Background: DNA lesions in different sequence contexts exhibit alternate conformations.
Results: DNA adduct conformation is differentially modulated by DNA structure, DNA polymerase, and the presence of an incoming nucleoside triphosphate.
Conclusion: Adduct conformation is influenced by binding of a non-hydrolysable nucleotide to pol β but not Klenow fragment.
Significance: Adduct conformation and coding potential is modulated by constraints imposed by the polymerase active site.
Keywords: DNA Damage, DNA Replication, DNA Structure, Mutagenesis Mechanisms, NMR
Abstract
The active site conformation of the mutagenic fluoroaminofluorene-deoxyguanine adduct (dG-FAF, N-(2′-deoxyguanosin-8-yl)-7-fluoro-2-aminofluorene) has been investigated in the presence of Klenow fragment of Escherichia coli DNA polymerase I (Kfexo−) and DNA polymerase β (pol β) using 19F NMR, insertion assay, and surface plasmon resonance. In a single nucleotide gap, the dG-FAF adduct adopts both a major-groove- oriented and base-displaced stacked conformation, and this heterogeneity is retained upon binding pol β. The addition of a non-hydrolysable 2′-deoxycytosine-5′-[(α,β)-methyleno]triphosphate (dCMPcPP) nucleotide analog to the binary complex results in an increase of the major groove conformation of the adduct at the expense of the stacked conformation. Similar results were obtained with the addition of an incorrect dAMPcPP analog but with formation of the minor groove binding conformer. In contrast, dG-FAF adduct at the replication fork for the Kfexo− complex adopts a mix of the major and minor groove conformers with minimal effect upon the addition of non-hydrolysable nucleotides. For pol β, the insertion of dCTP was preferred opposite the dG-FAF adduct in a single nucleotide gap assay consistent with 19F NMR data. Surface plasmon resonance binding kinetics revealed that pol β binds tightly with DNA in the presence of correct dCTP, but the adduct weakens binding with no nucleotide specificity. These results provide molecular insights into the DNA binding characteristics of FAF in the active site of DNA polymerases and the role of DNA structure and sequence on its coding potential.
Introduction
The genome requires constant surveillance to detect abnormalities that could alter its functional properties (e.g. gene expression, transcription, replication) (1). Endogenous and environmental chemicals can modify DNA bases thereby threatening the genome (2). Base modifications can result in an altered coding potential that could lead to deleterious mutations, depurination creating abasic sites that are mutagenic and an impediment to DNA synthesis, and bulky lesions that block DNA replication. Several DNA repair mechanisms protect cells from the deleterious effects of DNA lesions. The nucleotide excision repair pathway typically removes bulky DNA lesions, whereas abasic sites initiate the base excision repair pathway (3, 4). Persistent DNA lesions that escape repair and block DNA synthesis can be bypassed by translesional DNA synthesis. In this situation, a specialized DNA polymerase is recruited to the site of DNA damage and inserts a nucleotide opposite the blocking lesion permitting continued DNA synthesis (5). Translesion DNA synthesis can be error-free or error-prone.
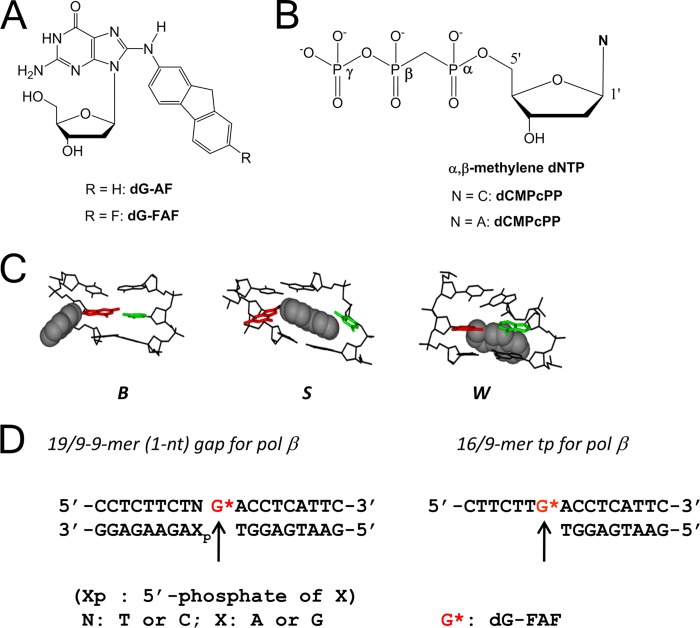
Arylamines are environmental mutagens, and aminofluorene is a model arylamine carcinogen due to its mutagenic and tumorigenic properties (6–8). It produces C8-substituted guanine adducts, e.g. [N-(2′-deoxyguanosin-8-yl)-2-aminofluorene (dG-AF; Fig. 1A) (9). The C8 adducts have been shown to adopt three conformational states depending on the location of the aminofluorene moiety; the major groove B-type (B), the base displaced stacked (S), and the minor groove wedge (W) type (Fig. 1C) (10–13). The arylamine-induced conformer heterogeneity is dependent on DNA sequence and expected to be influenced by DNA binding proteins such as a DNA polymerase.
FIGURE 1.
A, shown are structures of dG-AF (N-(2′-deoxyguanosin-8-yl)-2-aminofluorene) anddG-FAF adduct (N-(2′-deoxyguanosin-8-yl)-7-fluoro-2-aminofluorene). B, shown are structures of non-hydrolysable α,β-methylene-dNTP (dCMPcPP and dAMPcPP). C, shown are three major conformational motifs derived from FAF-modified fully paired DNA duplexes: major groove binding B-type (B), base displaced stacked (S), and minor groove binding wedge (W). D, shown are template-primer sequences (19/9/9-mer 1-nt gap and 16/9-mer DNA) used for 19F NMR and gel experiments.
DNA polymerase β (pol β)2 is the smallest eukaryotic polymerase (39 kDa), belongs to the X-family of DNA polymerases, and has been extensively characterized biologically, kinetically, dynamically, and structurally (14). It is composed of two domains that complement its biological function in base excision repair. The amino-terminal 8-kDa domain includes a lyase activity that removes the 5′-deoxyribose phosphate generated after incision by apurinic/apyrimidinic endonuclease during the repair of simple base lesions (15, 16). The lyase domain recognizes the 5′-phosphate in DNA gaps, thereby targeting the polymerase for gap-filling DNA synthesis (17). The nucleotidyltransferase activity of pol β resides in the 31-kDa polymerase domain. A helix-hairpin-helix structural motif is found in each domain and interacts with the DNA backbone in a non-sequence-dependent manner on opposite sides of gapped DNA. The helix-hairpin-helix motif is conserved in other members of the X-family DNA polymerases and is believed to facilitate DNA gap binding (18). DNA polymerase β lacks an intrinsic proofreading exonuclease activity, and accordingly, it has historically been described as an error-prone polymerase. This is supported by the observation that up-regulation or overexpression of pol β can result in genomic instability (19–21).
Although the primary cellular role of pol β is to provide lyase and nucleotidyltransferase activities during base excision repair, pol β has also been implicated in the replication bypass of a variety of bulky DNA lesions. Overexpression of pol β can result in decreased sensitivity to agents that generate bulky DNA adducts, such as cisplatin (19) or UV radiation (22). In vitro assays demonstrate the ability of pol β to bypass UV-induced lesions (22), bulky polycyclic aromatic hydrocarbon adducts (23, 24), and cisplatin-induced DNA lesions (25, 26). DNA polymerase β can bypass abasic sites (27) and 8-oxo-deoxyguanine (28) in short DNA gaps by utilizing the downstream templating base for coding and is consistent with its low deletion frameshift fidelity in gapped DNA (29). This observation suggests that lesion bypass might occur by employing the downstream templating base. In contrast, structures of pol β with active site mismatches indicate that the incorrect templating base is positioned upstream of the coding template base pocket, creating a pseudo-abasic site (i.e. coding potential of the templating pocket is lost) (30). Accordingly, an alternate mechanism for the bypass of a bulky lesion is to remove the modified nucleotide from the templating pocket through template-strand upstream translocation or, alternatively, expelling the lesion to an extra-helical position.
Because mechanisms of lesion bypass often rely on an altered DNA conformation, we have utilized several assays to study the conformation of the fluorine-tagged model arylamine DNA adduct dG-FAF (Fig. 1, A–C) in the confines of a polymerase active site. The results are interpreted in the framework of the influence of the DNA structure (gapped and non-gapped DNA) on adduct conformation and the effect of polymerase binding to the respective DNA structures on adduct conformational heterogeneity. Additionally, the results are contrasted with those observed with a model A-family DNA polymerase, Klenow fragment of Escherichia coli DNA polymerase I, that prefers non-gapped DNA.
EXPERIMENTAL PROCEDURES
Materials
Crude oligodeoxynucleotides (desalted, 10–15 μmol) were purchased from Operon (Eurofin, Huntsville, AL) and purified by reverse phase high performance liquid chromatography (HPLC). All HPLC solvents were purchased from Fisher and used as received. The HPLC system consisted of a Hitachi EZChrom Elite system with a L2450 diode array as a detector and a Clarity column (10 × 150 mm; 5 μm) (Phenomenex, Torrance, CA). The mobile phase was a 30-min linear gradient profile of 3–16% (v/v) acetonitrile, 10 mm ammonium acetate, pH 7.0, with a flow rate of 3.0 ml/min. T4 polynucleotide kinase was purchased from USB Corp. (Cleveland, OH). T4 DNA ligase was purchased from New England Biolabs (Ipswich, MA). [γ-32P]ATP (specific activity 6000 Ci/mmol) was purchased from PerkinElmer Life Sciences. The non-hydrolysable nucleotides dCMPcPP and dAMPcPP were purchased from Jena Biosciences (Jena, Germany) and used as received. 2′,3′-Dideoxythymidine triphosphate (ddTTP) and Illustra G-25 spin columns were obtained from GE Healthcare.
