Abstract
The repair of some types of DNA double-strand breaks is thought to proceed through DNA flap structure intermediates. A DNA flap is a bifurcated structure composed of double-stranded DNA and a displaced single-strand. To identify DNA flap cleaving activities in mammalian nuclear extracts, we created an assay utilizing a synthetic DNA flap substrate. This assay has allowed the first purification of a mammalian DNA structure-specific nuclease. The enzyme described here, flap endonuclease-1 (FEN-1), cleaves DNA flap strands that terminate with a 5' single-stranded end. As expected for an enzyme which functions in double-strand break repair flap resolution, FEN-1 cleavage is flap strand-specific and independent of flap strand length. Furthermore, efficient flap cleavage requires the presence of the entire flap structure. Substrates missing one strand are not cleaved by FEN-1. Other branch structures, including Holliday junctions, are also not cleaved by FEN-1. In addition to endonuclease activity, FEN-1 has a 5'-3' exonuclease activity which is specific for double-stranded DNA. The endo- and exonuclease activities of FEN-1 are discussed in the context of DNA replication, recombination and repair.
Full text
PDF
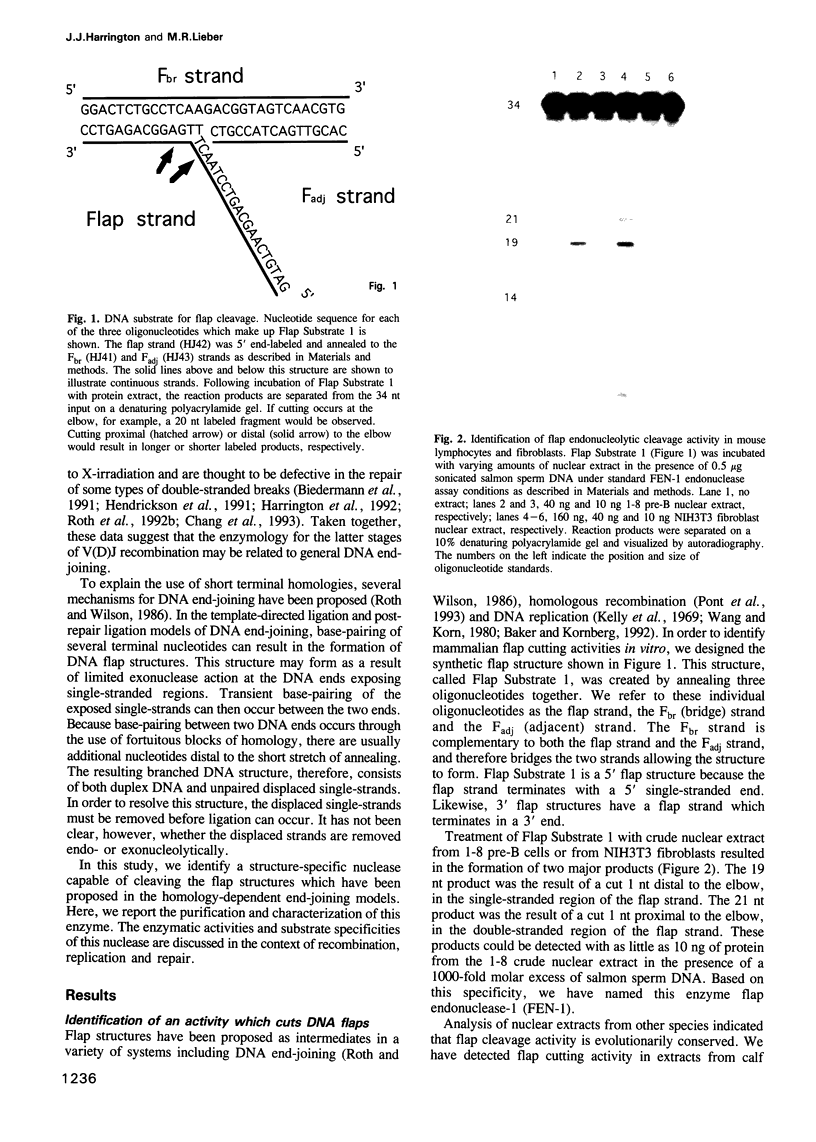
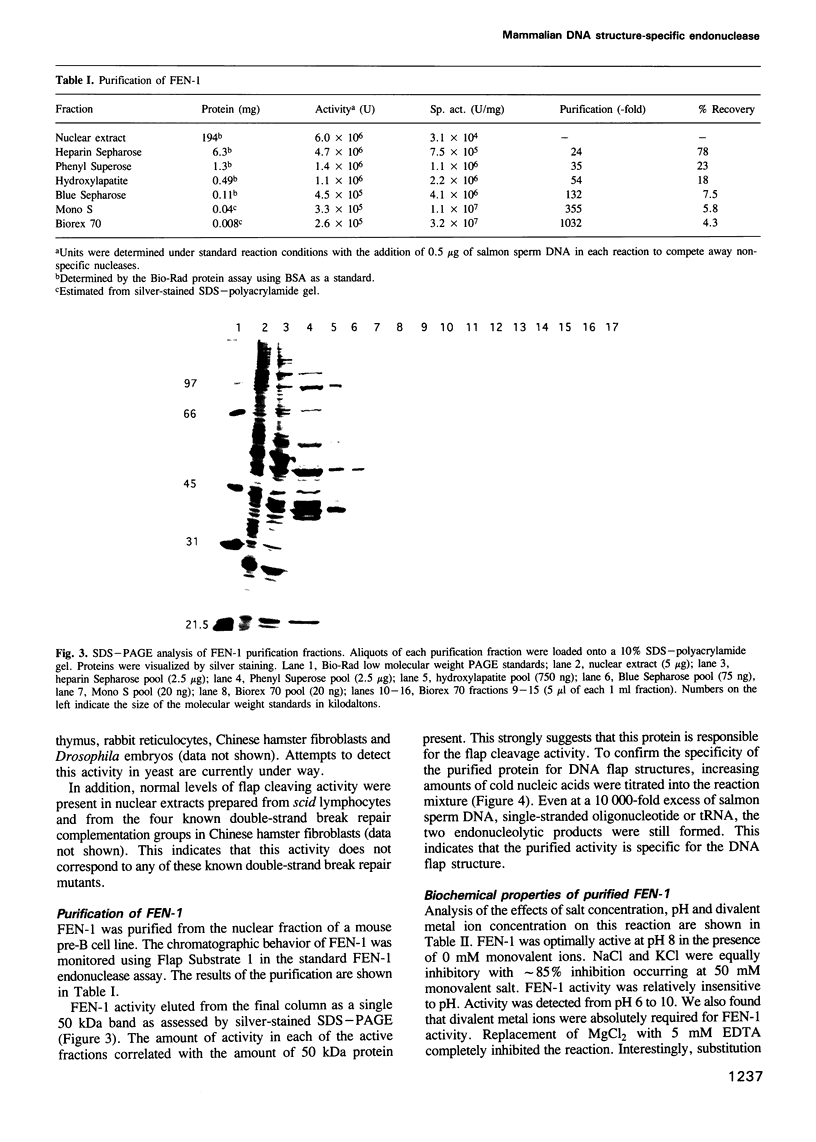







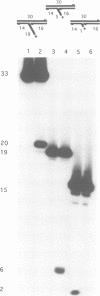
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt F. W., Baltimore D. Joining of immunoglobulin heavy chain gene segments: implications from a chromosome with evidence of three D-JH fusions. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4118–4122. doi: 10.1073/pnas.79.13.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt F. W., Yancopoulos G. D., Blackwell T. K., Wood C., Thomas E., Boss M., Coffman R., Rosenberg N., Tonegawa S., Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984 Jun;3(6):1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asarnow D. M., Cado D., Raulet D. H. Selection is not required to produce invariant T-cell receptor gamma-gene junctional sequences. Nature. 1993 Mar 11;362(6416):158–160. doi: 10.1038/362158a0. [DOI] [PubMed] [Google Scholar]
- Biedermann K. A., Sun J. R., Giaccia A. J., Tosto L. M., Brown J. M. scid mutation in mice confers hypersensitivity to ionizing radiation and a deficiency in DNA double-strand break repair. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1394–1397. doi: 10.1073/pnas.88.4.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosma G. C., Custer R. P., Bosma M. J. A severe combined immunodeficiency mutation in the mouse. Nature. 1983 Feb 10;301(5900):527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- Chang C., Biedermann K. A., Mezzina M., Brown J. M. Characterization of the DNA double strand break repair defect in scid mice. Cancer Res. 1993 Mar 15;53(6):1244–1248. [PubMed] [Google Scholar]
