Background: PTTH, a key insect neurohormone, regulates development by stimulating ecdysone synthesis and release.
Results: PRC2 protein ESC activates PTTH expression by meditating H3K27me3, which depends on methyltransferase activity of PRC2 complex.
Conclusion: PRC2 complex regulates insect developmental timing by controlling PTTH expression in the brain.
Significance: This is the first report suggesting PRC2 complex as an activator meditating developmental timing in insects.
Keywords: Epigenetics, Gene Expression, Gene Regulation, Histone Modification, Insect, Developmental Biology, Diapause, Longevity, Neurohormone
Abstract
The decision made by insects to develop into adults or halt development (enter diapause and prolong lifespan) is commonly based on environmental signals that provide reliable predictors of future seasons of adversity. For example, the short day lengths of early autumn accurately foretell the advent of winter, but little is known about the molecular mechanisms that preside over the hormonal events dictating whether the insect proceeds with development or enters diapause. In Helicoverpa armigera we show that day length affects H3K27me3 by affecting polycomb repressive complex 2 (PRC2) protein extra sex comb (ESC) and regulates the prothoracicotropic hormone (PTTH) gene, thus directly influencing developmental timing. ESC expression in brains of developing (nondiapause) pupae is higher than in brains from diapausing pupae. High ESC expression is localized in two pairs of PTTH neurosecretory cells, and H3K27me3 recruits on the PTTH promoter. Double strand ESC and PRC2 inhibitor (DzNep) treatment in vitro show that ESC triggers PTTH promoter activity, which in turn depends on PRC2 methyltransferase activity. Injection of DzNep into pupae programmed for development reduces the H3K27me3 mark and PTTH gene expression, thereby delaying development. Although ESC is best known as a transcriptional repressor, our results show that ESC prompts development and metamorphosis. We believe this is the first report showing that the PRC2 complex functions as an activator and that a low level of H3K27me3 can prolong lifespan (i.e. induce diapause) by controlling PTTH gene expression in insects.
Introduction
Environmental conditions are not always suitable to sustain insect development; many species enter a developmental arrest (diapause) in a specific stage during an unfavorable season and restart development when favorable environmental conditions return. Diapause is a “non-aging” state, similar to dauer in Caenorhabditis elegans, because it persists for months and results in extreme lifespan extension (1, 2). The insect brain perceives seasonal environmental signals such as day length and temperature and through processes that remain poorly understood, ultimately translates this information into neurohormonal signals that dictate either continuous development or diapause. One of the key neurohormones regulating insect development and longevity is prothoracicotropic hormone (PTTH)3 (3–5), a brain neuropeptide that stimulates the prothoracic gland to synthesize and release the steroid hormone, ecdysone (6). High ecdysone promotes insect development, whereas a low titer of the hormone results in diapause, a dramatic extension of lifespan due to developmental arrest (1, 6). The polycomb repressive complex 2 (PRC2) proteins are conserved chromatin modification factors best known for silencing gene expression by regulating chromatin through tri-methylation of lysine 27 on histone H3 (H3K27me3) (7, 8). PRC2 consists of three major components in insects: enhancer of zeste (E(z)), extra sex combs (ESC), and suppressor of zeste 12 (Su(z)12) (7). Hundreds of putative PRC2 and H3K27me3 target genes have been identified in Drosophila and mammals by genome-wide mapping, and more than half the target genes are developmental regulators (9–13). Although the repressor function of PRC2 complex proteins has been well documented, several recent reports also suggest an opposite effect; that is, the PRC2 protein Su(z)12 occupies actively transcribed regions in embryonic stem cells (14), H3K27me3 levels are higher in promoters of genes associated with active transcription (15), and a PRC2 protein, Ezh1, can activate mRNA transcription by promoting RNA polymerase II elongation during cell differentiation in mammalian cells in vitro (16). These reports imply that the PRC2 protein can also function as an activator in the regulation of gene expression.
PRC2 proteins are involved in a variety of physiological processes, including embryogenesis, stem cell pluripotency, and differentiation (17, 18). Most recently, several studies in nematodes and Drosophila demonstrated that PRC2 proteins are also associated with longevity and aging (19–21), but the mechanism is still unknown.
We speculate that the regulation of diapause by environmental signals may have an epigenetic basis and thus tested this idea by examining members of the polycomb family. The cotton bollworm, Helicoverpa armigera, is a species that overwinters in pupal diapause. This developmental arrest can be initiated in the laboratory by exposing 5th-6th instar larvae to short day lengths (10 h light:14 h dark) and 20 °C, and their lifespan is 3 times longer than in their nondiapause counterparts reared at long day lengths (14 h light:10 h dark) and 20 °C without diapause. Thus, this species is an excellent model for lifespan research. It is well known that short day length causes a reduction of PTTH gene expression and subsequently low ecdysone levels early in the pupal stage, resulting in diapause (22, 23). How environmental signals exert their control over PTTH gene expression and hence the decision to develop or enter diapause is not known (24, 25).
In this study we observed that a core component of the PRC2 complex, ESC, is differentially expressed in pupal brains of nondiapause- and diapause-destined individuals in H. armigera. Experiments from immunostaining, chromatin immunoprecipitation (ChIP) analysis, RNAi, and inhibitor treatment showed that the PTTH gene may be an ESC target. A high level of H3K27me3 in nondiapause pupal brain by PRC2 can promote insect development and metamorphosis by activating PTTH expression, whereas a low level of H3K27me3 can induce diapause by reduction of PTTH expression. Our results suggest that PRC2 functions as an activator of development, and the low level of H3K27me3 observed in diapausing individuals results in developmental arrest.
EXPERIMENTAL PROCEDUCES
Animals
H. armigera larvae were kindly provided by Dr. Jian-Ya Su, Nanjing Agriculture University (Nanjing, China), and reared on an artificial diet at 20 °C. When reared under a light-dark cycle of 14 h light:10 h dark all pupae developed into adults without entering diapause, whereas >95% entered pupal diapause when reared at 20 °C under a 10 h light:14 h dark photoperiod. Pupal brains were dissected in ice-cold 0.75% NaCl and stored at −80 °C until used.
RNA Extraction and Quantitative Real-time PCR
Total RNA was extracted from H. armigera pupal brains as reported (26, 27). Reverse transcription was performed on 1 μg of total RNA using a Prime Script RT-PCR kit (Takara). Quantitative real-time PCR was performed on a Light Cycler480 (Roche Applied Science) using SYBR Premix Ex TaqII (Takara). Gene-specific primers are listed in Table 1. RpL32 was used as an internal control.
TABLE 1.