Preparation of FAF-modified Oligonucleotides
For 19F NMR studies, an FAF-modified 19-mer oligonucleotide d(5′-CCTCTTCTNG*ACCTCATTC-3′) (N, C or T; G*, dG-FAF; Fig. 1D) was prepared according to a published procedure (31). Briefly, an unmodified 19-mer oligonucleotide was treated with N-acetoxy-N-trifluoroacetyl-7-fluoro-2-aminofluorene, an activated derivative of the carcinogen 7-fluoro-2-aminofluorene. The reaction was monitored over a 24-h period with reverse phase-HPLC using the linear gradient described above. The modified strand was purified by multiple injections on HPLC (supplemental Fig. S1) and characterized by MALDI-TOF in reflectron mode (supplemental Fig. S2). An exonuclease-deficient Klenow fragment (Kfexo−; exonuclease-deficient (D424A)) was generously provided by Prof. Catherine Joyce (Yale University).
19F NMR
One-dimensional 19F NMR spectra were recorded using a dedicated 5-mm 1H/19F dual probe on a Varian 500 spectrometer operating at 470.6 MHz or Bruker Avance spectrometer operating at 376.5 MHz and were referenced to CFCl3 by assigning external hexafluorobenzene (C6F6) in C6D6 at −164.90 ppm. Spectra were obtained at 20 °C with a recycle delay of 1.0 s between acquisitions. A total of 50,000 scans were acquired for each spectrum. The spectra were processed by zero-filling, exponential multiplication using 40-Hz line broadening and Fourier transformation.
Approximately 0.1 mm FAF-modified 19-mer oligonucleotide (5′-CCTCTTCTTG-FAF-ACCTCATTC-3′) was annealed with 0.1 mm upstream 9-mer (5′-GAATGAGGT-3′) and a 0.1 mm downstream 9-mer (5′-pAAGAAGAGG-3′) to create a 1-nt gapped template-primer by heating to 90 °C for 5 min and slowly cooling (3 h) before lyophilization (Fig. 1D). For Kfexo− studies, a 9-mer primer (5′-GAATGAGGT-3′) was annealed to a 16-mer template (5′-CTTCTTG-FAF-ACCTCATTC-3′) was similarly prepared (Fig. 1D) (32). For free DNA, samples were filtered by ultracentrifugation using a Pall Microsep MF centrifugal device (Mr cutoff = 1000). The centrifuged samples were dissolved in 300 μl of a neutral buffer (10% D2O, 90% H2O containing 100 mm NaCl, 10 mm sodium phosphate, pH 7.0, 100 μm Na4EDTA, and 5 mm MgCl2), filtered through a 0.2-μm membrane filter, and placed in a Shigemi tube for NMR experiments. For binary and ternary complex formation, 0.1 mm FAF-modified 1-nt gapped DNA was incubated with a 2-fold excess of pol β overnight at 20 °C in the presence or absence of a 20-fold excess (i.e. 2 mm) of a non-hydrolysable dCMPcPP or dAMPcPP (Fig. 1B). Similarly, 0.2 mm FAF-modified non-gapped template-primer was mixed with 0.3 mm Kfexo−. Ternary complexes were produced by the addition of 2 mm dCMPcPP or dAMPcPP. Line shape analysis was conducted using WINDNMR-Pro (Version 7.1.6; Journal of Chemical Education Software Series, Reich, H. J., University of Wisconsin, Madison, WI) (supplemental Fig. S3).
Single-nucleotide Gap-filling Assay
The 9-mer primer (5′-GAATGAGGT-3′) was 5′-radiolabeled with [γ-32P]ATP and T4 polynucleotide kinase following the manufacturer's protocol. The 5′-32P-labeled 9-mer primer (100 pmol) was annealed to either an unmodified or adducted template 19-mer oligonucleotide (120 pmol) along with a downstream 9-mer oligonucleotide with a 5′-PO43− to form a 1-nt gapped DNA by heating to 95 °C for 5 min and then slowly cooling to room temperature (Fig. 1D). A 1-nt gapped template-primer (100 nm) was incubated with varying concentrations of pol β for 5 min to form a binary complex. The reaction was then initiated by adding 250 μm dNTPs and 5 mm MgCl2, incubated at 37 °C for 20 min in a reaction containing 50 mm Tris-HCl, pH 7.4, 100 mm KCl, and 5% (v/v) glycerol, and terminated with 50 μl of 50 mm EDTA, pH 8.0, 95% formamide solution.
To study the effect of divalent metal ions on single-nucleotide gap filling, 5 mm Mn2+ or Co2+ was substituted for Mg2+. Quenched samples were heated to 95 °C for 5 min and immediately cooled on ice. The products were resolved on a denaturing polyacrylamide gel and quantified as described previously (33).
Determination of Nucleotide Insertion Kinetics Parameters
For pol β, steady-state kinetic parameters for nucleotide incorporation opposite unmodified and FAF-modified templates were determined at 37 °C as reported previously (34). Enzyme concentrations and reaction time intervals were optimized so that substrate depletion did not impact initial velocity measurements. The efficiency of nucleotide insertion by pol β was calculated as kcat/Km, and the relative insertion frequency (fins) is calculated as (kcat/Km)mismatch or lesion/(kcat/Km)unmodified control.
Kfexo− DNA Synthesis Kinetic Assays
The sequences (3′–5′) of the template and primer were GACGTCGACTACGCGCATGCCTAGGGGCCCATG (the templating base is underlined) and CTGCAGCTGATGCGC, respectively. The primer was 5′-labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs), and radioactive ATP was removed with a MicroSpin G-25 column.
Enzyme activities were determined at 22 °C using a standard reaction mixture (25 μl) containing 50 mm Tris-HCl, pH 7.4, 2.5 μg of bovine serum albumin, 10 mm MgCl2, and 500 nm template-primer DNA. Reactions were stopped with 20 μl of 0.5 m EDTA and mixed with an equal volume of formamide dye; the products were separated on 15% denaturing polyacrylamide gels and quantified in the dried gels by phosphorimaging. Steady-state kinetic parameters were determined by fitting the rate data to the Michaelis equation. Inhibition of nucleotide insertion by dCMPcPP was determined at various concentrations of inhibitor (I) and sub-saturating nucleotide concentration (S) as given in the figure legends. The inhibitor dissociation binding constant, Ki, was determined by fitting the inhibition data to Equation 1 for competitive inhibition by nonlinear regression methods.
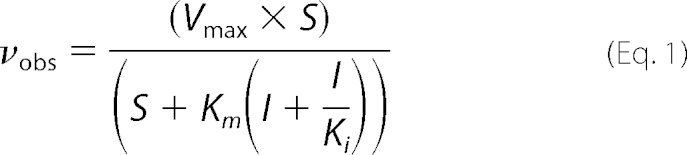 |
Surface Plasmon Resonance (SPR) Assay
A biotinylated 31-mer containing the same DNA sequence context as used in NMR experiments was employed in SPR analysis. An 83-mer hairpin-template-primer was prepared by following a previously reported protocol (34). Briefly, a biotinylated 31-mer (optical density of 5) was annealed with a 52-mer hairpin (optical density of 6) by heating to 95 °C for 5 min and cooling to room temperature. These were ligated with T4 DNA ligase (4000 units) in ligase buffer for 16 h at 20 °C. The ligated 83-mer oligonucleotides was purified in 10% denaturing polyacrylamide and extracted using the crush and soak method followed by desalting using G-25 spin columns. The template-dideoxy-primer terminus was obtained by incubating ddTTP (1 mm) in the presence of 1 μm Kfexo− and 5 mm MgCl2 for 12 h. The polymerase was precipitated with phenol-chloroform, and the 84-mer DNA was purified by reverse phase-HPLC using the solvent gradient described above. The biotinylated 31- and 84-mer-modified oligonucleotides were characterized with MALDI-TOF in reflectron and linear negative modes, respectively.
Immobilization of Streptavidin on CM5 Chip and DNA Coating
The immobilization of streptavidin on CM5 surface was carried out using l-ethyl-3-(3-dimethylaminopropyl)carbodiimide/N-hydroxysuccinimide amine coupling method by following the reported procedure (34). The biotinylated 84-mer DNA-hairpin with a dideoxy-terminated primer (0.25–0.3 nm) was injected for 120–240 s over the flow cell 2 and 4 to achieve 3–4 RU through manual injection. The unbound DNA was washed with running buffer. To ensure the DNA substrate has a 3′-dideoxyprimer terminus, 1 mm ddTTP and 1 μm Kfexo− with 5 mm MgCl2 was injected over the surface for 5 min and washed with 0.05% SDS. The drift in base line was monitored by passing running buffer over the surface for 0.5–1 h. For single-nucleotide gap filling analysis, an appropriate downstream oligonucleotide with a 5′-PO43− group (2 nm) was injected over the surface for 5 min, and an increase in response was observed. The free primers were washed with running buffer for 5 min.