- Connolly B., Parsons C. A., Benson F. E., Dunderdale H. J., Sharples G. J., Lloyd R. G., West S. C. Resolution of Holliday junctions in vitro requires the Escherichia coli ruvC gene product. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6063–6067. doi: 10.1073/pnas.88.14.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly B., West S. C. Genetic recombination in Escherichia coli: Holliday junctions made by RecA protein are resolved by fractionated cell-free extracts. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8476–8480. doi: 10.1073/pnas.87.21.8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elborough K. M., West S. C. Resolution of synthetic Holliday junctions in DNA by an endonuclease activity from calf thymus. EMBO J. 1990 Sep;9(9):2931–2936. doi: 10.1002/j.1460-2075.1990.tb07484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney A. J. Predominance of VH-D-JH junctions occurring at sites of short sequence homology results in limited junctional diversity in neonatal antibodies. J Immunol. 1992 Jul 1;149(1):222–229. [PubMed] [Google Scholar]
- Feeney A. J. Predominance of the prototypic T15 anti-phosphorylcholine junctional sequence in neonatal pre-B cells. J Immunol. 1991 Dec 15;147(12):4343–4350. [PubMed] [Google Scholar]
- Gerondakis S., Cory S., Adams J. M. Translocation of the myc cellular oncogene to the immunoglobulin heavy chain locus in murine plasmacytomas is an imprecise reciprocal exchange. Cell. 1984 Apr;36(4):973–982. doi: 10.1016/0092-8674(84)90047-3. [DOI] [PubMed] [Google Scholar]
- Gerstein R. M., Lieber M. R. Extent to which homology can constrain coding exon junctional diversity in V(D)J recombination. Nature. 1993 Jun 17;363(6430):625–627. doi: 10.1038/363625a0. [DOI] [PubMed] [Google Scholar]
- Goulian M., Richards S. H., Heard C. J., Bigsby B. M. Discontinuous DNA synthesis by purified mammalian proteins. J Biol Chem. 1990 Oct 25;265(30):18461–18471. [PubMed] [Google Scholar]
- Gu H., Förster I., Rajewsky K. Sequence homologies, N sequence insertion and JH gene utilization in VHDJH joining: implications for the joining mechanism and the ontogenetic timing of Ly1 B cell and B-CLL progenitor generation. EMBO J. 1990 Jul;9(7):2133–2140. doi: 10.1002/j.1460-2075.1990.tb07382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Kitamura D., Rajewsky K. B cell development regulated by gene rearrangement: arrest of maturation by membrane-bound D mu protein and selection of DH element reading frames. Cell. 1991 Apr 5;65(1):47–54. doi: 10.1016/0092-8674(91)90406-o. [DOI] [PubMed] [Google Scholar]
