Background: Variable lymphocyte receptors (VLRs) bind tumor-associated carbohydrates, such as Thomsen-Friedenreich antigen (TFα), with high selectivity.
Results: The crystal structure of a VLR-TFα complex coupled with thermodynamic analysis revealed the basis for selectivity.
Conclusion: VLRs utilize leucine-rich repeats to recognize glycans with affinity comparable to that of lectins and antibodies.
Significance: The VLR-TFα structure provides a template for engineering VLRs to bind biomedically relevant glycans.
Keywords: Antibodies, Biotechnology, Crystal Structure, Glycosylation, Immunology, Oligosaccharide, Protein Structure, Thermodynamics, Tumor Marker, X-ray Crystallography
Abstract
Variable lymphocyte receptors (VLRs) are leucine-rich repeat proteins that mediate adaptive immunity in jawless vertebrates. VLRs were recently shown to recognize glycans, such as the tumor-associated Thomsen-Friedenreich antigen (TFα; Galβ1–3GalNAcα), with a selectivity rivaling or exceeding that of lectins and antibodies. To understand the basis for TFα recognition by one such VLR (VLRB.aGPA.23), we measured thermodynamic parameters for the binding interaction and determined the structure of the VLRB.aGPA.23-TFα complex to 2.2 Å resolution. In the structure, four tryptophan residues form a tight hydrophobic cage encasing the TFα disaccharide that completely excludes buried water molecules. This cage together with hydrogen bonding of sugar hydroxyls to polar side chains explains the exquisite selectivity of VLRB.aGPA.23. The topology of the glycan-binding site of VLRB.aGPA.23 differs markedly from those of lectins or antibodies, which typically consist of long, convex grooves for accommodating the oligosaccharide. Instead, the TFα disaccharide is sandwiched between a variable loop and the concave surface of the VLR formed by the β-strands of the leucine-rich repeat modules. Longer oligosaccharides are predicted to extend perpendicularly across the β-strands, requiring them to bend to match the concavity of the VLR solenoid.
Introduction
The normal processes of glycosylation are often severely altered during oncogenic transformation, such that nearly all human tumors express glycoproteins with aberrant glycosylation patterns (1). These changes can affect interactions between tumor cell surface glycans and glycan binding receptors, which may determine the metastatic potential of the cancer cell. The best-studied tumor-associated carbohydrates are truncated glycans, such as T-nouvelle antigen (GalNAcα) and Thomsen-Friedenreich antigen (TFα4; Galβ1–3GalNAcα), which are linked to serine or threonine as O-glycans (1). These saccharides decorate mucin-type glycoproteins in ∼90% of human cancers but are absent from nearly all normal tissues (1, 2). A number of carbohydrate binding lectins and antibodies have been developed to detect expression of T-nouvelle antigen, TFα, and other cancer-specific glycans for diagnostic and prognostic applications (2–4). However, most of these reagents display either broad specificity or low affinity for their target glycans and, therefore, have limited clinical utility (5, 6).
A variety of approaches have been pursued for obtaining proteins that bind cancer-specific carbohydrates and other biomedically relevant glycans with high selectivity and affinity. Most often, monoclonal antibodies are isolated from mice immunized with the glycan of interest, but this process is laborious, and most human glycans are not particularly immunogenic in mice, probably due to their conserved structures in these species, which gives rise to self-tolerance. Other strategies include directed evolution of antibodies (7) or lectins (3, 8), carbohydrate binding peptides (9), and glycan-specific aptamers (10), but none has proven generally useful.
We recently described a rapid and cost-effective strategy using yeast surface display for isolating monoclonal variable lymphocyte receptors (VLRs) from lamprey that selectively bind various human glycans, including the pancarcinoma antigens T-nouvelle antigen and TFα (11). VLRs are leucine-rich repeat (LRR) proteins that mediate adaptive immunity in jawless vertebrates (lamprey and hagfish). Although VLRs are structurally unrelated to the immunoglobulin-based antibodies of jawed vertebrates, they are able to recognize as broad a spectrum of antigens as antibodies, with entirely comparable affinity and specificity (12). VLR genes are assembled from multiple LRR-encoding cassettes by DNA recombination in a process that generates a vast repertoire of >1014 unique receptors, which is sufficiently diverse to recognize most, if not all, antigens (13–15).
Among the lamprey VLRs isolated by screening naïve VLR libraries displayed on yeast for glycan binders was VLRB.aGPA.23, which recognized TFα (11). Glycan array profiling showed that VLRB.aGPA.23 possessed a high degree of selectivity for TFα. Indeed, its overall selectivity was superior to that of most lectins and antibodies (5, 6). Furthermore, a dimeric form of VLRB.aGPA.23 bound TFα with nanomolar affinity (11). In human tissue microarrays, VLRB.aGPA.23 selectively detected tumor-associated carbohydrate antigens in a variety of adenocarcinomas and squamous cell carcinomas. Moreover, lung cancer patients whose tumors stained with VLRB.aGPA.23 experienced a significantly worse survival rate compared with patients whose tumors did not (11). Thus, highly selective VLRs such as VLRB.aGPA.23 provide a powerful means for detecting aberrantly glycosylated proteins on tumor cells, which may lead to an entirely new class of diagnostic and prognostic reagents.