SPR Binding Kinetics Analysis
The surface performance, mass transport, and regeneration buffer scouting were performed as described previously (34). The binding kinetics for the interaction of pol β with DNA was performed by injecting pol β over the DNA surface using running buffer B (0.1 m Hepes, pH 7.4, 150 mm NaCl, 0.005% non-ionic surfactant P20, 5 mm MgCl2, and 100 μg/ml BSA) with a flow rate of 100 μl/min for 30 s followed by dissociation for 60 s. A different concentration range of pol β was necessary for double-stranded DNA and 1-nt gapped DNA due to the significantly different DNA binding affinities. Before kinetic experiments, the surface was conditioned by injecting running buffer followed by three startup steps with running buffer and four injections of zero concentration. The surface was regenerated with a 30-s injection of 0.05% SDS with a flow rate of 100 μl/min followed by an extra wash with running buffer. For the ternary complex system, individual dNTPs (100 μm) were added with pol β and injected over the surface. While steady-state affinity analysis was performed with unmodified DNA due to a diffusion-limited association rate constant (kon), association and dissociation rate constants were determined for FAF-modified sequences by fitting the sensorgrams with a Langmuir 1:1 fit using BIAevaluation software v2.0. The mass transport limitation factor has been included in all the fitting modules. The goodness of the fit was measured by χ-squared values that fall within 1% of Rmax.
RESULTS
Model Systems
We prepared the 19-mer (5′-CCTCTTCTTG-FAF-ACCTCATTC-3′) and 16-mer (5′-CTTCTTG-FAF-ACCTCATTC-3′) (Fig. 1D) oligonucleotides as templates for the pol β and Kfexo− binding experiments. The single guanines in these model sequences were adducted with fluoroaminofluorene adduct (dG-FAF). The utility of fluorine-tagged aromatic amine carcinogens as an effective conformational probe has been well documented (10, 35, 36). As shown in Fig. 1D, the modified templates were annealed with appropriate primers to create model duplexes.
19F NMR pol β Complex
A single nucleotide (1 nt) gapped DNA duplex was prepared by annealing the FAF-modified 19-mer template (5′-CCTCTTCTTG-FAF-ACCTCATTC-3′) with upstream and downstream oligonucleotides (9-mers) (Fig. 1D). It should be noted that the thermodynamic stability of the duplex was increased by adding CCT to the 5′-side of the usual 16-mer template sequence used in previous NMR studies (32). All experiments were carried out in a NMR buffer with 5 mm MgCl2 at 20 °C.
In the absence of pol β, the free 1-nt gapped substrate exhibited an equal ratio of two sharp peaks at −116.8 and −117.6 ppm (Fig. 2A, trace a). We have accumulated an extensive collection of 19F NMR data with FAF-modified duplexes in various sequence settings and found that in all cases that the S-type conformer fluorine is always shielded (upfield) relative to that of the external binding B-type conformer (35, 37). 19F shielding is a hallmark for the van der Waals interaction and the ring current effect caused by the carcinogen moiety within the stacked and bulge duplexes (S-type conformation). The same trend was observed with various fully paired and deletion duplexes modified with the bulky N-acetylated FAF analog that specifically revealed a mixture of B, S, and W conformations (Fig. 1C) in the −115.0 ∼ −115.5 ppm, −115.5 ∼ −117.0 ppm, and −116.5 ∼ −118.0 ppm ranges, respectively (38). Accordingly, the signals in Fig. 2A (trace a) were assigned as the B (green)- and S (red)-type conformers, respectively. We have used a similar strategy to assign the signals in the 19F NMR spectra of FAF-modified template-primer complexed with pol β and Kfexo− (see below).
FIGURE 2.
19F NMR spectra of the dG-FAF adduct (A) with pol β (B) Kfexo− in the presence and absence of dCMPcPP and dAMPcPP. T/P, template-primer.
The binary DNA and ternary DNA/dNTP enzyme complexes were produced by incubating the 1-nt gapped substrate with pol β to ensure complex formation (Fig. 1D). Upon the addition of 2 eq of pol β, the two 19F resonances persisted but were significantly broadened, consistent with an increase in molecular weight (Fig. 2A, trace b). The results suggest formation of a binary complex between the template-primer and the polymerase, resulting in an equal mix of B and S conformers within the polymerase binding site. The stacked S-type conformation (red) should be similar to the crystallographic structure exhibited by a benzo[c]phenanthrene diol epoxide-adducted gapped DNA bound to pol β, possibly accounting for the mutagenic potential of this bulky PAH lesion (23).
Upon the addition of the correct non-hydrolysable dCMPcPP (Fig. 1B), the peak at −116.8 ppm was broadened further and appears to split into two signals at −115.4 (B*, blue) and −115.6 (B, green) ppm, possibly at the expense of the S-conformer signal (Fig. 2A, trace c). In contrast, the addition of the mismatch non-hydrolysable dAMPcPP resulted in significant reduction of the B* signals (red). Instead, a new signal emerged at −119.5 ppm (Fig. 2A, trace d). This relatively sharp signal is shielded significantly, and its chemical shift range appears to be consistent with the so-called wedge (W)-type conformers, in which the carcinogenic aminofluorene moiety is accommodated in the narrow minor groove. The W-conformation has been observed in all dA-mismatched FAF-duplexes as well as many of the FAAF-modified duplexes (39). Previous dynamic 19F NMR studies further showed that the presence of either a 5′- or 3′-T to the dA-mismatched FAF adduct promoted conformational adduct heterogeneity compared with other flanking bases (39). Supplemental Fig. S3 shows computer line shape simulations of the NMR spectra of the pol β experiments.
19F NMR Klenow Complex
For comparison, we also compared the conformational response of the lesion to binding of an A-family DNA polymerase; i.e. Kfexo−. Fig. 2B, trace a, shows the 19F NMR spectrum of the FAF-16/9-mer template-primer where the FAF-modified guanine is situated at the first templating position in non-gapped DNA. We previously assigned the sharp peak at −117.5 ppm (Fig. 2B, trace a) as representing an interchangeable mix of B- and S-type conformers with a greater population of the S-conformer (32). Upon the addition of 1.5 eq of Kfexo− (Fig. 2B, trace b), the 19F resonance was significantly broadened and shielded by ∼1 ppm, indicating a slow motion associated with a binary complex with significant van der Waals protein-DNA interactions. This is in good agreement with EMSA (supplemental Fig. S4). It is also consistent with the structural features displayed by AF-adducted DNA bound to Bacillus fragment (Bf), i.e. the lesion adopts a syn conformation with the fluorene moiety sequestered in the preinsertion site (40).
The addition of the correct dCMPcPP (Fig. 1B) produced a conformational heterogeneity, a major signal (∼90%, green) at −117.5 ppm with a broad downfield shoulder and a minor signal (*, ∼10%, red) at −119.5 ppm (Fig. 2B, trace c). The addition of a large excess of free template-primer intensified the major signal (−116.8 ppm), confirming its B-type nature (not shown). The results suggest the formation of a ternary complex via specific protein-DNA interactions. An essentially similar 19F profile was observed upon the addition of the non-complementary dAMPcPP except that the major B signal (green) was slightly more homogeneous at the expense of an increased population (15%, red) of the minor signal. According to the structural characterization of Bf (40), FAF at the templating position of the binary complex is sterically constrained, deterring rotation about the glycosidic bond to an anti-B-type conformer to form either a Watson-Crick base pair with an incoming dCMPcPP or possibly a Hoogsteen pair with dAMPcPP. The primer extension data obtained by using a similar template-primer exhibited a preferential incorporation of correct dCTP opposite the FAF lesion followed by a small percentage of dATP insertion (33). Consequently, the major (green) and the upfield minor (red) signals in Fig. 2B (traces c and d) could be assigned to the B- and S-type conformers, respectively. However, unlike pol β, the non-hydrolysable dNMPcPP analogs do not bind as tightly to Kfexo− (see below). The broad 19F signals for the ternary complex may represent aspects of B/S-type heterogeneity, but it is certain that they do not induce a catalytically competent conformation (see below).