- Harrington J., Hsieh C. L., Gerton J., Bosma G., Lieber M. R. Analysis of the defect in DNA end joining in the murine scid mutation. Mol Cell Biol. 1992 Oct;12(10):4758–4768. doi: 10.1128/mcb.12.10.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson E. A., Qin X. Q., Bump E. A., Schatz D. G., Oettinger M., Weaver D. T. A link between double-strand break-related repair and V(D)J recombination: the scid mutation. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4061–4065. doi: 10.1073/pnas.88.10.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimi Y., Claude A., Bullock P., Hurwitz J. Complete enzymatic synthesis of DNA containing the SV40 origin of replication. J Biol Chem. 1988 Dec 25;263(36):19723–19733. [PubMed] [Google Scholar]
- Iwasaki H., Takahagi M., Shiba T., Nakata A., Shinagawa H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 1991 Dec;10(13):4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo P. A. Studies on mammalian mutants defective in rejoining double-strand breaks in DNA. Mutat Res. 1990 Jul;239(1):1–16. doi: 10.1016/0165-1110(90)90028-a. [DOI] [PubMed] [Google Scholar]
- Jensch F., Kemper B. Endonuclease VII resolves Y-junctions in branched DNA in vitro. EMBO J. 1986 Jan;5(1):181–189. doi: 10.1002/j.1460-2075.1986.tb04194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger R., Berg P. Repair of deletions and double-strand gaps by homologous recombination in a mammalian in vitro system. Mol Cell Biol. 1991 Jan;11(1):445–457. doi: 10.1128/mcb.11.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger R., Podust V., Hübscher U., Berg P. A mammalian protein complex that repairs double-strand breaks and deletions by recombination. J Biol Chem. 1993 Jul 15;268(20):15070–15079. [PubMed] [Google Scholar]
- Kleff S., Kemper B. Initiation of heteroduplex-loop repair by T4-encoded endonuclease VII in vitro. EMBO J. 1988 May;7(5):1527–1535. doi: 10.1002/j.1460-2075.1988.tb02972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleff S., Kemper B., Sternglanz R. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 1992 Feb;11(2):699–704. doi: 10.1002/j.1460-2075.1992.tb05102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopchick J. J., Stacey D. W. Differences in intracellular DNA ligation after microinjection and transfection. Mol Cell Biol. 1984 Feb;4(2):240–246. doi: 10.1128/mcb.4.2.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lewis S., Gellert M. The mechanism of antigen receptor gene assembly. Cell. 1989 Nov 17;59(4):585–588. doi: 10.1016/0092-8674(89)90002-0. [DOI] [PubMed] [Google Scholar]
- Lieber M. R., Hesse J. E., Lewis S., Bosma G. C., Rosenberg N., Mizuuchi K., Bosma M. J., Gellert M. The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell. 1988 Oct 7;55(1):7–16. doi: 10.1016/0092-8674(88)90004-9. [DOI] [PubMed] [Google Scholar]