Gene-specific primers
| qPCR primers |
| ESC-qF1, CGTGAAGAAACGGAGACGAGG |
| ESC-qR1, GTTGAATTGACAGCCGAATAGGG |
| PTTH-qF1, CAATCTTGTCGCCCGCCTTAC |
| PTTH-qR1, CAGACACAGCCTACGCTCAC |
| RpL32-qF1, CCCGTCACATGCTACCCAATGG |
| RpL32-qR1, CTCGCTCCACGATGGTCTTGC |
| Overexpression primers |
| ESC-ERF1, CGCGGATCCATGAACTTTTCTGACAATGAAG |
| ESC-ERF2, CGCGGATCCCTATTCGGCTGTCAATTC |
| ESC-ERR1, CCGCTCGAGCAGACACTGTTTGTGGTGA |
| ESC-ERR2, CCGCTCGAGAATAGGGGCTGGCCATG |
| ChIP primers |
| PTTH-ChIP-F1, AGGTTTGTCACCGACCTGC |
| PTTH-ChIP-R1, GCATGTTGATTGATGGCACCC |
| PTTH-ChIP-F2, GAGCGTTAGCGTACACAATGG |
| PTTH-ChIP-R2, AGCTTGTTCGCGTCCGTAG |
| PTTH-ChIP-F3, CCCTGCGCCTGTAAATTCAG |
| PTTH-ChIP-R3, CGACAAGATTGCTGACGTGC |
Protein Extraction and Western Blot Analysis
Brains were homogenized in radioimmune precipitation assay buffer (150 mm NaCl, 50 mm Tris-HCl (pH 8.0), 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm PMSF, 1 mm EGTA, 5 mm NaF, 10 mm Na3VO4, and 33 mm sodium butyrate); the lysate was shaken in a rotary shaker for 1 h at 4 °C and then centrifuged for 15 min at 16,000 × g at 4 °C (26, 27).
For histone extraction, brains were homogenized in nuclear extraction buffer (50 mm Tris-HCl (pH 7.5), 25 mm KCl, 250 mm sucrose, 1 mm PMSF, 1 mm EGTA, 5 mm NaF, 10 mm Na3VO4, and 33 mm sodium butyrate) using a Dounce homogenizer. Nuclei were pelleted by centrifugation for 1 min at 8000 × g at 4 °C. Pellets were thoroughly suspended in 500 μl of 0.2 m H2SO4, incubated on ice for 30 min, and centrifuged for 30 min at 16,000 × g at 4 °C. The supernatant was transferred to a 1.5-ml tube, and 132 μl of 100% trichloroacetic acid was added slowly followed by incubation on ice for 30 min. Then the solution was centrifuged for 30 min at 16,000 × g at 4 °C. Finally, the pellets were washed twice in cold acetone and redissolved in 40 μl of 10 mm Tris-HCl (pH 8.0).
Equal amounts of protein (30 μg) were separated on an SDS-PAGE gel and transferred to a PVDF membrane. The membrane was probed with an ESC antibody produced by injecting rabbits with purified recombinant Har-ESC protein. Antibody against full-length human EZH2 (SAB1405776, Sigma) was used against E(z) protein; H3 (ab1791), H3K27Ac (ab4729), H3K4me3 (ab8580) were purchased from Abcam, and H3K27me3 (17-622) was from Millipore. Recombinant Har-actin protein, which consists of 370 amino acids (GenBankTM number HM629442.1), was used to generate polyclonal antibodies in rabbits.
Phosphatase Assay
The phosphatase assay was performed as described with a slight modification (28). Total protein from nondiapausing pupal brains was extracted and homogenized in radioimmune precipitation assay buffer (150 mm NaCl, 50 mm Tris-HCl (pH 8.0), 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm PMSF). Samples (30 μg) were treated with or without 30 units of calf alkaline phosphatase (Takara, Japan) for 45 min at 37 °C.
Construction of the Overexpression System
Full-length ESC (1–413 amino acids), N-terminal ESC (1–67 amino acids), and C-terminal ESC (67–413 amino acids) were amplified with primers (Table 1) containing BamHI and XhoI restriction sites. PCR products were digested and subcloned into the pIZ/V5-GFP vector.
Cell Culture and Vector Transfection
HzAm1 cells from Helicoverpa zea and Sf9 cells from Spodoptera frugiperda were maintained at 27 °C in Grace's medium (Invitrogen) containing 10% fetal bovine serum (HyClone). Transfection of pIZ/V5 vector was performed with the Cellfectin II reagent (Invitrogen) as described in Bao et al. (29). For the cellular location assay, nuclei of cells were stained with Hoechst 33,342 (1 mm) for 10 min, and fluorescence of GFP and nuclei were visualized with a fluorescence microscope (Olympus IX71).
Co-immunoprecipitation and Immunoblot Analysis
HzAm1 cells were lysed in radioimmune precipitation assay buffer, and 400 μg of protein extract was used for coimmunoprecipitation. The lysate was immunoprecipitated overnight at 4 °C with 20 μl of Protein G Plus/Protein A-agarose suspension (Merck) and 1 μg of ESC antibody. The same amount of normal rabbit serum was used instead of the ESC antibody as a negative control. Immunoblotting was carried out with human EZH2 antibody followed by incubation of Clean-blot IP (HRP) (Thermo) 1:1000 and visualized by chemiluminescence detection on film.
Immunostaining in Insect Brain
Pupal brains were dissected from day 4 nondiapause- and diapause-destined pupae, fixed in 3.7% formaldehyde, PBT (PBS containing 0.3% Triton X-100) for 1–2 h at room temperature, and washed 6–8 times in PBT. After washing with PBT, brains were blocked in 10% normal goat serum, PBT for 30 min at room temperature. A Zenon tricolor rabbit IgG labeling kit #1 (Invitrogen) was used for dual-labeling experiments. Har-PTTH antibody (22) was labeled with Alexa Fluor 488, and Har-ESC antibody was labeled with Alexa Fluor 555. IgG labeling was performed according to the Zenon IgG labeling kit protocol. After labeling, the primary antibody/Alexa Fluor complex was diluted in PBT and incubated for 1 h at room temperature. Then the samples were washed 3 times in PBT and fixed in 3.7% formaldehyde, PBT for 15 min at room temperature. Finally, the sample was washed, mounted, and examined with a fluorescence microscope (ImagerZ1, Carl Zeiss). The negative-control sample was processed with the same protocol except normal rabbit serum was substituted for the primary antibody.