Non-hydrolysable Nucleotide Analog Binding to Kfexo−
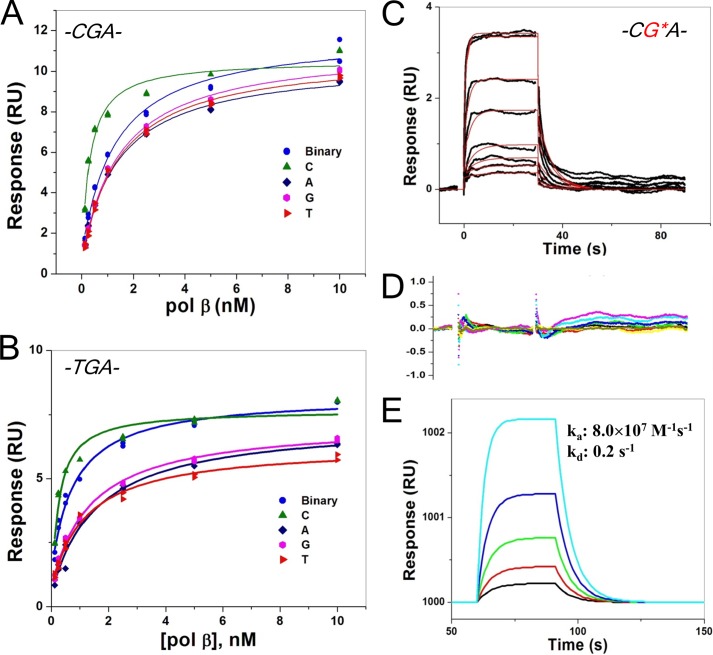
Inert non-hydrolysable nucleotide analogs are good inhibitors of pol β and have been successfully employed to trap pre-catalytic substrate/polymerase complexes for structure determination (23, 30). Replacing the bridging oxygen between Pα and Pβ with -NH- or -CH2- permits strong binding of the analog and prevents nucleotide insertion. Surprisingly, 1 mm dCMPcPP did not inhibit dCTP insertion by Kfexo− (Fig. 3A). Because DNA polymerase I (Klenow fragment) has been shown to display moderate processivity where the steady-state velocity is primarily determined by DNA dissociation (41), an alternate assay was employed to determine the binding affinity of the non-hydrolysable dCMPcPP analog. To prevent DNA dissociation from becoming rate-limiting, misinsertion of dTTP opposite the templating guanine was assessed, and its sensitivity toward dCMPcPP was measured (supplemental Fig. S5). The kinetic parameters for dTTP misinsertion opposite guanine were determined (kcat = 0.034 1/s, Km = 235 μm; means of three determinations). A titration of the reaction velocity with dCMPcPP in the presence of 5 μm dTTP indicated that binding of the analog was weak (Kd ∼ 375 μm).
FIGURE 3.
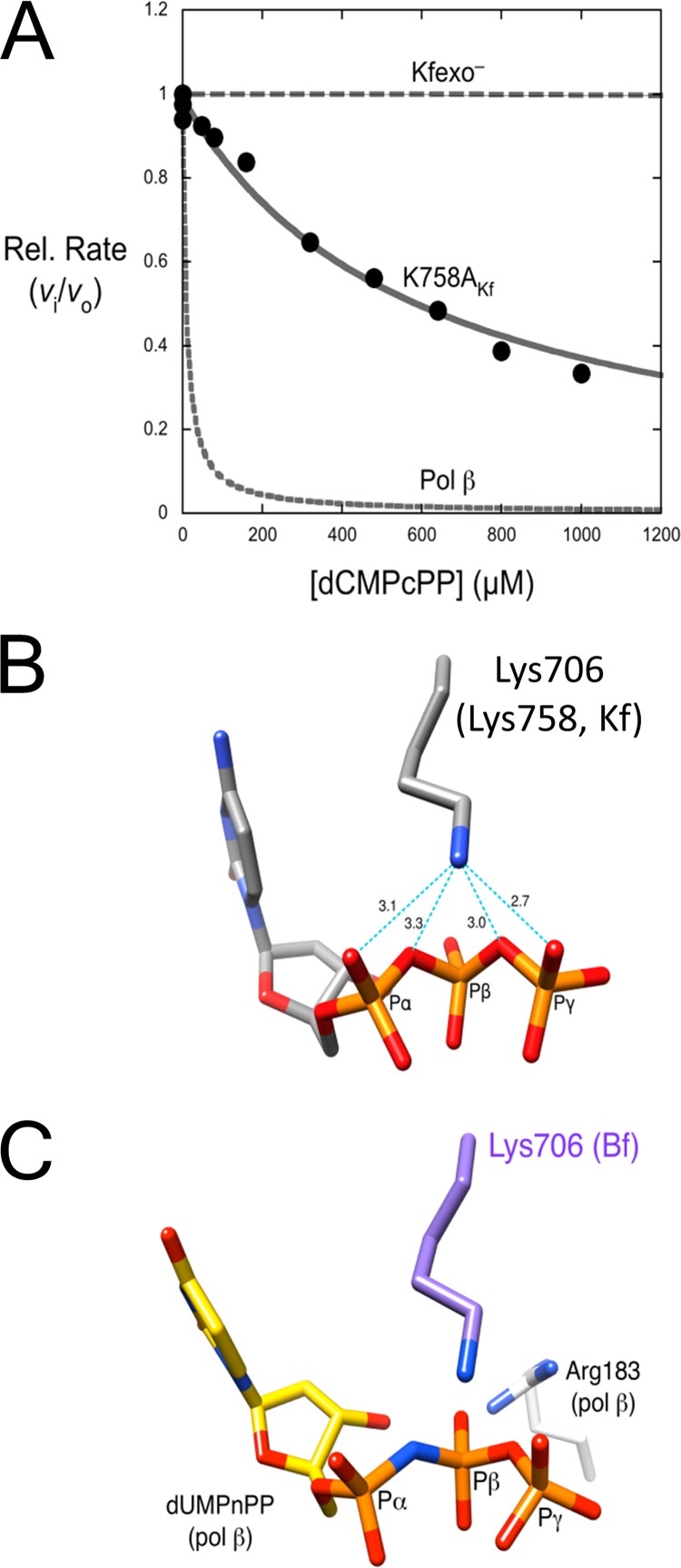
Inhibition of dCTP insertion opposite guanine. A, shown is a non-hydrolysable nucleotide inhibitor titration of relative activity (vi/v0) for dCMP insertion with K758A Kfexo− (filled circles). The data were fitted to Equation 1 for competitive inhibition (solid line). indicating weak binding (Ki = 310 ± 20 μm). A simulated data set for inhibition of wild-type Kfexo− is also shown (dashed line), indicating the lack of inhibition when binding of the correct incoming nucleotide is tight and the affinity of dCMPcPP binding is weak. In contrast, the binding of this analog to pol β is tight (Ki = 0.9 μm; dashed line) (57). The concentration of dCTP for these experiments/simulations was 20 μm. B, shown is the interaction of a Bf active site lysine residue with the incoming nucleotide. Key distances (Å) from Nζ of Lys-706 are shown (light blue dashed lines). This lysine is equivalent to Lys-758 of Kfexo− (Kf). C, shown is an overlay of the pol β and Bf ternary substrate complexes (PDB ID codes 2FMS and 1LV5, respectively) aligned using the triphosphate moiety of the incoming nucleotides. The non-hydrolysable 2′-deoxy-uridine-5′-(α,β)-imido triphosphate (dUMPnPP) analog and Arg-183 from the pol β structure are shown. Only Lys-706 from the Bf structure is shown situated near the Pα–Pβ bridging atom.
X-family DNA polymerases lack a basic side chain that interacts with the pro-SP oxygen on Pα (42). A-family DNA polymerases have a conserved lysine residue that interacts with this oxygen as well as the Pα–Pβ bridging oxygen (Fig. 3, B and C). This lysine residue corresponds to Lys-706 of Bf and Lys-758 of Kfexo−. Previously, an alanine mutant of this residue of Kfexo− was shown to dramatically reduce insertion efficiency when using a homopolymeric template-primer system (43). Using a heteropolymeric template-primer, single-nucleotide extension reactions suggest that the rate of dCTP insertion is dramatically decreased, whereas dNTP binding affinity is moderately decreased (supplemental Table S1; Kd ∼ 5 μm for wild-type enzyme (41)). In contrast to wild-type enzyme, dCMPcPP inhibition of correct dCTP insertion opposite dG can easily be measured with the K758A mutant (Fig. 3A). The affinity for the analog is weak (Ki = 300 μm) but similar to that observed for wild-type enzyme determined through competition with incorrect nucleotide insertion (supplemental Fig. S5B).
Single-nucleotide Insertion Assay
To examine the effect of conformational heterogeneity of aminofluorene adduct in the specificity of the nucleotide insertion, qualitative 1-nt gap insertion assays were conducted in the presence of individual nucleotides with pol β. As shown in Fig. 4A, correct dCMP insertion was readily observed over other dNTPs with unmodified guanine. Although some misinsertion of other nucleotides was observed in a -CGA- sequence, DNA synthesis was very faithful in the -TGA- sequence. In contrast, whereas insertion of dCMP with the FAF-modified gapped substrate was seen (Fig. 4, B and C, Mg2+), there was little or no incorporation of the other nucleotides.
FIGURE 4.

Single nucleotide gap assay of pol β. A, shown is a comparison of 1-nt gap assay between -CGA- and -TGA- in the presence of Mg2+. Shown is a comparison of nucleotide insertion opposite -CG*A- (B) and opposite -TG*A- (C) in the presence of Mg2+, Mn2+, and Co2+.
Steady-state Kinetics
To quantify the influence of FAF-induced conformational heterogeneity on nucleotide insertion, steady-state kinetic parameters were determined with the lesion positioned in the 1-nt gapped DNA substrate with pol β. From the kinetics experiments, insertion efficiency (kcat/Km) of dCMP was strongly inhibited; 270-fold in the case of -CG*A- and 200-fold with -TG*A- (Table 1). The loss in efficiency was primarily due to an increase in Km (dCTP) in both sequence contexts. The efficiency of misinsertion of dAMP was stronger in the -TGA- than the -CGA- unmodified DNA sequence context, resulting in a 24-fold higher misinsertion frequency with the -TGA- sequence. Due to the poor incorporation of dAMP opposite the FAF-modified guanine, steady-state kinetic parameters could not be reliably determined.
TABLE 1.