- Lieber M. R. Site-specific recombination in the immune system. FASEB J. 1991 Nov;5(14):2934–2944. doi: 10.1096/fasebj.5.14.1752360. [DOI] [PubMed] [Google Scholar]
- Lutze L. H., Cleaver J. E., Morgan W. F., Winegar R. A. Mechanisms involved in rejoining DNA double-strand breaks induced by ionizing radiation and restriction enzymes. Mutat Res. 1993 May;299(3-4):225–232. doi: 10.1016/0165-1218(93)90099-y. [DOI] [PubMed] [Google Scholar]
- Lyamichev V., Brow M. A., Dahlberg J. E. Structure-specific endonucleolytic cleavage of nucleic acids by eubacterial DNA polymerases. Science. 1993 May 7;260(5109):778–783. doi: 10.1126/science.7683443. [DOI] [PubMed] [Google Scholar]
- Malynn B. A., Blackwell T. K., Fulop G. M., Rathbun G. A., Furley A. J., Ferrier P., Heinke L. B., Phillips R. A., Yancopoulos G. D., Alt F. W. The scid defect affects the final step of the immunoglobulin VDJ recombinase mechanism. Cell. 1988 Aug 12;54(4):453–460. doi: 10.1016/0092-8674(88)90066-9. [DOI] [PubMed] [Google Scholar]
- Morzycka-Wroblewska E., Lee F. E., Desiderio S. V. Unusual immunoglobulin gene rearrangement leads to replacement of recombinational signal sequences. Science. 1988 Oct 14;242(4876):261–263. doi: 10.1126/science.3140378. [DOI] [PubMed] [Google Scholar]
- Mueller J. E., Newton C. J., Jensch F., Kemper B., Cunningham R. P., Kallenbach N. R., Seeman N. C. Resolution of Holliday junction analogs by T4 endonuclease VII can be directed by substrate structure. J Biol Chem. 1990 Aug 15;265(23):13918–13924. [PubMed] [Google Scholar]
- Müller B., Jones C., Kemper B., West S. C. Enzymatic formation and resolution of Holliday junctions in vitro. Cell. 1990 Jan 26;60(2):329–336. doi: 10.1016/0092-8674(90)90747-3. [DOI] [PubMed] [Google Scholar]
- Müller B., Jones C., West S. C. T7 endonuclease I resolves Holliday junctions formed in vitro by RecA protein. Nucleic Acids Res. 1990 Oct 11;18(19):5633–5636. doi: 10.1093/nar/18.19.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons C. A., Kemper B., West S. C. Interaction of a four-way junction in DNA with T4 endonuclease VII. J Biol Chem. 1990 Jun 5;265(16):9285–9289. [PubMed] [Google Scholar]
- Parsons C. A., Murchie A. I., Lilley D. M., West S. C. Resolution of model Holliday junctions by yeast endonuclease: effect of DNA structure and sequence. EMBO J. 1989 Jan;8(1):239–246. doi: 10.1002/j.1460-2075.1989.tb03369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons C. A., West S. C. Specificity of binding to four-way junctions in DNA by bacteriophage T7 endonuclease I. Nucleic Acids Res. 1990 Aug 11;18(15):4377–4384. doi: 10.1093/nar/18.15.