Recent structural studies of VLRs bound to protein or carbohydrate ligands have begun to reveal the features that endow these LRR-based adaptive immune receptors with specificity and affinity for diverse antigens (12). However, the structural database for VLRs, which at present comprises only four VLR-antigen complexes (16–19), is far smaller than that for antibodies, for which hundreds of structures have been determined. Here we report the structure of VLRB.aGPA.23 bound to its tumor-associated ligand TFα. This structure, in conjunction with a thermodynamic analysis of glycan binding by this VLR, provides a basis for understanding the remarkable selectivity of VLRB.aGPA.23 at the atomic level and for engineering VLRs as highly specific reagents for recognizing tumor-associated and other biomedically important carbohydrates.
EXPERIMENTAL PROCEDURES
Glycans
Galβ1–3GalNAcα (TFα) was purchased from Toronto Research Chemicals. Galβ1–3GalNAcα-polyacrylamide-biotin (TFα-PAA) was purchased from Glycotech. Galβ1–3GalNAcα-Ser (TFα-Ser) and Galβ1–3GalNAcα-Thr (TFα-Thr) were prepared by deprotection of peracetylated TF-Fmoc (N-(9-fluorenyl)methoxycarbonyl)-Ser-OH and TF-Fmoc-Thr-OH (Sussex Research Laboratories) with sodium methoxide in methanol followed by neutralization with HCl and purification by sequential silica gel and reverse phase silica gel chromatography in 75 and 88% yields, respectively. Products were characterized by 1H NMR and mass spectrometry. Fucα1–2Galβ (BG-H) was provided by the Consortium for Functional Glycomics. Human glycophorin A (GPA) was purchased from Sigma. Asialo human glycophorin A (aGPA) was prepared by desialylating GPA by mild acid treatment using 0.025 n H2SO4 in 0.85% NaCl for 2 h at 80 °C, then neutralized with 1 m Tris-HCl (pH 8.8) and dialyzed overnight against PBS (pH 7.4) (Quality Biological)
Protein Expression and Purification
VLRB.aGPA.23 was isolated from a VLRB library (1.2 × 108 independent clones) generated from lymphocyte cDNA of unimmunized lampreys and displayed on the surface of yeast (11). The library was screened by alternating rounds of magnetic-activated and fluorescent-activated cell sorting with TFα or aGPA. The diversity region of VLRB.aGPA.23, from LRRNT to LRRCT (residues 1–219), was cloned into the expression vector pT7–7 (Novagen) and expressed as inclusion bodies in BL21-CodonPlus(DE3)-RIL Escherichia coli cells (Stratagene). Bacteria were grown at 37 °C in LB medium to an absorbance of 0.6–0.8 at 600 nm and induced with 1 mm isopropyl-β-d-thiogalactoside. After incubation for 3 h, the bacteria were harvested by centrifugation and resuspended in 50 mm Tris-HCl (pH 8.0) containing 0.1 m NaCl and 2 mm EDTA; cells were disrupted by sonication. Inclusion bodies were washed with 50 mm Tris-HCl (pH 8.0), 0.1 m NaCl, and 0.5% (v/v) Triton X-100, then solubilized in 8 m urea and 100 mm Tris-HCl (pH 8.5). For in vitro folding, inclusion bodies were diluted to a final concentration of 10 mg/liter into 0.8 m arginine, 100 mm Tris-HCl (pH 8.5), 2 mm EDTA, 3 mm reduced glutathione, and 0.3 mm oxidized glutathione. After 3 days at 4 °C, the folding mixture was concentrated, dialyzed against 20 mm Tris-HCl (pH 8.5), and applied to a MonoQ column (GE Healthcare). Further purification was carried out using a Superdex 75 HR column.
Isothermal Titration Calorimetry
ITC measurements were carried out using a MicroCal iTC200 titration microcalorimeter. Purified VLRB.aGPA.23 was exhaustively dialyzed against 10 mm phosphate (pH 7.2), 136 mm NaCl, and 4 mm KCl. In a typical experiment, 1- or 2-μl aliquots of 2.46–7.40 mm glycan solution were injected from a 40-μl rotating syringe into the sample cell containing 200 μl of 0.157–0.377 mm VLRB.aGPA.23 solution at 10 °C. For each titration experiment, an identical buffer dilution correction was conducted; these heats of dilution were subtracted from the corresponding binding experiment. Equilibrium binding constants (Kb) were obtained by non-linear least-squares fit of the ITC data to a single-site binding model. Data acquisition and analysis were performed with the software package ORIGIN.