Steady-state kinetics parameters for insertion of nucleotides in a single-nucleotide gap gapped DNA substrate
| Sequence Contexta | Incoming dNTP | kcat | Km | kcat/Km | fins |
|---|---|---|---|---|---|
| min−1 | μm | μm−1 min−1 | |||
| -CGA- | dCTP | 2.96 (0.15)b | 0.78 (0.19) | 3.79 (0.94) | 1 |
| dATP | 0.06 (0.01) | 360 (119) | 0.00016 (0.00006) | 4.2 × 10−5 | |
| -CG*A- | dCTP | 0.49 (0.13) | 34 (27) | 0.014 (0.012) | 3.7 × 10−3 |
| dATP | NDc | ND | ND | ND | |
| -TGA- | dCTP | 1.58 (0.21) | 1.94 (1.03) | 0.81 (0.44) | 1 |
| dATP | 0.01 (0.002) | 8.94 (5.98) | 0.0011 (0.0008) | 1.0 × 10−3 | |
| -TG*A- | dCTP | 0.68 (0.14) | 164 (65) | 0.004 (0.001) | 5.0 × 10−3 |
| dATP | ND | ND | ND | ND |
a The underlined G (G) represents the templating base, and the asterisk (*) indicates an FAF-adducted guanine.
b Values represents the mean (S.E.) of at least two determinations.
c ND, not determined.
Effect of Divalent Metals on Nucleotide Insertion
Although Mg2+ is the primary metal cofactor for DNA synthesis, other divalent metals can support nucleotide insertion and are often found to decrease polymerase fidelity (44, 45). To investigate whether other divalent metals can promote nucleotide insertion opposite the dG-FAF adduct with pol β, three metal ions (Mg2+, Mn2+, and Co2+) were compared. With unadducted gapped DNA, Mn2+ and Co2+ exhibited reduced substrate specificity (data not shown). In contrast to Mg2+, where dCMP was preferentially incorporated opposite the lesion, Mn2+ and Co2+ supported the incorporation/misincorporation of all four nucleotides (Fig. 4, B and C). Interestingly, the misinsertion of thymidine was not as strong as the misincorporation of purines.
Ribonucleotide Insertion
Because ribonucleotides are present at significantly higher concentrations in the cell (46, 47), DNA polymerases are predicted to insert many more nucleotides with the wrong sugar than with a non-complementary base (48, 49). To assess the effect of FAF-modified adducts during insertion of ribonucleotides, individual rNTPs were assayed in the presence of different divalent metal ions. With unmodified 1-nt gapped DNA controls, CTP and GTP were inserted, whereas ATP or UTP were not (Fig. 5). In contrast to dNMP insertion, Mn2+ and Co2+ only supported misinsertion of GMP with the unmodified gapped DNA substrates. With the TG*A substrate, Mn2+ supported weak CMP insertion and GMP misinsertion opposite the adducted guanine. In the case of the CG*A sequence, no ribonucleotide insertion was observed in the presence of any of the divalent metals tested.
FIGURE 5.

Single nucleotide gap assay of pol β with the -CGA-, -CG*A-, -TGA-, and -TG*A- contexts in the presence of Mg2+, Mn2+, and Co2+ and individual NTPs.
Pol β DNA Binding Kinetics
DNA polymerase binding to modified and lesion-containing DNA was monitored by SPR as described previously (34). The biotinylated DNA substrates were immobilized on streptavidin coated on a dextran surface. The amount of DNA immobilized on the chip was monitored with an increase in response units (RU) by varying oligonucleotide concentration and injection time. The surface performance, regeneration buffer, and mass transport limitation was optimized to perform the kinetic experiments using previously reported literature (34).
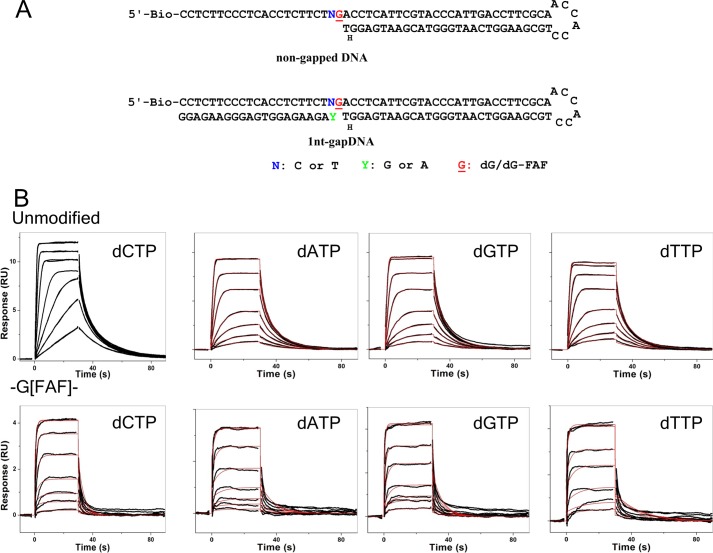
Polymerase binding to non-gapped and single-nucleotide gapped DNA was analyzed in the absence (binary) and presence of nucleoside triphosphates (ternary) by varying the concentration of pol β (Fig. 6). For unmodified DNA, in the presence of complementary or non-complementary nucleotides, the rate of association was rapid. In the presence of nucleotides, the mass transfer constant (kt) for the binding of pol β with unmodified guanine has also been determined and found to be 109 RU-m−1·s−1, which is close to a diffusion limited process (108 RU-m−1·s−1) (supplemental Fig. S6). So instead of the Langmuir 1:1 fit to determine Kd values, 1:1 affinity analysis was carried out by plotting steady-state binding levels (Req) versus varying concentrations of the polymerase. When the plot reaches the plateau, the Kd values were determined from 0.5 Rmax values (Figs. 6B and 7, A and B). In contrast, the weaker binding of the polymerase with dG-FAF-adducted DNA was determined by using a 1:1 Langmuir model due to the absence of mass transport limited data, as the mass transfer constant (kt) was around 1011-1013 RU-m−1·s−1 (Fig. 6B and supplemental Fig. S7). The fitted curves to extract association and dissociation rate constants are shown with experimental data illustrating the robustness of the fit (Fig. 7, C–E). From the binding kinetics, pol β binds open unmodified non-gapped DNA weakly (Kd ∼800 nm). However, in the presence of 1-nt unmodified gapped DNA, the binding of pol β increases ∼1000-fold compared with non-gapped DNA (Table 2). Upon the addition of the complementary incoming nucleotide (i.e. dCTP) to 1-nt gapped DNA, pol β binds with a 4-fold higher affinity than DNA alone (binary complex) and ∼3000-fold tighter than dsDNA. The presence of an incorrect incoming nucleotide weakens pol β binding by ∼10-fold than with a correct incoming nucleotide in the -CGA- context and 4–5 fold in the -TGA- sequence. Overall, pol β bound to the -TGA- sequence ∼1.5-fold tighter than the -CGA- sequence. In contrast to unmodified 1-nt gapped DNA, the FAF-modified oligonucleotides weakened the binding of pol β 3-fold (Table 3). The binding of pol β with the FAF-adducted DNA was similar in the presence of either correct or incorrect dNTPs. The binding affinities of pol β with the lesion-containing DNA was only slightly tighter in the presence of nucleotides (up to 2-fold) with the -TG*A- sequence.
FIGURE 6.
A, shown is the oligonucleotide constructs used in SPR assay. B, shown are SPR binding curves of unmodified -CGA- and -CG*A- in the presence of individual dNTPs (fitted curves are shown in red lines).
FIGURE 7.
Shown are steady-state affinity analyses of interaction of pol β and CGA (A) and TGA (B), an SPR sensorgram of binary complex (pol β and CG*A sequence) (C), and residuals of the fit (D) and simulated data (E).
TABLE 2.
SPR binding kinetics of pol β with gapped unmodified DNA
| Sequence contexta | Incoming nucleotide | Kdb | χ2 |
|---|---|---|---|
| nm | |||
| -CGA- | None | 0.9 (0.10) | 0.06 |
| dCTP | 0.2 (0.04) | 0.14 | |
| dATP | 2.1 (0.09) | 0.01 | |
| dGTP | 2.1 (0.16) | 0.02 | |
| dTTP | 1.8 (0.20) | 0.04 | |
| -TGA- | None | 0.8 (0.17) | 0.08 |
| dCTP | 0.3 (0.09) | 0.16 | |
| dATP | 1.6 (0.30) | 0.06 | |
| dGTP | 1.4 (0.15) | 0.02 | |
| dTTP | 1.2 (0.20) | 0.03 |
a DNA sequence is written 5′ to 3′ with the templating G in the DNA gap.
b Kd values were measured using steady-state affinity analysis (1:1) with BIAevaluation software. Error limits are provided in parentheses.
TABLE 3.
SPR binding kinetics of pol β with single-nucleotide dG-FAF-gapped DNA using 1:1 Langmuir kinetic model
Error limits are provided in parentheses.
| Sequence contexta | Incoming nucleotide | kon | koff | Kd | χ2 |
|---|---|---|---|---|---|
| 108/m·s | s−1 | n | |||
| -CG*A- | None | 0.8 (0.03) | 0.2 (0.006) | 2.50 | 0.012 |
| dCTP | 0.9 (0.02) | 0.3 (0.006) | 3.33 | 0.005 | |
| dATP | 0.5 (0.01) | 0.1 (0.001) | 2.00 | 0.008 | |
| dGTP | 0.7 (0.02) | 0.1 (0.001) | 1.43 | 0.012 | |
| dTTP | 0.7 (0.16) | 0.1 (0.001) | 1.43 | 0.013 | |
| -TG*A- | None | 0.9 (0.03) | 0.2 (0.005) | 2.22 | 0.004 |
| dCTP | 4.2 (0.26) | 0.7 (0.005) | 1.66 | 0.005 | |
| dATP | 0.9 (0.18) | 0.2 (0.003) | 2.22 | 0.005 | |
| dGTP | 1.5 (0.13) | 0.2 (0.002) | 1.33 | 0.009 | |
| dTTP | 1.1 (0.05) | 0.2 (0.005) | 1.8 | 0.008 |
a DNA sequence is written 5′ to 3′ with the modified templating G* in the DNA gap.