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergola F., Zdzienicka M. Z., Lieber M. R. V(D)J recombination in mammalian cell mutants defective in DNA double-strand break repair. Mol Cell Biol. 1993 Jun;13(6):3464–3471. doi: 10.1128/mcb.13.6.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picksley S. M., Parsons C. A., Kemper B., West S. C. Cleavage specificity of bacteriophage T4 endonuclease VII and bacteriophage T7 endonuclease I on synthetic branch migratable Holliday junctions. J Mol Biol. 1990 Apr 20;212(4):723–735. doi: 10.1016/0022-2836(90)90233-C. [DOI] [PubMed] [Google Scholar]
- Pont-Kingdon G., Dawson R. J., Carroll D. Intermediates in extrachromosomal homologous recombination in Xenopus laevis oocytes: characterization by electron microscopy. EMBO J. 1993 Jan;12(1):23–34. doi: 10.1002/j.1460-2075.1993.tb05628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottmeyer S., Kemper B. T4 endonuclease VII resolves cruciform DNA with nick and counter-nick and its activity is directed by local nucleotide sequence. J Mol Biol. 1992 Feb 5;223(3):607–615. doi: 10.1016/0022-2836(92)90977-r. [DOI] [PubMed] [Google Scholar]
- Roth D. B., Menetski J. P., Nakajima P. B., Bosma M. J., Gellert M. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992 Sep 18;70(6):983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- Roth D. B., Nakajima P. B., Menetski J. P., Bosma M. J., Gellert M. V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor delta rearrangement signals. Cell. 1992 Apr 3;69(1):41–53. doi: 10.1016/0092-8674(92)90117-u. [DOI] [PubMed] [Google Scholar]
- Roth D. B., Porter T. N., Wilson J. H. Mechanisms of nonhomologous recombination in mammalian cells. Mol Cell Biol. 1985 Oct;5(10):2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Wilson J. H. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986 Dec;6(12):4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatino R. D., Myers T. W., Bambara R. A. Substrate specificity of the exonuclease associated with calf DNA polymerase. Cancer Res. 1990 Sep 1;50(17):5340–5344. [PubMed] [Google Scholar]
- Salganik R. I., Dianov G. L. Molecular mechanisms of the formation of DNA double-strand breaks and induction of genomic rearrangements. Mutat Res. 1992 Apr;266(2):163–170. doi: 10.1016/0027-5107(92)90183-3. [DOI] [PubMed] [Google Scholar]
- Schuler W., Weiler I. J., Schuler A., Phillips R. A., Rosenberg N., Mak T. W., Kearney J. F., Perry R. P., Bosma M. J. Rearrangement of antigen receptor genes is defective in mice with severe combined immune deficiency. Cell. 1986 Sep 26;46(7):963–972. doi: 10.1016/0092-8674(86)90695-1. [DOI] [PubMed] [Google Scholar]
