysiological mechanism resulting in the disease.
Keywords: Inflammatory bowel disease, Phagocytosis, Polymorphism, Monocytes, Granulocytes, Nucleotide-binding ligomerization domain-containing protein 2, Immunity-related guanosine triphosphatase gene, Autophagy related like 1
INTRODUCTION
Inflammatory bowel disease (IBD) is comprised of two major disorders: ulcerative colitis (UC) and Crohn’s disease (CD). IBD is a complex genetic and immunological disease, wherein antigens in the lumen of the gut initiate an inadequate immune response in a genetically susceptible host. In normal healthy individuals the immune response to commensals in the intestine is kept under strict regulation. When these regulatory mechanisms fail, for instance when bacterial clearance is impaired, an inflammatory response in the intestines can result in IBD. For effective extracellular bacterial clearance there should be accurate phagocytic activity in the gut by phagocytes like monocytes, macrophages, dendritic cells (DC) and granulocytes. Mononuclear phagocytes are able to present bacterial-derived antigens after phagocytosis via major histocompatibility complex (MHC) class II complex to CD4+ T cells to initiate an adaptive immune response. However, several pathogens are capable of evading this mechanism and survive in the cytoplasm. For intracellular bacterial clearance there exists a similar efficient pathway of antigen delivery for MHC class II presentation called (macro) autophagy[1,2]. This is a complex cellular process, present in all eukaryotic cells in which intracellular components including organelles but also bacteria, are sequestered in double-membrane vesicles or vacuoles called autophagosomes that eventually fuse with lysosomes, resulting in the degradation of their contents.
In 2001 NOD2 was the first gene to be identified as being associated with CD. The discovery led to extensive genetic research of this gene[3]. Nucleotide-binding ligomerization domain-containing protein 2 (NOD2) encodes an intracellular receptor that recognizes the bacterial component muramyl dipeptide (MDP) of both gram positive and negative bacteria. NOD2 thus detects invading pathogens and plays a central role in the production of cytokines and antimicrobial peptides[4] via the receptors for inacitive C-kinase/phosphatidylinositol-4,5-bisphosphate, nuclear factor kappa B and mitogen-activated protein kinases[5]. Three major NOD2 variants are associated with CD: R702W, G908R and L1007fsinsC. The latter, being a frameshift mutation, causes an incapability of transcription activation in response to MDP. It has also been shown that this mutation is involved in other functional abnormalities such as enhanced cytokine expression[6] and decreased expression of interleukin (IL)-10[7]. It has been shown in several studies that in cells of patients carrying the NOD2 mutations, there is an impaired immune response to microbial infections and bacterial ligands and thus a loss of function. Furthermore, NOD2 deficiency in mice caused exaggerated intestinal inflammation as a result of disrupted immune responses[8,9].
The genome-wide association studies performed in the first half of this decade, have identified over 30 susceptibility loci that are independently associated with CD, of which two are involved in autophagy[10-13]. One of the strongest associations was found in the variant of the autophagy related like 1 (ATG16L1) gene[14]. ATG16L1 encodes an autophagy pathway protein that forms a non-covalent protein complex with ATG5 and ATG12 (800 kD). This complex is essential for the autophagosome formation. A threonine to alanine substitution at position 300 (T300A) of the WD domain is associated with CD. The second autophagy gene associated with CD is Immunity-related GTPase family M protein (IRGM)[15]. IRGM belongs to a family of genes encoding interferon-inducible guanosine triphosphatases involved in newly recognised forms of pathogen clearance. The single-nucleotide polymorphisms (SNPs) associated with CD in IRGM are located in the flanking region of the gene[16]. Sequencing of the gene in both CD patients as well as healthy controls did not identify any non-synonymous variation in linkage disequilibrium with the associated allele. It has been suggested by Xavier et al[17] that susceptibility to CD could operate via modulation of IRGM gene expression. Because dysregulated host responses to intracellular organisms could contribute to the development of CD we hypothesize that one of the underlying mechanisms of this inadequate response is an impaired innate immune response showing in either a disabled or overly active phagocytic uptake of antigens by granulocytes and monocytes. It has recently been shown that there is a physical interaction between NOD2 and ATG16L1 and that this interaction is required for autophagic clearance of intracellular pathogens[18]. In this study we first assessed the differential responses in phagocytosis by measuring phagocytic activity and the percentage of active phagocytic monocytes and granulocytes in IBD patients as well as healthy controls (HC). Secondly, we correlated phagocytic capacity to the known associated variant in ATG16L1, IRGM and NOD2 (C-ins). This revealed an impaired phagocytic reaction in IBD patients that carried the mutant alleles.