DISCUSSION
The classical hypothesis proposed by Loechler and co-workers (50) stated that each DNA adduct can adopt sequence dependent multiple conformations and that each conformation may lead to multiple alternate mutational outcomes. In the present study we have attempted to understand the role of adduct-induced conformational heterogeneity on mutagenic potential. The heterogeneity was modulated by placing a model adduct in different DNA structures (1-nt gapped and non-gapped DNA), sequence contexts, and DNA polymerase active sites.
Crystallographic structures of pol β bound to gapped DNA and Bf bound to non-gapped DNA provides molecular insights into similarities and differences on how these enzymes interact with substrate DNA. Bf is a prototypical A-family DNA polymerase that has been extensively characterized by crystallography (40, 51, 52). Upon binding DNA, both polymerases form an open conformation where the N-subdomain is positioned so that the active site is solvent-exposed. The N-subdomain of A-family DNA polymerases is also referred to as the “fingers” (53). Upon binding a nucleotide, the N-subdomain closes around the nascent base pair. A key difference in the way each polymerase interacts with their respective DNA substrates is how the templating (i.e. coding) nucleotide is positioned in the binary complex. For pol β, the templating base is situated within the DNA helix (54); in contrast, A-family DNA polymerases bind the templating base in an extra-helical position that has been termed the “pre-insertion site” (55). In addition to closing of the N-subdomain upon nucleotide binding, the templating nucleotide is also positioned in the DNA helix and is referred to as the “insertion site.” Thus, it is expected that the conformational heterogeneity of the FAF adduct should be sensitive to DNA sequence as well as the polymerase that binds to the respective substrates.
19F NMR Studies of Binary and Ternary Complexes
We have shown previously (12, 32, 33, 35, 39) that purine bases 3′ to the FAF-adducted site favors the S-conformation, whereas the presence of pyrimidine bases 5′ to the lesion site favored the major groove B-conformer (36). In full duplexes, a 5′-T favors a higher percentage of the major groove B-type conformer, whereas a 5′-C favors the stacked conformation. At a replication fork with the adduct positioned at the first templating position, the aminofluorene adducted to guanine is flexible and thus oscillates between two conformers (B and S), which are in equilibrium and observed as a single 19F NMR signal (32). In contrast to replication fork DNA, the adduct in a single-nucleotide gapped DNA substrate exhibited two well-defined peaks corresponding to B- and S-conformers (Fig. 2A). Even in the presence of pol β, the dG-FAF adduct exhibits similar NMR shifts as in a buffered solution, indicating the existence of conformational heterogeneity in or near the active site of pol β.
To capture a polymerase DNA-dNTP ternary complex, we used a non-hydrolysable dNMPcPP (Fig. 1B). These methylene analogs bound with high affinity to pol β with no active site distortions permitting catalytic metal binding (56, 57). dCMPcPP binds tightly to pol β with Ki of 0.9 μm in unmodified DNA (57). In the presence of dCMPcPP, the dG-FAF adduct exhibited two conformers B and B*, which indicates that the adducted nucleotide adopts an anti conformation, placing the aminofluorene moiety in the major groove. The present NMR study further supports the previous crystallographic studies where the correct incoming nucleotide (dCTP) forms a Watson-Crick base pair with the dG-AF adduct in which the aminofluorene moiety has adopted a major groove conformation resulting in less perturbation of the polymerase active site geometry (40). Interestingly, in the presence of an incorrect dAMPcPP, the population of the major groove conformation is reduced slightly compared with the dCMPcPP addition (Fig. 2, supplemental Fig. S3). In contrast to pol β, the A-family polymerase Kfexo− modulates the conformation of the dG-FAF adduct mainly to the anti conformation. Even in the presence of complementary dCMPcPP and non-complementary dAMPcPP, the adduct exhibits a single major peak as observed for free template-primer, indicating that the incoming nucleotide did not affect the conformation of the adduct even in the polymerase active site. The observed results were in good agreement with previously reported crystal structure data on the pre- and post-insertion site of AF adduct in Bf, in which the presence of correct base (dCTP) the dG-AF adopts the major groove conformation with minimal perturbations to the active site of polymerase (40). Importantly, the correct nucleotide kept the polymerase in a closed conformation compared with the open conformation with an incorrect nucleotide opposite the dG-AF adduct (40).
Surprisingly, we found that the dCMPcPP analogs did not bind to Kfexo−/DNA as effectively as compared with that of pol β (Fig. 3A). The lower binding affinity with Kfexo− might be explained in terms of a key amino acid residue that interacts near the α- and β-phosphates. Although X-family DNA polymerases lack a basic side chain that interacts with the pro-SP oxygen on Pα of the incoming nucleotide, A-family DNA polymerases have a conserved lysine residue that interacts with this oxygen as well as the Pα–Pβ non-bridging oxygen (Fig. 3, B and C) (42). This lysine residue corresponds to Lys-758 of Klenow fragment, and loss of this interaction results in a greater apparent inhibition with dCMPcPP due to a lower binding affinity for natural nucleotides. Thus, the weak binding of the non-hydrolysable analog suggests that Kfexo− remains in an open conformation when the analog binds.
Single-nucleotide Gap-filling
Although correct dCTP was readily inserted opposite unadducted guanine, dG-FAF greatly diminishes insertion. Whereas the misinsertion of dAMP and dTMP were readily observed opposite guanine, misinsertion opposite dG-FAF was not observed (Fig. 4). Similar with a previous study of Kfexo− (34), the insertion of correct dCTP was more efficient in the -TG*A- sequence compared with that of the -CG*A- context. This could be explained in terms of the predominant B-conformation of the adduct in the -TG*A- sequence compared with the S-conformation in the -CG*A- sequence context. However, the lack of dAMP misinsertion opposite the adducted site in both sequences suggests that the adduct stabilizes a catalytically inactive state such as an open polymerase conformation that would deter incorrect nucleotide binding.
To further investigate whether other divalent metal ions might promote insertion of nucleotides opposite dG-FAF, alternate divalent metals were substituted for Mg2+. Previous studies have demonstrated that Mn2+ and Co2+ support DNA synthesis and lower substrate specificity (44, 45). Mn2+ serves as an excellent metal substitute for Mg2+ due to similar ionic radii and coordination geometries (58). The results reported in this study demonstrate that these metals decreased polymerase fidelity, as insertion of the non-complementary nucleotides were significantly higher in the presence of these alternate divalent cations. As with Mg2+, the adduct conformation plays a significant role in nucleotide specificity as -CG*A- with a higher S-conformer population deters misinsertion relative to the -TG*A- sequence in the presence of Mn2+ and Co2+.
As observed previously (48, 49), weak rCTP and rGTP insertion opposite template dG with all divalent metal ions was observed. The weak insertion of ribonucleotides by pol β with unmodified DNA has been attributed to steric and electrostatic deterrents with Tyr-271 (59). Although in the -CG*A- sequence the insertion of rNTPs was not observed, weak insertion of CTP opposite dG-FAF in -TG*A- could be clearly seen (Fig. 5). The ability to insert a ribonucleotide opposite guanine or an adducted guanine can be correlated with the difference in the geometry of the nucleotides in the active site as well as an altered hydrogen bonding between the Tyr-271 side chain and the minor groove edge of the terminal primer base in the closed ternary substrate conformation (54). In contrast to -TG*A- sequence context, the -CG*A- sequence exhibiting a higher portion of the S-conformation may alter interactions with the primer terminus, resulting in the loss of hydrogen bonding with tyrosine hydroxyl group.
DNA Binding by SPR
SPR is a chip-based, label-free flow-through technique that allows real-time monitoring of macromolecular bindings in solution (34). Unlike fluorescence, gel mobility shift, and footprinting assays, samples of interest will be in chemical equilibrium and are amenable to testing across various temperatures or concentrations.
Pol β was found to bind poorly on non-gapped dsDNA. The introduction of a 1-nt gapped DNA increased the binding affinity of pol β 1000-fold. This is in agreement with previous work indicating a preference for short gaps with 5′-phosphorylated margin (17, 60, 61). Significantly, the addition of dCTP enhanced the DNA binding affinity of the polymerase to the gapped substrate, whereas incorrect nucleotides modestly decreased binding affinity (Table 2). These results are consistent with an induced-fit mechanism for polymerase specificity where correct nucleotides stabilize the closed polymerase conformation and incorrect nucleotides destabilize the closed active site complex. This result is similar to that reported previously for Kfexo− using a similar approach (34).