- Siegal G., Turchi J. J., Myers T. W., Bambara R. A. A 5' to 3' exonuclease functionally interacts with calf DNA polymerase epsilon. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9377–9381. doi: 10.1073/pnas.89.20.9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G. R., Wahl G. M. Gene amplification. Annu Rev Biochem. 1984;53:447–491. doi: 10.1146/annurev.bi.53.070184.002311. [DOI] [PubMed] [Google Scholar]
- Subramani S., Berg P. Homologous and nonhomologous recombination in monkey cells. Mol Cell Biol. 1983 Jun;3(6):1040–1052. doi: 10.1128/mcb.3.6.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switzer R. C., 3rd, Merril C. R., Shifrin S. A highly sensitive silver stain for detecting proteins and peptides in polyacrylamide gels. Anal Biochem. 1979 Sep 15;98(1):231–237. doi: 10.1016/0003-2697(79)90732-2. [DOI] [PubMed] [Google Scholar]
- Taccioli G. E., Rathbun G., Oltz E., Stamato T., Jeggo P. A., Alt F. W. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993 Apr 9;260(5105):207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- Taccioli G. E., Rathbun G., Shinkai Y., Oltz E. M., Cheng H., Whitmore G., Stamato T., Jeggo P., Alt F. W. Activities involved in V(D)J recombination. Curr Top Microbiol Immunol. 1992;182:107–114. doi: 10.1007/978-3-642-77633-5_13. [DOI] [PubMed] [Google Scholar]
- Turchi J. J., Bambara R. A. Completion of mammalian lagging strand DNA replication using purified proteins. J Biol Chem. 1993 Jul 15;268(20):15136–15141. [PubMed] [Google Scholar]
- Wang T. S., Korn D. Reactivity of KB cell deoxyribonucleic acid polymerases alpha and beta with nicked and gapped deoxyribonucleic acid. Biochemistry. 1980 Apr 29;19(9):1782–1790. doi: 10.1021/bi00550a009. [DOI] [PubMed] [Google Scholar]
- Wang Z., Wu X., Friedberg E. C. DNA repair synthesis during base excision repair in vitro is catalyzed by DNA polymerase epsilon and is influenced by DNA polymerases alpha and delta in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Feb;13(2):1051–1058. doi: 10.1128/mcb.13.2.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. H., Berget P. B., Pipas J. M. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol Cell Biol. 1982 Oct;2(10):1258–1269. doi: 10.1128/mcb.2.10.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdzienicka M. Z., van Wessel N., van der Schans G. P. A fourth complementation group among ionizing radiation-sensitive Chinese hamster cell mutants defective in DNA double-strand break repair. Radiat Res. 1992 Sep;131(3):309–314. [PubMed] [Google Scholar]
- de Massy B., Weisberg R. A., Studier F. W. Gene 3 endonuclease of bacteriophage T7 resolves conformationally branched structures in double-stranded DNA. J Mol Biol. 1987 Jan 20;193(2):359–376. doi: 10.1016/0022-2836(87)90224-5. [DOI] [PubMed] [Google Scholar]