MATERIALS AND METHODS
Patients and electronic data collection
In this study 99 IBD patients (65 CD and 34 UC) were included, along with 8 healthy controls. All patients were recruited through the outpatient clinic at the department of Gastroenterology of the Academic Medical Centre (AMC) Amsterdam, the Netherlands as part of the Elephant Study. This study was initially designed to associate the immunological phenotype of IBD patients to the genotype of the known susceptibility genes and clinical phenotype. All clinical phenotypic patient data is available in an electronic patient file, part of which is excerpted and listed in Table 1. All patients and controls gave informed consent and the Elephant Study was approved by the ethics review committee of the AMC.
Table 1.
Clinical phenotypic characteristics of study population n (%)
| CD (n = 65) | UC (n = 34) | |
| Gender: M/F | 29/36 | |
| Montreal classification | ||
| Age at diagnosis (yr) | 27 (9-68) | |
| < 17 | 10 (15.4) | |
| 17-40 | 44 (67.7) | |
| > 40 | 9 (13.8) | |
| Disease localisation | ||
| Terminal ileum (L1) | 17 (26.2) | |
| Colonic (L2) | 8 (12.3) | |
| Ileocolonic (L3) | 23 (35.4) | |
| Upper gastrotractinal (L4) | 0 | |
| Disease behavior | ||
| Non-stricturing /non-penetrating | 20 (30.8) | |
| Stricturing | 15 (23.1) | |
| Penetrating | 11 (16.9) | |
| Missing | 19 (29.2) | |
| Age at diagnosis (yr) | - | 32 (19-74) |
| < 40 | - | 25 (73.5) |
| > 40 | - | 7 (20) |
| Disease localisation | ||
| Proctitis (L1) | 3 (8.8) | |
| Left sided (L2) | 11 (32.4) | |
| Pancolitis (L3) | 10 (29.4) | |
| Missing | 10 (29.4) | |
| Disease activity (Y/N) | 17/47 | 14/10 |
| Positive family history | 12 | 8 |
| Operated on | 40 | 7 |
CD: Crohn’s disease; UC: Ulcerative colitis; M/F: Male/female; Y/N: Yes/no.
Genotyping with restriction fragment length polymorphisms
The Elephant Study cohort was genotyped for three of the CD-associated genes ATG16L1, IRGM and NOD2. DNA was isolated from venous blood, which was collected within the framework of the Elephant Study. Genotyping for the SNPs was performed by polymerase chain reaction restriction fragment length polymorphisms assay. Designed primers, thermal cycling and restriction enzymes (New England BioLabs, Ipswich, MA, United States) are listed in Table 2. Restriction fragments were separated and visualised using 3% agarose gel containing ethydium bromide.
Table 2.
Designed primers and restriction enzymes for genotyping by restriction fragment length polymorphisms
| Gene | Forward | Reverse | Restriction enzyme |
| ATG16L1 | GGTACCCTCACTTCTTTACCAGAA CCAGGAAGAG | TGGAGTCCACAGGTTAGTGTGCAGGAGAGTAAGG | Sap1 |
| rs2241880 | |||
| IRGM | CCCGTGTCGTACCCAAGCAGAGTGTGCTTGAAGA | CTTTACCATTGTACTCCTTGTGCCCAGCAGGTG | MboI |
| rs13361189 | |||
| NOD2 | ATGTGTCTAAGGGACAGGTG | AACTGAGGTTCGGAGAGCTA | NlaIV |
| rs2066847 | |||
| rs5743293 |
rs2066847 and rs5743293 are the same single-nucleotide polymorphism (SNP) in nucleotide-binding ligomerization domain-containing protein 2 (NOD2). ATG16L1: Autophagy related like 1; IRGM: Immunity-related GTPase family M protein.