For pol β, the presence of the adduct in the gap modestly weakened DNA binding (Table 3). The binding affinity was similar to that observed for non-adducted DNA in the presence of incorrect nucleotides. In this case, however, the addition of correct or incorrect nucleotides to form a ternary complex did not appreciably alter apparent DNA binding. In contrast, Kfexo− binds more tightly to adducted DNA (34). Because Kfexo− binds DNA primarily through the upstream duplex, this may provide the adduct more freedom to be accommodated near the active site. Additionally, A-family DNA polymerases bind the templating nucleotide outside the DNA helix in the binary DNA complex. Because pol β binds to both upstream and downstream duplex, the templating nucleotide is usually situated within the DNA helix in the binary DNA complex. These features would constrain the adduct that could weaken overall binding. The observation that correct and incorrect nucleotides alter adduct binding with Kfexo− (34), but not pol β, is consistent with this suggestion.
In conclusion, we used 19F NMR to characterize the various states of the mutagenic dG-FAF adduct bound to pol β and Kfexo− in the presence and absence of various non-hydrolysable nucleotide analogs. These NMR data are nicely complemented by kinetic data derived from surface plasmon resonance and primer kinetic studies. The dG-FAF adduct in a single nucleotide gap adopts both a major-groove-oriented and base-displaced stacked conformation, and this heterogeneity is retained upon binding pol β. The addition of a non-hydrolysable dCMPcPP nucleotide analog to the binary complex increased the major groove conformation at the expense of the stacked conformation. Similar results were obtained with the addition of a non-complementary dAMPcPP analog but with an additional minor groove binding conformer. dG-FAF adduct at the replication fork for the Kfexo− complex, however, adopts a mix of the major and minor groove conformers with minimal effect upon the addition of non-hydrolysable nucleotides. For pol β, the insertion of dCTP was preferred opposite the dG-FAF adduct in a single nucleotide gap assay, consistent with 19F NMR data. SPR binding kinetics revealed that pol β binds tightly with DNA in the presence of correct dCTP. We have discussed these data in terms of adduct-induced conformational heterogeneity (B, S, W), sequence effect (TGA versus CGA), and the nature of a substrate (single/double-stranded versus gapped junction) and a polymerase (pol β versus Klenow). The present results provide valuable molecular insights into the binding characteristics of a bulky DNA lesion in the active site of DNA polymerases and their coding potentials.
Supplementary Material
Acknowledgments
We thank Professor Catherine Joyce and Olga Potapova of Yale University for providing Kfexo− (D424A) for 19F NMR experiments and the K758A mutant.
This work was supported, in whole or in part, by National Institutes of Health (NIH) Grants CA098296 (NCI) and P20 RR016457 (Center for Research Resources) (to B. P. C.) and Research Projects Z01-ES050158 and Z01-ES050161 in the Intramural Research Program of NIEHS, NIH, and in association with NIH Grant 1U19CA105010 (to S. H. W.).

This article contains supplemental Table S1 and Figs. S1--S7.
- pol β
- human DNA polymerase β
- FAF
- fluoroaminofluorene
- AF
- aminofluorene
- dG-FAF
- N-(2′-deoxyguanosin-8-yl)-7-fluoro-2-aminofluorene
- Kfexo−
- Klenow fragment exonuclease deficient
- SPR
- surface plasmon resonance
- dCMPcPP
- 2′-deoxycytosine-5′-[(α,β)-methyleno]triphosphate
- dAMPcPP
- 2′-deoxyadenosine-5′-[(α,β)-methyleno]triphosphate
- Bf
- Bacillus fragment
- RU
- response units
- nt
- nucleotide.
REFERENCES
- 1. Vijg J., Suh Y. (2013) Genome instability and aging. Annu Rev. Physiol. 75, 645–668 [DOI] [PubMed] [Google Scholar]
- 2. Stone M. P., Huang H., Brown K. L., Shanmugam G. (2011) Chemistry and structural biology of DNA damage and biological consequences. Chem. Biodivers. 8, 1571–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schärer O. D. (2011) Multistep damage recognition, pathway coordination, and connections to transcription, damage signaling, chromatin structure, cancer, and aging. Current perspectives on the nucleotide excision repair pathway. DNA Repair 10, 667. [DOI] [PubMed] [Google Scholar]
- 4. Hübscher U., Maga G. (2011) DNA replication and repair bypass machines. Curr. Opin. Chem. Biol. 15, 627–635 [DOI] [PubMed] [Google Scholar]
- 5. Klarer A. C., McGregor W. (2011) Replication of damaged genomes. Crit. Rev. Eukaryot. Gene Expr. 21, 323–336 [DOI] [PubMed] [Google Scholar]
- 6. Turesky R. J., Le Marchand L. (2011) Metabolism and biomarkers of heterocyclic aromatic amines in molecular epidemiology studies. Lessons learned from aromatic amines. Chem. Res. Toxicol. 24, 1169–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Skipper P. L., Kim M. Y., Sun H. L., Wogan G. N., Tannenbaum S. R. (2010) Monocyclic aromatic amines as potential human carcinogens. Old is new again. Carcinogenesis 31, 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Neumann H. G. (2007) Aromatic amines in experimental cancer research. Tissue-specific effects, an old problem, and new solutions. Crit. Rev. Toxicol. 37, 211–236 [DOI] [PubMed] [Google Scholar]
- 9. Beland F. A., Kadlubar F. F. (1990) Handbook of Experimental Pharmacology, pp. 281–285, Spring-Verlag, Heidelberg, Germany [Google Scholar]
- 10. Cho B. (2010) in The Chemical Biology of DNA Damage, pp. 217–238, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany [Google Scholar]
- 11. Cho B. P. (2004) Dynamic conformational heterogeneities of carcinogen-DNA adducts and their mutagenic relevance. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 22, 57–90 [DOI] [PubMed] [Google Scholar]
- 12. Lukin M., de Los Santos C. (2006) NMR structures of damaged DNA. Chem. Rev. 106, 607–686 [DOI] [PubMed] [Google Scholar]
- 13. Patel D. J., Mao B., Gu Z., Hingerty B. E., Gorin A., Basu A. K., Broyde S. (1998) Nuclear magnetic resonance solution structures of covalent aromatic amine-DNA adducts and their mutagenic relevance. Chem. Res. Toxicol. 11, 391–407 [DOI] [PubMed] [Google Scholar]
- 14. Beard W. A., Wilson S. H. (2006) Structure and mechanism of DNA polymerase β. Chem. Rev. 106, 361–382 [DOI] [PubMed] [Google Scholar]
- 15. Matsumoto Y., Kim K. (1995) Excision of deoxyribose phosphate residues by DNA polymerase β during DNA repair. Science 269, 699–702 [DOI] [PubMed] [Google Scholar]
- 16. Piersen C. E., Prasad R., Wilson S. H., Lloyd R. S. (1996) Evidence for an imino intermediate in the DNA polymerase β deoxyribose phosphate excision reaction. J. Biol. Chem. 271, 17811–17815 [DOI] [PubMed] [Google Scholar]
- 17. Prasad R., Beard W. A., Wilson S. H. (1994) Studies of gapped DNA substrate binding by mammalian DNA polymerase β. Dependence on 5′-phosphate group. J. Biol. Chem. 269, 18096–18101 [PubMed] [Google Scholar]
- 18. Moon A. F., Garcia-Diaz M., Batra V. K., Beard W. A., Bebenek K., Kunkel T. A., Wilson S. H., Pedersen L. C. (2007) The X family portrait. Structural insights into biological functions of X family polymerases. DNA Repair 6, 1709–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Canitrot Y., Cazaux C., Fréchet M., Bouayadi K., Lesca C., Salles B., Hoffmann J.-S. (1998) Overexpression of DNA polymerase β in cell results in a mutator phenotype and a decreased sensitivity to anticancer drugs. Proc. Natl. Acad. Sci. U.S.A. 95, 12586–12590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bergoglio V., Pillaire M.-J., Lacroix-Triki M., Raynaud-Messina B., Canitrot Y., Bieth A., Garès M., Wright M., Delsol G., Loeb L. A., Cazaux C., Hoffmann J.-S. (2002) Deregulated DNA polymerase β induces chromosome instability and tumorigenesis. Cancer Res. 62, 3511–3514 [PubMed] [Google Scholar]
- 21. Canitrot Y., Capp J.-P., Puget N., Bieth A., Lopez B., Hoffmann J.-S., Cazaux C. (2004) DNA polymerase β overexpression stimulates the Rad51-dependent homologous recombination in mammalian cells. Nucleic Acids Res. 32, 5104–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Servant L., Cazaux C., Bieth A., Iwai S., Hanaoka F., Hoffmann J.-S. (2002) A role for DNA polymerase β in mutagenic UV lesion bypass. J. Biol. Chem. 277, 50046–50053 [DOI] [PubMed] [Google Scholar]
- 23. Batra V. K., Shock D. D., Prasad R., Beard W. A., Hou E. W., Pedersen L. C., Sayer J. M., Yagi H., Kumar S., Jerina D. M., Wilson S. H. (2006) Structure of DNA polymerase β with a benzo[c]phenanthrene diol epoxide-adducted template exhibits mutagenic features. Proc. Natl. Acad. Sci. U.S.A. 103, 17231–17236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chary P., Beard W. A., Wilson S. H., Lloyd R. S. (2012) DNA polymerase β gap-filling translesion DNA synthesis. Chem. Res. Toxicol. 25, 2744–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoffmann J. S., Pillaire M. J., Garcia-Estefania D., Lapalu S., Villani G. (1996) In vitro bypass replication of the cisplatin-d(GpG) lesion by calf thymus DNA polymerase β and human immunodeficiency virus type I reverse transcriptase is highly mutagenic. J. Biol. Chem. 271, 15386–15392 [DOI] [PubMed] [Google Scholar]