ChIP
ChIP assays were performed as described (29, 30). Briefly, 60 pupal brains from day 4 nondiapause- or diapause-destined pupae were manually homogenized in 1 ml of nuclei extraction buffer (10 mm Tris-HCl (pH 7.5), 3 mm CaCl2, 0.5% Triton X-100, 0.25 m sucrose, 1 mm PMSF, and 1 mm DTT). Then 37% formaldehyde was added to a 1% final concentration, and the tube was rotated for 15 min at room temperature. After sonication, the DNA fragments were immunoprecipitated with anti-ESC, anti-H3K27me3, anti-H3K27Ac, and anti-H3 antibodies. Immunoprecipitated DNA and input DNA were subjected to quantitative real-time PCR analysis. The gene-specific primers are listed in Table 1.
RNA Knockdown in Vitro
dsRNAs of Har-ESC (519 bp of coding region) and GFP (700 bp of coding region) were synthesized using the T7 RiboMAX express RNAi system (Promega) and inserted into HzAm1 cells. Gene-specific primers are listed in Table 1. Transfection of dsRNA was performed with the Cellfectin II reagent (Invitrogen) as described in the Cellfectin II protocol. Cells were collected 24–72 h after RNAi. At least three independent dsRNA treatments were carried out to confirm the RNAi effects, and dsGFP served as a negative control.
Reporter Gene Assays
Reporter gene assays were performed as described (29). In brief, Har-PTTH gene promoter was amplified with specific primers (Table 1), digested with XhoI and Hind III, and subcloned into the digested pGL3-Basic vector (Promega). The recombinant pGL3-PTTH promoter vector was transfected or co-transfected with dsRNA. The pRL-TK vector (Promega) was used as an internal control for variations in transfection efficiency.
RESULTS
Molecular Characterization, Developmental Expression of ESC in H. armigera
An orthologous gene of ESC was found in H. armigera EST libraries (LIBEST_027843, LIBEST_027842), and a full-length ESC cDNA was obtained by rapid amplification of cDNA ends (GenBankTM number JQ744271). The cDNA encodes 413 amino acids with two conserved structural domains, the nuclear location signal and WD40 domain (a short structural motif of ∼40 amino acids, often terminating in a tryptophan-aspartic acid dipeptide). The amino acid sequence has high identity with ESCs of other species: Danaus plexippus (95%), Bombyx mori (93%), Drosophila melanogaster (56%), Mus musculus (60%), and Homo sapiens (63%) (Fig. 1). Hence, we refer to the cDNA as Har-ESC cDNA.
FIGURE 1.
Homology comparison to other known ESC proteins. Black shading represents ≥50% sequence identity. H. armigera, GenBankTM number JQ744271; D. plexippus, GenBankTM number EH_J72379.1; B. mori, NP_001188366.1; D. melanogaster, NP_477431.1); H. sapiens, NP_694536.1; M. musculus, NP_068676.1.
Quantitative real-time PCR (qPCR) and Western blots demonstrate that both Har-ESC mRNA (Fig. 2A) and protein (Fig. 2B) are more abundant in nondiapause-destined pupal brains than in brains from diapause-destined pupae. Western blots show two distinct bands for ESC; evidence from Drosophila indicates that the upper band is phosphorylated and the lower is non-phosphorylated ESC (28, 31). To further test this possibility, we used a phosphatase assay to determine if the upper band was phosphorylated ESC. Treatment of total protein with calf alkaline phosphatase eliminated the upper band (Fig. 2C), a result consistent with the observation that the N-terminal of ESC possesses a phosphorylation site (28). The lower band thus appears to be unphosphorylated ESC, as in Drosophila. This result implies that high expression of Har-ESC correlates with continuous development rather than diapause.
FIGURE 2.
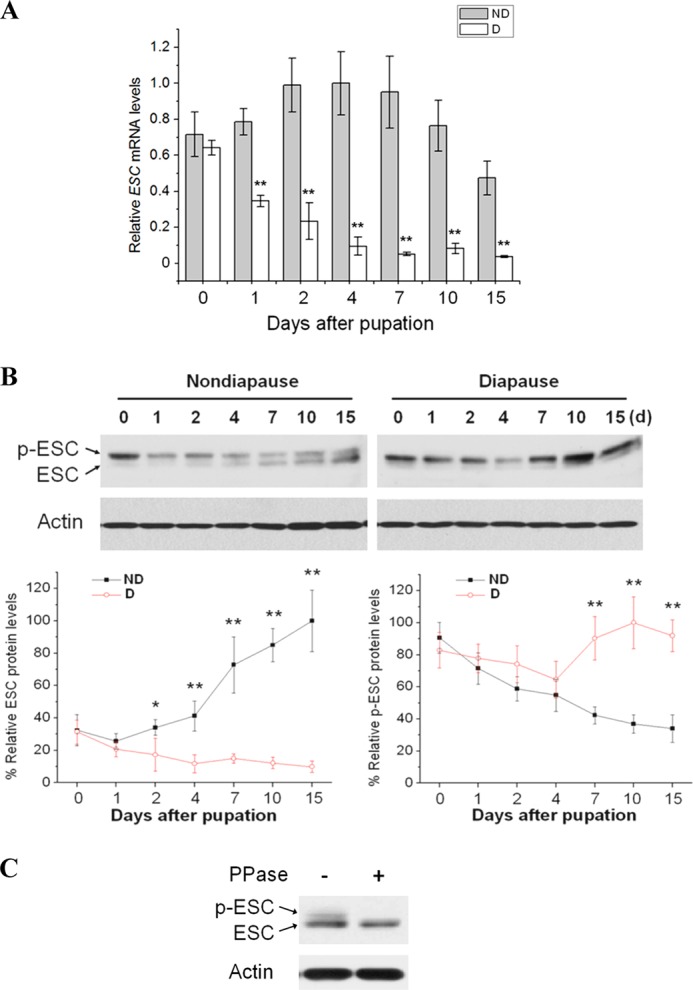
Developmental expression and molecular characterization of ESC and the methylation or acetylation of histone H3. A, qPCR analysis was used to monitor levels of ESC in brains of nondiapause (ND)- and diapause (D)-destined pupae, and expression levels were normalized to RPL32. Each point represents the mean ± S.D. of three independent replicates. **, p < 0.01 (determined by one-way ANOVA). B, shown is Western blot analysis of ESC and phosphorylated ESC (p-ESC). ESC protein was detected in brains of nondiapause (ND)- and diapause-destined (D) pupae using an ESC antibody; actin was the internal control. Protein bands were quantified and normalized to the level of Har-actin. Numbers represent days of pupal stage. Each point represents the mean ± S.D. of three independent replicates. * indicates a p value <0.05, and ** indicates a p value <0.01 (determined by one-way ANOVA). C, shown is a ESC phosphorylation assay in pupal brains. Protein extracted from the brains of day-10 nondiapause-destined pupae was treated with (+) or without (−) protein phosphatase (PPase). A Western blot performed using the ESC antibody.