Phrodo assay and flow cytometry
The pHrodo™ Escherichia coli BioParticles® Phagocytosis Kit for Flow Cytometry from Invitrogen Molecular Probes (Eugene, OR, United States) was used to assess the phagocytic capacity of our Elephant Study cohort and HCs. The dye is nonfluorescent at neutral pH and bright red fluorescent in acidic environments. The advantage of this is that actual phagocytosis and lysosomal acidification is measured while extracellular adherent particles are not detected. The pHrodo™ Escherichia coli BioParticles® Phagocytosis assay was performed according to the manufacturer’s protocol using heparinized blood from all included patients and HCs. Granulocytes and monocytes were discriminated on the basis of their forward and site scatter profiles by flow cytometry using the FACSscan from BD Biosciences (Erembodegem, Belgium). Phagocytic activity of monocytes and granulocytes was measured as the ratio of the mean of the mean fluorescent intensity of the positive control at 4 °C compared to the one at 37 °C. Percentage phagocytic cells were calculated using the ratio of gated cells in M1 at 4 °C and 37 °C.
Statistical analysis
Statistical analyses were performed using SPSS (SPSS Inc., Chicago, IL, United States) or GraphPad (GraphPad Software Inc., La Jolla, CA, United States). All comparisons with phagocytic activity and the percentage of phagocytic cells were tested between the different groups using non-parametric Mann-Whitney and Kruskall-Wallis tests. Significance level was set at 0.05. Association of the different genotypes with the phagocytic activity or cells was tested using a one-way ANOVA and linear regression analysis. P-values of 0.05 or less were considered significant.
RESULTS
Phagocytosis in granulocytes and monocytes in IBD patients
To test for phagocytic capacity in both granulocytes and monocytes the phagocytic activity and the percentage of phagocytic cells was measured by flow cytometry. Analysis of both the granulocyte population and the monocyte population showed a comparable phagocytic activity between the patient group and the healthy controls (Figure 1). When comparing the amount of active phagocytic granulocytes of both the IBD patient group and the healthy controls, similar results were obtained for both groups (Figure 2). Interestingly, a significant difference was found in the percentage of active phagocytic monocytes of IBD patients when compared to the healthy controls (P < 0.04). When comparing all three groups a significant difference was found between the CD patient group and the healthy controls (P = 0.05) (Figure 2). A similar pattern was seen in the group of the UC patients when compared with the healthy controls. As expected, both CD and UC patient groups showed a comparable percentage of active phagocytic monocytes and granulocytes (see Figure 2 and data not shown).
Figure 1.
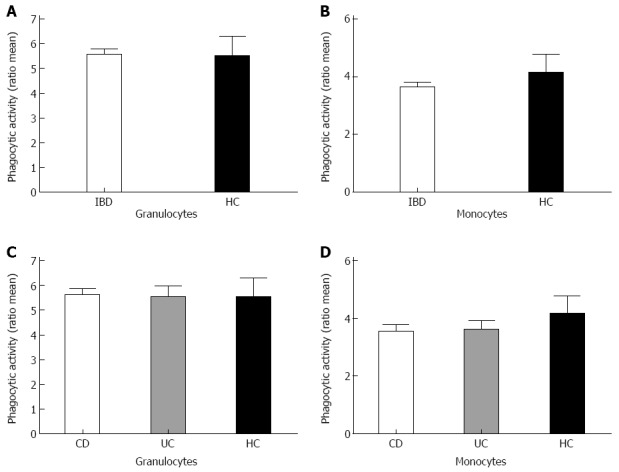
Phagocytic activity of the disease groups compared to healthy controls. Bars represent ratio of the mean phagocytic activity. Total inflammatory bowel disease patients (IBD) patients (A, B for granulocytes and monocytes respectively) were analyzed as well as for separate Crohn’s disease (CD) and ulcerative colitis (UC) patients compared to healthy controls (C, D). HC: Healthy controls.