- 26. Vaisman A., Chaney S. G. (2000) The efficiency and fidelity of translesion synthesis past cisplatin and oxaliplatin GpG adducts by human DNA polymerase β. J. Biol. Chem. 275, 13017–13025 [DOI] [PubMed] [Google Scholar]
- 27. Efrati E., Tocco G., Eritja R., Wilson S. H., Goodman M. F. (1997) Abasic translesion synthesis by DNA polymerase β violates the “A-rule.” Novel types of nucleotide incorporation by human DNA polymerase β at an abasic lesion in different sequence contexts. J. Biol. Chem. 272, 2559–2569 [DOI] [PubMed] [Google Scholar]
- 28. Efrati E., Tocco G., Eritja R., Wilson S. H., Goodman M. F. (1999) “Action-at-a-Distance” mutagenesis. 8-Oxo-7,8-dihydro-2′-deoxyguanosine causes base substitution errors at neighboring template sites when copied by DNA polymerase β. J. Biol. Chem. 274, 15920–15926 [DOI] [PubMed] [Google Scholar]
- 29. Osheroff W. P., Beard W. A., Yin S., Wilson S. H., Kunkel T. A. (2000) Minor groove interactions at the DNA polymerase β active site modulate single-base deletion error rates. J. Biol. Chem. 275, 28033–28038 [DOI] [PubMed] [Google Scholar]
- 30. Batra V. K., Beard W. A., Shock D. D., Pedersen L. C., Wilson S. H. (2008) Structures of DNA polymerase β with active site mismatches suggest a transient abasic site intermediate during misincorporation. Mol. Cell 30, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liang F., Cho B. P. (2007) Probing the thermodynamics of aminofluorene-induced translesion DNA synthesis by differential scanning calorimetry. J. Am. Chem. Soc. 129, 12108–12109 [DOI] [PubMed] [Google Scholar]
- 32. Liang F., Cho B. P. (2011) Conformational and thermodynamic impact of bulky aminofluorene adduction on simulated translesion DNA synthesis. Chem. Res. Toxicol. 24, 597–605 [DOI] [PubMed] [Google Scholar]
- 33. Vaidyanathan V. G., Cho B. P. (2012) Sequence effects on translesion synthesis of an aminofluorene-DNA adduct. Conformational, thermodynamic, and primer extension kinetic studies. Biochemistry 51, 1983–1995 [DOI] [PubMed] [Google Scholar]
- 34. Vaidyanathan V. G., Xu L., Cho B. P. (2012) Binary and ternary binding affinities between exonuclease-deficient Klenow fragment (Kf-exo−) and various arylamine DNA lesions characterized by surface plasmon resonance. Chem. Res. Toxicol. 25, 1568–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meneni S., Liang F., Cho B. P. (2007) Examination of the long-range effects of aminofluorene-induced conformational heterogeneity and its relevance to the mechanism of translesional DNA synthesis. J. Mol. Biol. 366, 1387–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meneni S. R., Shell S. M., Gao L., Jurecka P., Lee W., Sponer J., Zou Y., Chiarelli M. P., Cho B. P. (2007) Spectroscopic and theoretical insights into sequence effects of aminofluorene-induced conformational heterogeneity and nucleotide excision repair. Biochemistry 46, 11263–11278 [DOI] [PubMed] [Google Scholar]
- 37. Zhou L., Rajabzadeh M., Traficante D. D., Cho B. P. (1997) Conformational heterogeneity of arylamine-modified DNA. 19F NMR evidence. J. Am. Chem. Soc. 119, 5384–5389 [Google Scholar]
- 38. Patnaik S., Cho B. P. (2010) Structures of 2-acetylaminofluorene modified DNA revisited. Insight into conformational heterogeneity. Chem. Res. Toxicol. 23, 1650–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jain N., Meneni S., Jain V., Cho B. P. (2009) Influence of flanking sequence context on the conformational flexibility of aminofluorene-modified dG adduct in dA mismatch DNA duplexes. Nucleic Acids Res. 37, 1628–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hsu G. W., Kiefer J. R., Burnouf D., Becherel O. J., Fuchs R. P., Beese L. S. (2004) Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase. J. Biol. Chem. 279, 50280–50285 [DOI] [PubMed] [Google Scholar]
- 41. Kuchta R. D., Mizrahi V., Benkovic P. A., Johnson K. A., Benkovic S. J. (1987) Kinetic mechanism of DNA polymerase I (Klenow). Biochemistry 26, 8410–8417 [DOI] [PubMed] [Google Scholar]
- 42. Beard W. A., Wilson S. H. (2003) Structural insights into the origins of DNA polymerase fidelity. Structure 11, 489–496 [DOI] [PubMed] [Google Scholar]
- 43. Astatke M., Grindley N. D., Joyce C. M. (1995) Deoxynucleoside triphosphate and pyrophosphate binding sites in the catalytically competent ternary complex for the polymerase reaction catalyzed by DNA polymerase I (Klenow fragment). J. Biol. Chem. 270, 1945–1954 [DOI] [PubMed] [Google Scholar]
- 44. Kunkel T. A., Loeb L. A. (1979) On the fidelity of DNA replication. Effect of divalent metal ion activators and deoxyribonucleoside triphosphate pools on in vitro mutagenesis. J. Biol. Chem. 254, 5718–5725 [PubMed] [Google Scholar]
- 45. Sirover M. A., Loeb L. A. (1977) On the fidelity of DNA replication. Effect of metal activators during synthesis with avian myeloblastosis virus DNA polymerase. J. Biol. Chem. 252, 3605–3610 [PubMed] [Google Scholar]
- 46. Traut T. W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell Biochem. 140, 1–22 [DOI] [PubMed] [Google Scholar]
- 47. Ferraro P., Franzolin E., Pontarin G., Reichard P., Bianchi V. (2010) Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res. 38, e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nick McElhinny S. A., Watts B. E., Kumar D., Watt D. L., Lundström E.-B., Burgers P. M., Johansson E., Chabes A., Kunkel T. A. (2010) Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U.S.A. 107, 4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cavanaugh N. A., Beard W. A., Wilson S. H. (2010) DNA polymerase β ribonucleotide discrimination, Insertion, misinsertion, extension, and coding. J. Biol. Chem. 285, 24457–24465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Seo K.-Y., Jelinsky S. A., Loechler E. L. (2000) Factors that influence the mutagenic patterns of DNA adducts from chemical carcinogens. Mutat. Res. 463, 215–246 [DOI] [PubMed] [Google Scholar]
- 51. Kiefer J. R., Mao C., Braman J. C., Beese L. S. (1998) Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature 391, 304–307 [DOI] [PubMed] [Google Scholar]
- 52. Wang W., Wu E. Y., Hellinga H. W., Beese L. S. (2012) Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides. J. Biol. Chem. 287, 28215–28226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Beard W. A., Shock D. D., Yang X.-P., DeLauder S. F., Wilson S. H. (2002) Loss of DNA polymerase β stacking interactions with templating purines, but not pyrimidines, alters catalytic efficiency and fidelity. J. Biol. Chem. 277, 8235–8242 [DOI] [PubMed] [Google Scholar]
- 54. Sawaya M. R., Prasad R., Wilson S. H., Kraut J., Pelletier H. (1997) Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA, Evidence for an induced fit mechanism. Biochemistry 36, 11205–11215 [DOI] [PubMed] [Google Scholar]
- 55. Johnson S. J., Taylor J. S., Beese L. S. (2003) Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl. Acad. Sci. U.S.A. 100, 3895–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Upton T. G., Kashemirov B. A., McKenna C. E., Goodman M. F., Prakash G. K., Kultyshev R., Batra V. K., Shock D. D., Pedersen L. C., Beard W. A., Wilson S. H. (2009) α,β-Diflurormethylene deoxynucleoside 5′-triphosphates. A convenient synthesis of useful probes for DNA polymerase β structure and function. Org. Lett. 11, 1883–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liang F., Jain N., Hutchens T., Shock D. D., Beard W. A., Wilson S. H., Chiarelli M. P., Cho B. P. (2008) α,β-Methylene-2′-deoxynucleoside 5′-triphosphates as noncleavable substrates for DNA polymerases. Isolation, characterization, and stability studies of novel 2′-deoxycyclonucleosides, 3,5′-cyclo-dG, and 2,5′-cyclo-dT. J. Med. Chem. 51, 6460–6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Harding M. M., Nowicki M. W., Walkinshaw M. D. (2010) Metals in protein structures. A review of their principal features. Crystallogr. Rev. 16, 247–302 [Google Scholar]
- 59. Cavanaugh N. A., Beard W. A., Batra V. K., Perera L., Pedersen L. G., Wilson S. H. (2011) Molecular insights into DNA polymerase deterrents for ribonucleotide insertion. J. Biol. Chem. 286, 31650–31660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Beard W. A., Wilson S. H. (2000) Structural design of a eukaryotic DNA repair polymerase. DNA polymerase β. Mutat. Res. 460, 231–244 [DOI] [PubMed] [Google Scholar]
- 61. Singhal R. K., Wilson S. H. (1993) Short gap-filling synthesis by DNA polymerase β is processive. J. Biol. Chem. 268, 15906–15911 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.