Cellular Localization of ESC, Interaction between ESC and E(z), and Methylation Levels of Histone H3 in Brains of Nondiapause- and Diapause-destined Pupae
Har-ESC possesses the nuclear location signal and WD40 domains found in the N-terminal region (35–42 amino acids) and C-terminal region of ESC (132–406 amino acids), respectively. In preliminary experiments we found low efficiency of vector transfection in HzAm1 cells but higher efficiency in Sf9 cells. To determine the location of these key domains on ESC, full-length ESC (1–413 amino acids), N-terminal ESC (1–67 amino acids), and C-terminal ESC (67–413 amino acids) were constructed, fused with a pIZ/V5-GFP plasmid, and expressed in Sf9 cells. Full-length ESC and N-terminal ESC were mainly localized in the nucleus, but C-terminal ESC was only present in the cytoplasm (Fig. 3, A and B). These results imply that Har-ESC can enter the nucleus to exert its biological function.
FIGURE 3.
Expression and cellular localization of ESC, interaction between ESC and E(z), and methylation levels of histone H3 in brains of nondiapause- and diapause-destined pupae. A, shown is a Western blot analysis of Har-ESC transiently expressed in Sf9 cells. This is a schematic diagram of full-length ESC (ESC-FL), N-terminal ESC (ESC-N), and C-terminal ESC (ESC-C), which were used for transfection, respectively. NLS, nuclear location site, is in red; WD domain is in blue; full-length ESC-GFP at 1, 2, and 5 μg, N-terminal-GFP at 0.5 and 1 μg, and C-terminal-GFP at 1 and 2 μg transiently expressed in Sf9 cells. B, cellular localization of transiently expressed recombinant ESC. Hoechst 33342 staining shows the cell nuclei. Full-length ESC-GFP, N-terminal ESC-GFP, and C-terminal ESC-GFP were transfected into Sf9 cells; pIZ-V5-GFP (EGFP) was used as a control. Scale bar, 20 μm. C, ESC is physically associated with E(z). The HzAm1 cell extracts were immunoprecipitated (IP) with anti-ESC antibody, and the immunoblot (IB) was performed with anti-EZH2 antibody. Rabbit serum (IgG) served as a negative control. The 84-kDa band is E(z), and the 52-kDa band is IgG heavy chain. D, a Western blot analysis of H3K27me3, H3K4me3, and H3K27Ac detects methylation or acetylation of histone H3. The antibody against total H3 served as the loading control.
Previous reports indicate that ESC directly interacts with E(z) to form the PRC2 complex (32, 33). To confirm this in our system, a co-immunoprecipitation analysis was used to detect the ESC-E(z) complex; the result showed that ESC interacts with E(z), suggesting that Har-ESC and E(z) form the expected complex within the pupal brain of H. armigera (Fig. 3C).
Because ESC is a core component of the PRC2 complex and regulates trimethylation of H3K27 (32, 34), methylation levels of histone H3 were also examined in the two types of pupal brains. H3K27me3 was more abundant in nondiapause-type brains than in brains from diapausing pupae, whereas the opposite mark, H3K27Ac, decreased gradually in brains of nondiapausing pupae during pupal-adult development but was elevated in brains of diapausing pupae (Fig. 3D). Another active histone mark, H3K4me3, was not detected in either brain type. Our results show a consistently high level of H3K27me3 in the brains of developing individuals.
High Expression of Har-ESC in Two Pairs of PTTH Neurosecretory Cells and H3K27 Modification Recruited on the PTTH Promoter
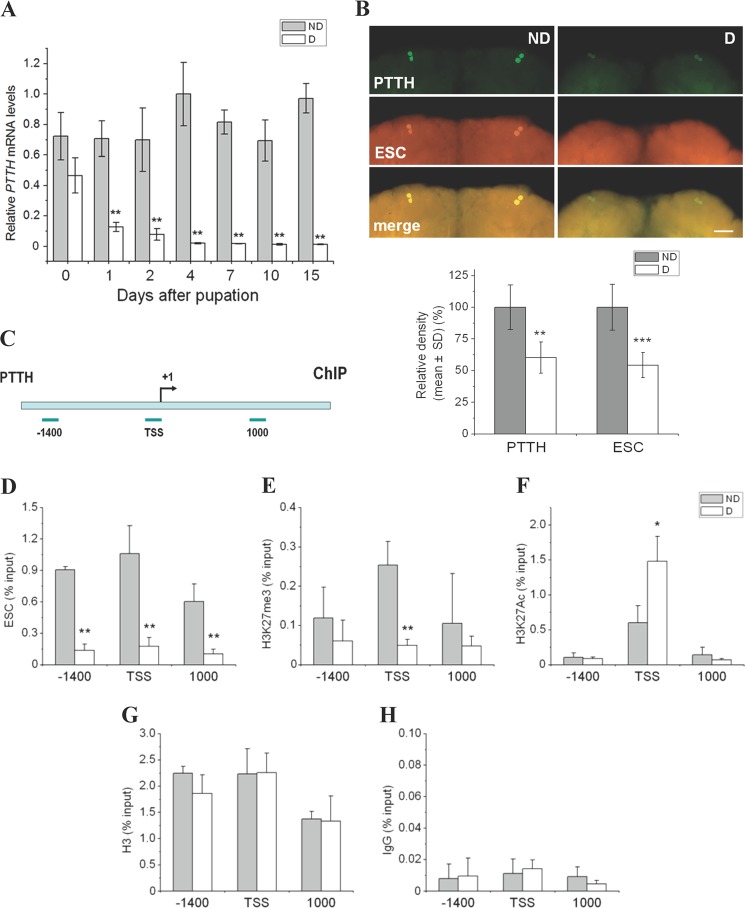
The differences in ESC expression and methyl modification we noted in nondiapause- and diapause-type pupal brains suggested that ESC may regulate PcG target genes to dictate the development fate of the pupa. We performed qPCR to detect PTTH mRNA, and the data were consistent with previous results showing higher expression of the PTTH gene in nondiapause-destined pupae compared with diapause-destined pupae (Fig. 4A). We thus tested whether ESC regulates PTTH gene expression. Co-immunostaining showed high levels of ESC in two pairs of PTTH neurosecretory cells in nondiapause pupal brains and low levels in PTTH cells of diapausing pupae (Fig. 4B), thus suggesting that Har-ESC may regulate Har-PTTH gene expression directly or indirectly.
FIGURE 4.