Figure 2.
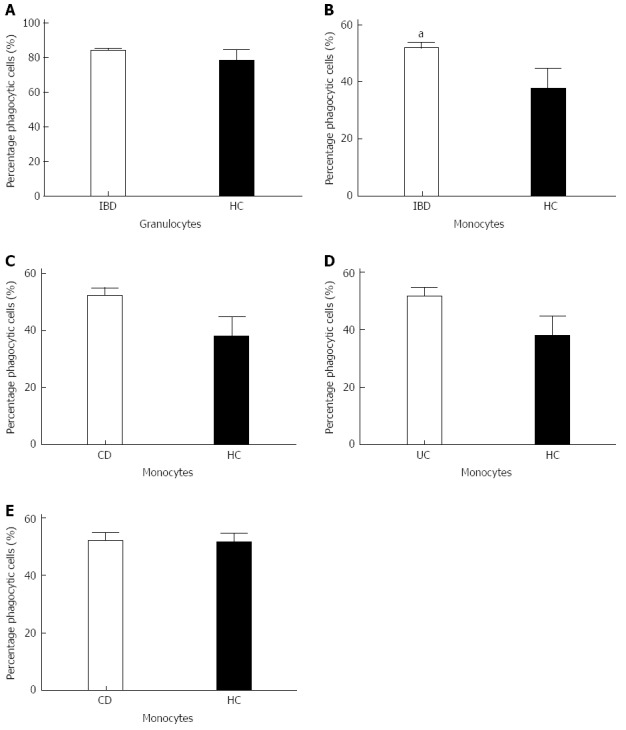
Percentage of phagocytic cells in inflammatory bowel disease patients patients and healthy controls. A: Percentages of active granulocytes are comparable between both groups; B: Significantly higher amount of active phagocytic monocytes was found in the inflammatory bowel disease (IBD) patient group compared to healthy controls (P = 0.0408), aP < 0.05 vs HC group; C: Crohn’s disease (CD) patients show a significantly higher percentage of active phagocytic monocytes compared to healthy controls; D, E: This was not seen when compared to ulcerative colitis (UC) patients. Bars represent means.
Phagocytosis correlated with disease activity
Since monocytes display an enhanced amount of active phagocytic cells in comparison with the healthy controls, disease activity and possibly the extend of the disease might be of influence. Disease activity was defined as the patient having a leucocytosis, elevated C-reactive protein level and clinical view of the gastroenterologist. The extent of the disease was established and defined using the Montreal Classification (Table 1). Analysis of the influence of the disease activity on phagocytic activity did not reveal any differences between the different categories in UC or CD (Figure 3). The extent of the disease also does not influence on both the phagocytic activity and the percentage of active phagocytic cells (results not shown).
Figure 3.

Phagocytic activity and percentage of phagocytic cells and the correlation with disease activity in Crohn’s disease patients and ulcerative colitis patients. A, B: Crohn’s disease (CD); C, D: Ulcerative colitis (UC). 0: No disease activity; 1: Disease activity.
Phagocytosis associated with variants in IRGM, ATG16L1 and NOD2
We next investigated whether the genotypes of our CD patient cohort for ATG16L1, IRGM and NOD2 were associated with the overall phagocytic activity and the percentage of active phagocytic cells (Figure 4). Figure 4C shows a positive association of the variants of ATG16L1 and the phagocytic activity of the monocytes (P < 0.009). A P-value of 0.08 was found for the amount of active phagocytic monocytes when all three groups were compared. Although we do see the same trend for the variant in IRGM, no significant association has been found (Figure 4A, B). The homozygous mutant genotype of the variant 3020 C-ins of NOD2 influences both phagocytic activity and the percentage of active monocytes, the latter showing a significant difference (P = 0.05) (Figure 4E, F).
Figure 4.
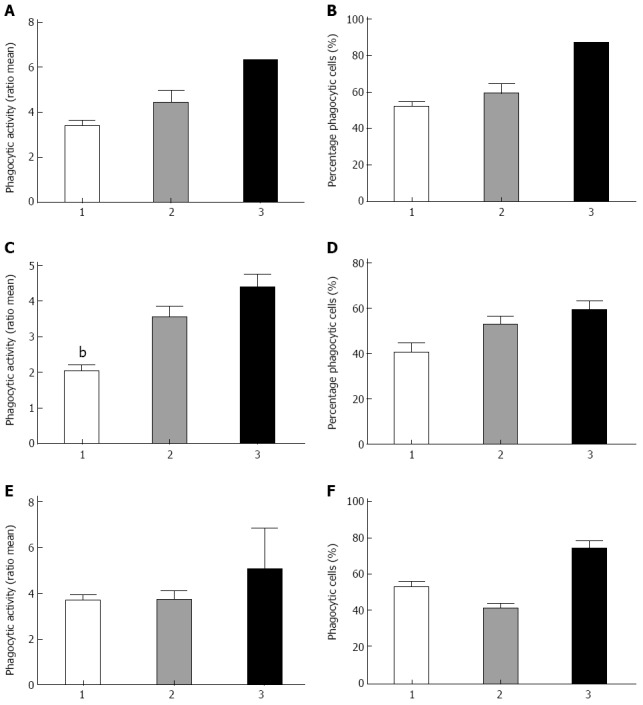
Phagocytosis and variants in Crohn’s disease associated genes autophagy related like 1, immunity-related guanosine triphosphatase gene and nucleotide-binding ligomerization domain-containing protein 2. A, B: Genotypes in immunity-related guanosine triphosphatase variant and association with either phagocytic activity or the percentage of active monocytes, respectively; C, D: Genotypes of the autophagy related like 1 variant associated with enhanced phagocytic activity (P = 0.009) and percentage of active monocytes respectively. bP < 0.01 vs genetype 3; E, F: The genotype of the variant 3020 C-ins of nucleotide-binding ligomerization domain-containing protein 2 and phagocytic activity as well as the percentage of active monocytes (P = 0.05) respectively. Genotypes 1: Wild type; Genotypes 2: Heterozygous variant; Genotypes 3: Homozygous variant.
DISCUSSION
Monocytes are of importance in extracellular bacterial clearance and therefore play a role in the regulation of the innate immune response of IBD patients. We demonstrate that monocytes of CD patients show enhanced phagocytosis. In addition we show that the enhanced phagocytosis is not influenced by disease activity and is associated with the disease-related variants of both ATG16L1 and NOD2. The ex vivo model we use gives us the opportunity to study the functional consequences of polymorphisms in IBD approximate to the natural situation. The increased phagocytosis in CD patients can result in an accumulation of bacterial products in the cell and secondarily lead to an increased inflammatory reaction.
Phagocytosis and autophagy are processes that are of major importance regarding bacterial clearance. Phagocytosis by professional phagocytes like macrophages, monocytes and granulocytes is responsible for clearing the extracellular compartment; autophagy plays its role in intracellular bacterial clearance. Just a few years ago it has been shown that impaired bacterial clearance is a potential pathogenic factor in IBD. Monocytes are immediate effector cells and produce cytokines and perform phagocytosis during inflammation[19,20]. Monocytes derive from a myeloid precursor and in mice two subsets of monocytes, Ly-6C+ and Ly-6C–, leave the bone marrow to enter the blood. The Gr1+/Ly-6C+ monocytes give rise to macrophages and DCs during different infections, such as Listeria monocytogenes and Toxoplasma gondii, but are also found in tumors as suppressor cells, as well as contribute to the recovery of spinal cord injury. Monocytes can therefore have activating as well as inhibiting functions in the immune response[20].