Co-localization of ESC and PTTH and histone modification of the PTTH gene in the pupal brain. A, quantitative PCR analysis was used to monitor levels of PTTH in brains of nondiapause- and diapause-destined pupae, and expression levels were normalized to RPL32. Each point represents the mean ± S.D. from three independent replicates. **, p < 0.01 (determined by one-way ANOVA). B, co-localization of ESC and PTTH in the brains of day 4 nondiapause- and diapause-destined pupae using antibodies against PTTH (green) and ESC (orange). Scale bar = 100 μm. Fluorescence of PTTH and ESC in two pairs of PTTH neurosecretory cells were quantified by Image-Pro Plus 6.0 from six independent replicates; PTTH and ESC values were determined by subtracting the background (fluorescence signal generated by the pre-immune serum, which was used as the negative control); diapause values were expressed relative to nondiapause values, which were set at 100%. **, p < 0.01; ***, p < 0.001 (determined by one-way ANOVA). C, shown is a schematic diagram of the PTTH promoter and the location of primers used for ChIP analysis. Primers were within 150 bp of the PTTH gene enhancer position −1400 bp (−1400), TSS, and PTTH gene body (from the TSS 1000 bp) (1000), respectively. The bent arrow indicates the transcription initiation site. D--H, ESC and two histone modification marks H3K27me3 and H3K27Ac were recruited on PTTH promoter or body; histone H3 and the pre-immune serum (IgG) were used as a control and a negative control, respectively. Antibodies against ESC, H3K27me3, H3K27Ac, and H3 were used for ChIP analysis. Enrichment of individual antibodies was plotted as % input DNA. ND, nondiapause-destined pupa; D, diapause-destined pupa. Each point represents the mean ± S.D. of three independent replicates. *, p < 0.05; **, p < 0.01 (determined by one-way ANOVA).
To test how ESC may regulate PTTH expression, chromatin immunoprecipitation was performed on the PTTH promoter in both nondiapause- and diapause-type pupal brains. ESC was more highly recruited by the PTTH promoter in nondiapause-type brains than in diapause-type brains regardless of the site, e.g. PTTH enhancer position (−1400), transcriptional start site (TSS), and at site 1000 on the gene body (1000) (Fig. 4, C and D). The response of H3K27me3 on the PTTH promoter was similar to ESC, but in this case the only significant difference was at the PTTH TSS position (Fig. 4E). By contrast, the opposing histone mark, H3K27Ac, was less recruited at the PTTH TSS position in nondiapause-type brains (Fig. 4F). The recruiting state of histone H3, which serves as an internal control, was similar in the two types of brains (Fig. 4G). Rabbit serum (pre-immune serum) was used as a negative control (Fig. 4H). This finding implies that Har-ESC can directly regulate PTTH expression by influencing H3K27me3 levels within the pupal brain of H. armigera.
ESC Knockdown Reduces H3K27me3 Mark and PTTH Promoter Activity
To further test the hypothesis that ESC and H3K27me3 regulate PTTH gene expression, RNAi was used to knock down ESC expression in HzAm1 cells. After treatment with dsESC, expression of the ESC gene and abundance of the protein decreased (Fig. 5, A and B). H3K27me3 was reduced by dsESC treatment after 24–72 h, whereas levels of E(z) protein and H3K27Ac were not altered (Fig. 5C). This result demonstrates that dsESC only knocks down the target ESC and its downstream histone mark. In addition, a recombinant pGL3-PTTH promoter-Luc vector was constructed and co-transfected with dsESC into HzAm1 cells, resulting in a decline of PTTH promoter activity (Fig. 5D). Taken together, the RNAi and luciferase activity experiments show that Har-ESC can activate the PTTH gene promoter.
FIGURE 5.
Effect of ESC knockdown on the H3K27me3 mark and promoter activity of the PTTH gene. A, shown are ESC mRNA levels after application of ESC dsRNA. HzAm1 cells were treated with indicated doses of ESC dsRNA, and expression of the endogenous ESC gene was examined by qPCR after 48 h. ESC mRNA levels were normalized to an internal control, RPL32. Each point represents the mean ± S.D. of three independent replicates. *, p < 0.05; **, p < 0.01 (determined by one-way ANOVA). B, shown is a Western blot for ESC protein in response to ESC dsRNA. Cells were treated with the indicated doses of ESC dsRNA, and the endogenous ESC protein was examined after 48 h. C, shown is a Western blot of PRC2 complex proteins ESC and E(z), and histone modification marks H3K27me3 and H3K27Ac in response to ESC dsRNA. Cells were treated with 2 μg of dsRNA, and levels of ESC, E(z), H3K27me3, and H3K27Ac were examined after treatment using the indicated antibodies. D, promoter activity of the PTTH gene by ESC dsRNA is shown. Shown is a schematic diagram of PTTH promoter containing enhancer region and intron 1 for luciferase activity (top). Recombinant pGL3-PTTH promoter-Luc vector was constructed and co-transfected into HzAm1 cells. Cells were treated with dsESC (1, 2, and 5 μg), and the luciferase assay was performed 48 h after ESC dsRNA treatment; dsGFP (5 μg) was used as a negative control. Luciferase activity from the pRL-TK vector was used as an internal control to normalize promoter activity. Each point represents the mean ± S.D. of three independent replicates. *, p < 0.05 (determined by one-way ANOVA).
DzNep Treatment Reduces H3K27me3 Mark, PTTH Promoter Activity, and Delays Pupal Development
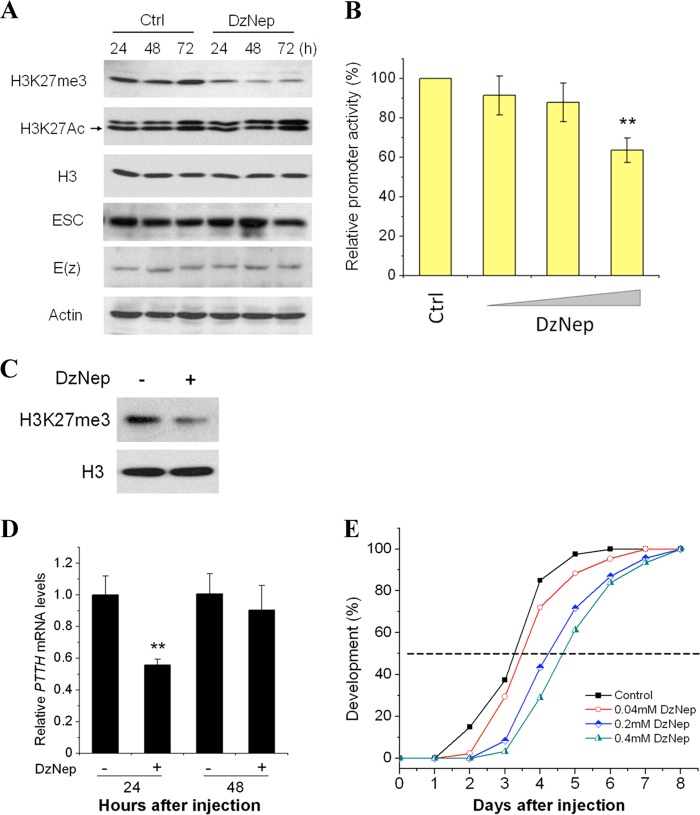
Because ESC is a core subunit in the PRC2 complex, we still did not know whether ESC activates the PTTH gene promoter via PRC2 mediation of methyl modification of histone. To test this possibility we used 3-deazaneplanocin A (DzNep) to inhibit methyltransferase activity of the PRC2 complex (35). After treatment of HzAm1 cells with 10 μm DzNep, H3K27me3 was significantly reduced, but another histone mark, H3K27Ac, was not (data not shown). Reduction of H3K27me3 persisted 72 h after treatment, whereas levels of ESC, E(z), and H3K27Ac were not affected (Fig. 6A).