Not only genes involved in primary recognition of bacterial compounds such as NOD2, but also genes involved in autophagy were found to be of interest due to the genome-wide association studies, which identified several autophagy genes as susceptibility genes for CD. Autophagy serves its purpose in the innate immune response by clearing several intracellular pathogens, such as Salmonella enterica, Mycoplasma tuberculosis and Listeria monocytogenes[16]. When this system fails, pathogens are able to expand and an adaptive response should start to remove these pathogens. The failure of the innate response, called the theory of innate immune deficiency, has been proposed as responsible for the elevated immune activation seen in CD patients carrying this SNP[21]. This is in concordance with our data. Patients carrying the mutant allele have higher phagocytic activity (P < 0.009). This can be explained by the fact that when the innate immune response is inadequate, because of diminished autophagy due to impaired autophagosome formation, the adaptive immune response is overactivated, resulting in a higher percentage of activated monocytes. However, this does not mean that these monocytes have a more effective phagocytic capacity, measured in phagocytic activity. This was shown to be somewhat higher in the healthy controls. Two other studies looked at the phagocytosis of monocytes in relation to NOD2 polymorphisms. Henckaerts et al[22] showed that NOD2 variants were associated with reduced phagocytosis and bacteremia in critically ill patients and Glubb et al[23] demonstrated that NOD2 was shown to affect the elimination of the Mycobacterium avium subspecies paratuberculosis from peripheral blood monocytes, whereas ATG16L1 polymorphism showed increased expression of IL-10 and IL-6. Concerning the effect of the ATG16L1 polymorphism on cytokine production, Plantinga et al[24] demonstrate that the genetic variation in ATG16L1 is associated with higher production of pro-inflammatory cytokines (IL-1β and IL-6) upon NOD2 stimulation in CD patients that could drive the chronic inflammation.
A physical interaction between NOD2 and ATG16L1 appears to be essential for autophagic clearance of intracellular pathogens; NOD2 seems to be an autophagy inducer[18,25]. Our data are in accordance with this observation. We have shown that IBD patients carrying both risk alleles show a significantly higher phagocytic capacity in both ATG16L1 and NOD2. In the case of ATG16L1 this can be due to loss of response showing that carrying one or more risk alleles interferes with autophagy and enhances the phagocytic capacity as a backup mechanism. The same holds true for the NOD2 variant as this causes a loss of function. When patients carry this risk allele they have a dysfunctional protein causing a direct problem in the formation of the autophagosome, thereby impairing autophagy. In this case an enhanced phagocytic capacity is also observed, strengthening the hypothesis of a backup mechanism. Our data also demonstrate the same trend in the patients who carry the variety in IRGM (P < 0.08), however the sample size was too small to show significant differences. Therefore, future studies with a higher number of participants must be undertaken to explore the role of IRGM in phagocytosis of monocytes.
COMMENTS
Background
Single-nucleotide polymorphisms (SNPs) in susceptibility genes in inflammatory bowel disease (IBD) can contribute to the disease. The autophagy related like 1 (ATG16L1) and nucleotide-binding ligomerization domain-containing protein 2 (NOD2) susceptibility genes and their role in autophagy and elimination of intracellular bacteria are extensively studied. Most studies were done in mice and cell lines. Authors demonstrate that in humans with variants of ATG16L1 and NOD2 the clearance of bacteria by phagocytosis is enhanced compared to controls.
Research frontiers
There are at the moment 163 different SNPs that are associated with IBD described. The focus on the current research is to analyse the relationship between genotypes and phenotypes for all IBD-associated polymorphisms.
Innovations and breakthroughs
The analysis of the relationship between genotypes and phenotypes is currently mainly focussed on elucidating the correlation with the clinical phenotype. The research toward the determination of functional differences in the innate and adaptive immune responses that could play a role in the IBD risk is the challenge for the coming years.