FIGURE 6.
Effect of the inhibitor DzNep on the H3K27me3 mark, promoter activity of PTTH, developmental delay. A, shown is the effect of DzNep on PRC2 complex proteins ESC and E(z) and histone modification marks H3K27me3 and H3K27Ac. Cells were treated with 10 μm DzNep. After treatment, levels of ESC, E(z), H3K27me3, and H3K27Ac were examined by Western blot using corresponding antibodies. B, shown is promoter activity of PTTH after DzNep treatment. The luciferase assay was performed 48 h after treatment with 10 μm DzNep. Ethanol was used as a negative control (Ctrl). Each point represents the mean ± S.D. of six independent replicates. Luciferase activity from the pRL-TK vector was used as an internal control to normalize promoter activity. *, p < 0.05; **, p < 0.01 (determined by one-way ANOVA). C, shown is the effect of DzNep on H3K27me3 mark in vivo. Day-1 nondiapause pupae were injected with 5 μl of 20% ethanol (−) or 0.2 mm DzNep (+); pupal brains were collected 24 h after injection, and levels of H3K27me3 were measured by Western blots; H3 was used as a loading control. D, PTTH expression after injection of DzNep is shown. Day-1 nondiapausing pupae were injected with 0.2 mm DzNep, and expression levels of PTTH in pupal brains were detected by qPCR and normalized to the internal control, RPL32, 24 and 48 h after injection. Each point represents the mean ± S.D. of three independent replicates. **, p < 0.01 (determined by one-way ANOVA). E, shown is a developmental delay caused by injection of DzNep. Day-1 nondiapause pupae were injected with DzNep, and the developmental delay was determined by checking location of the pupal stemmata. 20% ethanol (Control), n = 40; 0.04 mm DzNep, n = 43; 0.2 mm DzNep, n = 46; 0.4 mm DzNep (pupae were injected with 0.2 mm DzNep on day 1 and 2, respectively), n = 31.
Because DzNep could repress H3K27me3 levels through reduction of PRC2 activity, we tested whether DzNep could eliminate promoter activity of the PTTH gene in vitro. After transfection of the pGL3-PTTH promoter and DzNep treatment in HzAm1 cells, luciferase activity decreased significantly compared with the control (Fig. 6B). These results suggest that Har-ESC activates the PTTH promoter by trimethylation of H3K27.
Because our results indicated that Har-ESC can activate Har-PTTH gene expression through trimethylation of H3K27 in vitro, we asked whether Har-ESC can also activate PTTH gene expression in vivo. Because RNAi does not work well in Lepidoptera and especially in neural systems, DzNep was injected into day 1 nondiapausing pupae to detect H3K27me3 levels and PTTH gene expression in the brain. The H3K27me3 level in the pupal brain was significantly reduced (Fig. 6C), and expression of the PTTH gene also dropped to approximately half that of the control 24 h after injection (Fig. 6D). As previously reported, migration of the pupal stemmata is a good landmark for pupal development (36); thus we injected a range of doses of DzNep into day-1 pupae and monitored the timing of the disappearance of pupal stemmata. In controls injected with solvent, stemmata disappeared in 3 days, whereas stemmata disappearance was delayed 1 day in pupae injected with 0.2 mm DzNep (Fig. 6E). In pupae injected twice with 0.2 mm DzNep (days 1 and 2 for a total of 0.4 mm) stemmata disappearance was delayed ∼1.5 days. These delays in pupal development are consistent with reduced PTTH gene expression by inhibition of H3K27me3.
DISCUSSION
Previous studies showed that lifespan can be controlled epigenetically via the deacetylase Sir2 family (37, 38). Recently, histone methylation was also implicated as a regulator of longevity: loss of demethylase UTX-1 in C. elegans increases the global H3K27me3 level and extends lifespan by down-regulation of the daf-2 gene (19, 20); reduction of the H3K27me3 level in Drosophila E(z) and esc mutants also increases longevity (21). Thus, there are obvious differences in the effect of methylation on longevity in these two species.
In H. armigera, developmental arrest is induced by the short day length perceived during the larval stage, and a reduction of PTTH expression prompts pupal diapause, thus prolonging the lifespan (1). But, the regulatory mechanism for PTTH expression is unknown. The results we present here demonstrate that these environmental factors likely regulate PTTH gene expression by PRC2 mediation of trimethylation of H3K27 and result in a great protraction of the pupal stage. This conclusion is based on the fact that both Har-ESC mRNA and protein are down-regulated in diapause-type pupal brains, suggesting that high Har-ESC expression is associated with an active brain programmed for continuous development rather than diapause. Intriguingly, phosphorylated ESC protein, which appears to be the inactive form of ESC (28, 33), was high in both nondiapause- and diapause-type pupae at pupation (day 0) but then decreased gradually by day 4 after pupation. In nondiapause-type pupae, a low level of phosphorylated ESC was maintained from days 7 to 15, accompanying pupal-adult development, whereas the level increased gradually from days 7 to 10 in diapausing pupae and remained high through day 15. This species enters pupal diapause on day 10 after pupation; thus the elevation of phosphorylated ESC may contribute to the developmental arrest.
High levels of ESC are present in two pairs of PTTH neurosecretory cells in nondiapause-type pupal brains but not in the brains of diapause-destined pupae, a finding that suggests ESC regulates PTTH expression. Our ChIP-qPCR experiments showed that ESC binds to the PTTH promoter, modifies trimethylation of histone H3K27, and up-regulates PTTH gene expression in nondiapausing individuals. H3K27me3 is the substrate for the PRC2 complex and can thus affect transcriptional expression (34, 39), but H3K27me3 levels do not completely correspond with the abundance of PRC2 complex proteins in whole genome study (9). In this study we have shown that DzNep, a specific inhibitor of PRC2 activity, reduced levels of H3K27me3, resulting in a decrease of PTTH promoter activity in vitro and repression of PTTH gene expression in vivo. Based on these results, we suggest that PRC2 activity is significantly activated at TSS to regulate PTTH transcription, and indeed the entire PTTH gene is bound by ESC. In contrast, a low level of H3K27me3 results in low PTTH gene expression and consequently the pupae enter diapause, thereby dramatically extending their lifespan.
In addition, we showed that ESC interacts with E(z), as evidenced by the fact that both dsESC and DzNep treatments reduced H3K27me3 levels. Previous studies have shown that the PRC2 complex does not show enzyme activity unless all subunits are activated (32, 34, 40). Our results suggest that the two other PRC2 subunits, E(z) and Su(z)12, are likely to be activated along with ESC.
In conclusion, we found that day length, the dominant environmental stimulus for diapause, regulates ESC and PRC2 activity. We speculate that long day length stimulates photoperiod-sensitive proteins and/or signals (27, 41), and then these signals elevate ESC and PRC2 activity to mediate trimethylation of H3K27 for PTTH transcription, and this in turn leads to uninterrupted insect development, whereas short day length results in low activity of ESC and PRC2 and low levels of histone methylation, leading to developmental arrest (Fig. 7). Many details of this molecular mechanism, however, remain unknown. PcG proteins are best known as transcriptional repressors that control developmental genes by eliciting high levels of histone methylation (9, 42, 43). But in this case Har-ESC does not act as a repressor but instead activates PTTH gene expression in nondiapause individuals to regulate insect development by PRC2 mediation of the trimethylation of H3K27. In addition, our finding demonstrates that low levels of histone methylation caused by a reduction of the PRC2 protein ESC can induce diapause by repressing PTTH gene expression. This result is consistent with observations in Drosophila showing that a reduction of H3K27me3 level in E(z) and esc mutants leads to longevity (21). We believe this is the first report suggesting that a PRC2 complex protein functions as an activator and that low levels of histone methylation can prolong development in insects by controlling PTTH gene expression.
FIGURE 7.

Schematic representation of the epigenetic mechanism regulating development and diapause in H. armigera. Larvae were cultured at 20 °C and 14 h light:10 h dark (long day length) or 10 h light:14 h dark (short day length). Pupae develop without interruption and emerge as adults within 3 weeks under long-day conditions, whereas pupae enter diapause for over 3 months when reared under short-day conditions. In developing (nondiapausing) pupae, long-day length activates PTTH gene expression by stimulating photoperiod-sensitive (PPS) proteins and/or signals, which in turn activate PRC2-mediation of the methyl modification of histone H3 at lysine 27 (the green dot represents the trimethyl site) and initiate adult development. Short-day length represses PTTH gene expression with low activity of PRC2 and low level of the methyl modification, causing pupal diapause and lifespan extension.
This work was supported by Natural Scientific Foundation of China Grant-in-aid 31230066, National Basic Research Program of China, Ministry of Science and Technology Grant 2012CB114101 (to W.-H. X.), and United States Department of Agriculture National Institute of Food and Agriculture Grant 2011-67013-30199 (D. L. D.).
- PTTH
- prothoracicotropic hormone
- ESC
- extra sex combs
- PRC2
- polycomb repressive complex 2
- H3K27me3
- trimethylation of lysine 27 on histone H3
- Har-ESC
- Helicoverpa armigera ESC
- E(z)
- enhancer of zeste
- Su(z)12
- suppressor of zeste 12
- DzNep
- 3-deazaneplanocin A
- qPCR
- quantitative real-time PCR
- TSS
- transcriptional start site.
REFERENCES
- 1. Denlinger D. L., Yocum G. D., Rinehart J. P. (2005) in Comprehensive Insect Molecular Science (Gilbert L. I., Iatrou K., Gill S., eds.) pp. 615–650, Elsevier, Amsterdam [Google Scholar]
- 2. Hu P. J. (2007) Dauer. WormBook 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kawakami A., Kataoka H., Oka T., Mizoguchi A., Kimura-Kawakami M., Adachi T., Iwami M., Nagasawa H., Suzuki A., Ishizaki H. (1990) Molecular cloning of the Bombyx mori prothoracicotropic hormone. Science 247, 1333–1335 [DOI] [PubMed] [Google Scholar]
- 4. McBrayer Z., Ono H., Shimell M., Parvy J. P., Beckstead R. B., Warren J. T., Thummel C. S., Dauphin-Villemant C., Gilbert L. I., O'Connor M. B. (2007) Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev. Cell 13, 857–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rewitz K. F., Yamanaka N., Gilbert L. I., O'Connor M. B. (2009) The insect neuropeptide PTTH activates receptor-tyrosine kinase torso to initiate metamorphosis. Science 326, 1403–1405 [DOI] [PubMed] [Google Scholar]
- 6. Gilbert L. I., Rybczynski R., Warren J. T. (2002) Control and biochemical nature of the ecdysteroidogenic pathway. Annu. Rev. Entomol. 47, 883–916 [DOI] [PubMed] [Google Scholar]
- 7. Ringrose L., Paro R. (2004) Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 38, 413–443 [DOI] [PubMed] [Google Scholar]
- 8. Campos E. I., Reinberg D. (2009) Histones. Annotating chromatin. Annu. Rev. Genet. 43, 559–599 [DOI] [PubMed] [Google Scholar]
- 9. Schwartz Y. B., Kahn T. G., Nix D. A., Li X. Y., Bourgon R., Biggin M., Pirrotta V. (2006) Genome-wide analysis of polycomb targets in Drosophila melanogaster. Nat. Genet. 38, 700–705 [DOI] [PubMed] [Google Scholar]
- 10. Tolhuis B., de Wit E., Muijrers I., Teunissen H., Talhout W., van Steensel B., van Lohuizen M. (2006) Genome-wide profiling of PRC1 and PRC2 polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 38, 694–699 [DOI] [PubMed] [Google Scholar]
- 11. Lee T. I., Jenner R. G., Boyer L. A., Guenther M. G., Levine S. S., Kumar R. M., Chevalier B., Johnstone S. E., Cole M. F., Isono K., Koseki H., Fuchikami T., Abe K., Murray H. L., Zucker J. P., Yuan B., Bell G. W., Herbolsheimer E., Hannett N. M., Sun K., Odom D. T., Otte A. P., Volkert T. L., Bartel D. P., Melton D. A., Gifford D. K., Jaenisch R., Young R. A. (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., Bell G. W., Otte A. P., Vidal M., Gifford D. K., Young R. A., Jaenisch R. (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 [DOI] [PubMed] [Google Scholar]
- 13. Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. (2007) Genome regulation by polycomb and trithorax proteins. Cell 128, 735–745 [DOI] [PubMed] [Google Scholar]