Applications
IBD patients that have the risk alleles ATG16L1 and/or NOD2 have a different immunologic reactivity and might benefit from a treatment regimen focused on the specific disease variant effects
Peer review
The authors present an interesting study showing that in Crohn’s disease (CD) patients phagocytic activity of monocytes is enhanced. Moreover they could demonstrate that this increased activity is associated with the genotype. This information is interesting for all of those who try to uncover the pathophysiological mechanism resulting in CD.
Footnotes
P- Reviewer: Ukena SN S- Editor: Gou SX L- Editor: A E- Editor: Liu XM
References
- 1.Schmid D, Münz C. Immune surveillance via self digestion. Autophagy. 2007;3:133–135. doi: 10.4161/auto.3591. [DOI] [PubMed] [Google Scholar]
- 2.Schmid D, Münz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 4.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 5.Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology. 2010;139:1630–1641, 1641.e1-2. doi: 10.1053/j.gastro.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeda S, Hsu LC, Liu H, Bankston LA, Iimura M, Kagnoff MF, Eckmann L, Karin M. Nod2 mutation in Crohn’s disease potentiates NF-kappaB activity and IL-1beta processing. Science. 2005;307:734–738. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]
- 7.Noguchi E, Homma Y, Kang X, Netea MG, Ma X. A Crohn’s disease-associated NOD2 mutation suppresses transcription of human IL10 by inhibiting activity of the nuclear ribonucleoprotein hnRNP-A1. Nat Immunol. 2009;10:471–479. doi: 10.1038/ni.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn’s disease associated ATG16L1 variant. PLoS One. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fava F, Danese S. Intestinal microbiota in inflammatory bowel disease: friend of foe? World J Gastroenterol. 2011;17:557–566. doi: 10.3748/wjg.v17.i5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 15.Roberts RL, Hollis-Moffatt JE, Gearry RB, Kennedy MA, Barclay ML, Merriman TR. Confirmation of association of IRGM and NCF4 with ileal Crohn’s disease in a population-based cohort. Genes Immun. 2008;9:561–565. doi: 10.1038/gene.2008.49. [DOI] [PubMed] [Google Scholar]
- 16.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 17.Xavier RJ, Huett A, Rioux JD. Autophagy as an important process in gut homeostasis and Crohn’s disease pathogenesis. Gut. 2008;57:717–720. doi: 10.1136/gut.2007.134254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fritz T, Niederreiter L, Adolph T, Blumberg RS, Kaser A. Crohn’s disease: NOD2, autophagy and ER stress converge. Gut. 2011;60:1580–1588. doi: 10.1136/gut.2009.206466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu Rev Immunol. 2009;27:669–692. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- 20.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casanova JL, Abel L. Revisiting Crohn’s disease as a primary immunodeficiency of macrophages. J Exp Med. 2009;206:1839–1843. doi: 10.1084/jem.20091683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henckaerts L, Nielsen KR, Steffensen R, Van Steen K, Mathieu C, Giulietti A, Wouters PJ, Milants I, Vanhorebeek I, Langouche L, et al. Polymorphisms in innate immunity genes predispose to bacteremia and death in the medical intensive care unit. Crit Care Med. 2009;37:192–201, e1-3. doi: 10.1097/CCM.0b013e31819263d8. [DOI] [PubMed] [Google Scholar]
- 23.Glubb DM, Gearry RB, Barclay ML, Roberts RL, Pearson J, Keenan JI, McKenzie J, Bentley RW. NOD2 and ATG16L1 polymorphisms affect monocyte responses in Crohn’s disease. World J Gastroenterol. 2011;17:2829–2837. doi: 10.3748/wjg.v17.i23.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plantinga TS, Crisan TO, Oosting M, van de Veerdonk FL, de Jong DJ, Philpott DJ, van der Meer JW, Girardin SE, Joosten LA, Netea MG. Crohn’s disease-associated ATG16L1 polymorphism modulates pro-inflammatory cytokine responses selectively upon activation of NOD2. Gut. 2011;60:1229–1235. doi: 10.1136/gut.2010.228908. [DOI] [PubMed] [Google Scholar]
- 25.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]