- 14. Pasini D., Bracken A. P., Hansen J. B., Capillo M., Helin K. (2007) The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 27, 3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Young M. D., Willson T. A., Wakefield M. J., Trounson E., Hilton D. J., Blewitt M. E., Oshlack A., Majewski I. J. (2011) ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res. 39, 7415–7427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mousavi K., Zare H., Wang A. H., Sartorelli V. (2012) Polycomb protein Ezh1 promotes RNA polymerase II elongation. Mol Cell 45, 255–262 [DOI] [PubMed] [Google Scholar]
- 17. Sparmann A., van Lohuizen M. (2006) Polycomb silencers control cell fate, development, and cancer. Nat. Rev. Cancer 6, 846–856 [DOI] [PubMed] [Google Scholar]
- 18. Schwartz Y. B., Pirrotta V. (2008) Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 20, 266–273 [DOI] [PubMed] [Google Scholar]
- 19. Jin C., Li J., Green C. D., Yu X., Tang X., Han D., Xian B., Wang D., Huang X., Cao X., Yan Z., Hou L., Liu J., Shukeir N., Khaitovich P., Chen C. D., Zhang H., Jenuwein T., Han J. D. (2011) Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 14, 161–172 [DOI] [PubMed] [Google Scholar]
- 20. Maures T. J., Greer E. L., Hauswirth A. G., Brunet A. (2011) The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell 10, 980–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Siebold A. P., Banerjee R., Tie F., Kiss D. L., Moskowitz J., Harte P. J. (2010) Polycomb repressive complex 2 and trithorax modulate Drosophila longevity and stress resistance. Proc. Natl. Acad. Sci. U.S.A. 107, 169–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wei Z. J., Zhang Q. R., Kang L., Xu W. H., Denlinger D. L. (2005) Molecular characterization and expression of prothoracicotropic hormone during development and pupal diapause in the cotton bollworm, Helicoverpa armigera. J. Insect Physiol. 51, 691–700 [DOI] [PubMed] [Google Scholar]
- 23. Hou C. L., Xu W. H. (2007) Synthesis dynamic and developmental profile of prothoracicotropic hormone between diapause- and nondiapause-destined individuals in Helicoverpa armigera. Chin. Sci. Bull. 52, 2095–2099 [Google Scholar]
- 24. Wei Z. J., Yu M., Tang S. M., Yi Y. Z., Hong G. Y., Jiang S. T. (2011) Transcriptional regulation of the gene for prothoracicotropic hormone in the silkworm, Bombyx mori. Mol. Biol. Rep. 38, 1121–1127 [DOI] [PubMed] [Google Scholar]
- 25. Ohtsuka K., Atsumi T., Fukushima Y., Shiomi K. (2011) Identification of a cis-regulatory element that directs prothoracicotropic hormone gene expression in the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 41, 356–361 [DOI] [PubMed] [Google Scholar]
- 26. Lu Y. X., Xu W. H. (2010) Proteomic and phosphoproteomic analysis at diapause initiation in the cotton bollworm, Helicoverpa armigera. J. Proteome Res. 9, 5053–5064 [DOI] [PubMed] [Google Scholar]
- 27. Zhang Q., Lu Y. X., Xu W. H. (2012) Integrated proteomic and metabolomic analysis of larval brain associated with diapause induction and preparation in the cotton bollworm, Helicoverpa armigera. J. Proteome Res. 11, 1042–1053 [DOI] [PubMed] [Google Scholar]
- 28. Tie F., Siebold A. P., Harte P. J. (2005) The N terminus of Drosophila ESC mediates its phosphorylation and dimerization. Biochem. Biophys. Res. Commun. 332, 622–632 [DOI] [PubMed] [Google Scholar]
- 29. Bao B., Hong B., Feng Q. L., Xu W. H. (2011) Transcription factor fork head regulates the promoter of diapause hormone gene in the cotton bollworm, Helicoverpa armigera, and the modification of SUMOylation. Insect Biochem. Mol. Biol. 41, 670–679 [DOI] [PubMed] [Google Scholar]
- 30. Zhang Y., Lu Y. X., Liu J., Yang C., Feng Q. L., Xu W. H. (2013) A regulatory pathway, ecdysone-transcription factor relish-cathepsin L, is involved in insect fat body dissociation. PLoS Genet. 9, e1003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ng J., Hart C. M., Morgan K., Simon J. A. (2000) A Drosophila ESC-E(Z) protein complex is distinct from other polycomb group complexes and contains covalently modified ESC. Mol. Cell. Biol. 20, 3069–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Müller J., Hart C. M., Francis N. J., Vargas M. L., Sengupta A., Wild B., Miller E. L., O'Connor M. B., Kingston R. E., Simon J. A. (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111, 197–208 [DOI] [PubMed] [Google Scholar]
- 33. Tie F., Furuyama T., Harte P. J. (1998) The Drosophila polycomb group proteins ESC and E(Z) bind directly to each other and co-localize at multiple chromosomal sites. Development 125, 3483–3496 [DOI] [PubMed] [Google Scholar]
- 34. Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V. (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell 111, 185–196 [DOI] [PubMed] [Google Scholar]
- 35. Fiskus W., Wang Y., Sreekumar A., Buckley K. M., Shi H., Jillella A., Ustun C., Rao R., Fernandez P., Chen J., Balusu R., Koul S., Atadja P., Marquez V. E., Bhalla K. N. (2009) Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood 114, 2733–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Phillips J. R., Newsom L. D. (1966) Diapause in Heliothis zea and Heliothis virescens (Lepidoptera: Noctuidea). Annals Entomological Society of America 59, 154–159 [Google Scholar]
- 37. Dang W., Steffen K. K., Perry R., Dorsey J. A., Johnson F. B., Shilatifard A., Kaeberlein M., Kennedy B. K., Berger S. L. (2009) Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459, 802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Blander G., Guarente L. (2004) The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73, 417–435 [DOI] [PubMed] [Google Scholar]
- 39. Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D. (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16, 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nekrasov M., Wild B., Müller J. (2005) Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep 6, 348–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tian K., Xu W. H. (2013) High expression of PP2A-Aα is associated with diapause induction during the photoperiod-sensitive stage of the cotton bollworm, Helicoverpa armigera. J. Insect Physiol. 59, 588–594 [DOI] [PubMed] [Google Scholar]
- 42. Sawarkar R., Paro R. (2010) Interpretation of developmental signaling at chromatin. The polycomb perspective. Dev. Cell 19, 651–661 [DOI] [PubMed] [Google Scholar]
- 43. Simon J. A., Kingston R. E. (2009) Mechanisms of polycomb gene silencing. Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10, 697–708 [DOI] [PubMed] [Google Scholar]