Surface Plasmon Resonance (SPR) Analysis
All SPR binding experiments were performed at 25 °C in phosphate-buffered saline (pH 7.4) using a BIAcore T100 biosensor. Biotinylated TFα-PAA (33 nm), aGPA (2.8 nm), and GPA (4.6 nm) were immobilized on streptavidin-coated BIAcore SA chips followed by blocking the remaining streptavidin sites with 1 μm biotin solution. An additional flow cell was injected with only free biotin to serve as a blank control. Solutions containing different concentrations of VLRB.aGPA.23 were then serially injected over the chips with immobilized ligands and the blank until SPR signals reached a plateau. Equilibrium affinity measurements were performed at different flow rates (30–50 μl/min). Specific binding data were fitted with a 1:1 Langmuir binding model using BIAevaluation 4.1 software (BIAcore) to calculate Kb values.
Crystallization and Data Collection
For crystallization of the VLRB.aGPA.23-TFα complex, VLRB.aGPA.23 (15 mg/ml) was mixed with TFα-Ser in a 1:10 molar ratio. Crystals grew at 25 °C in hanging drops in 20% (w/v) polyethylene glycol (PEG) 3350, 4% (v/v) glycerol, and 0.1 m Tris-HCl (pH 7.0). Crystals of the VLRB.aGPA.23-BG-H complex grew in 28.5% (w/v) PEG 3350, 0.6 m NaCl, and 0.1 m Tris-HCl (pH 7.0) from solutions containing a 5-fold molar excess of BG-H. For data collection, both complexes were cryoprotected in 100% paraffin oil before flash-cooling. X-ray diffraction data were recorded in-house at 100 K using a Rigaku R-axis IV2+ image plate detector. All data were indexed, integrated, and scaled with the program CrystalClear (Rigaku). Both crystals contain four molecules in the asymmetric unit. Data collection statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
| VLRB.aGPA.23−TFα | VLRB.aGPA.23−BG-H | |
|---|---|---|
| Data collection | ||
| Space group | P1211 | P1 |
| Unit cell (Å) (º) | a = 56.68, b = 68.68, c = 99.92, α = γ = 90.00, β = 93.58 | a = 57.71, b = 62.78, c = 63.37, α = 72.03, β = 71.39, γ = 84.35 |
| Resolution range (Å)a | 33.66–2.20 (2.28–2.20) | 59.72–1.70 (1.76–1.70) |
| Unique reflectionsa | 38816 (3744) | 82662 (7776) |
| Completeness (%)a | 99.1 (95.8) | 93.3 (87.8) |
| I/ σIa | 23.3 (6.3) | 12.4 (3.3) |
| Rmerge (%)a,b | 9.3 (20.9) | 6.9 (20.5) |
| Average redundancya | 6.31 (4.74) | 3.95 (3.90) |
| Refinement statistics | ||
| Resolution range (Å) | 33.66–2.20 | 59.72–1.70 |
| Rwork/Rfree (%)c | 20.1/25.1 | 20.6/24.7 |
| No. reflections | 36760 | 78270 |
| Number of protein atoms | 6656 | 6642 |
| Number of ligand atoms | 81 | 48 |
| Number of water atoms | 281 | 384 |
| Root mean squared deviation from ideality | ||
| Bond lengths (Å) (º) | 0.018 | 0.028 |
| Bond angles (º) | 1.657 | 2.220 |
| Mean B-factors (Å2) | 28.6 | 22.4 |
| Ramachandran plot statistics | ||
| Most favored (%) | 96.2 | 96.8 |
| Additionally allowed (%) | 3.8 | 3.2 |
| Outliers (%) | 0 | 0 |
a Values in parentheses refer to the highest resolution shell.
b Rmerge = Σ|Ij − 〈I〉|/ΣIj, where Ij is the intensity of an individual reflection, and 〈I〉 is the average intensity of that reflection.
c Rwork = Σ‖Fo| − |Fc‖/Σ|Fo|, where Fc is the calculated structure factor. Rfree is as for Rwork but calculated for a randomly selected 5.0% of reflections not included in the refinement.
Structure Determination and Refinement
The structure of the VLRB.aGPA.23-TFα complex was solved by molecular replacement with the Phaser program (20) using VLRB.RBC36 (PDB accession code 3E6J) (16) as a search model. Refinement was performed with RefMac 5.0 (21). Modeling and rebuilding were accomplished with COOT (22) using σA-weighted 2Fo − Fc and Fo − Fc electron density maps. The structure of the VLRB.aGPA.23-BG-H complex was solved by molecular replacement with Phaser (20). The search model used in the calculations was the refined VLRB.aGPA.23-TFα structure. Final refinement statistics are presented in Table 1.
Protein Data Bank Accession Codes
Coordinates and structure factors for the VLRB.aGPA.23-TFα and VLRB.aGPA.23-BG-H complexes have been deposited under accession codes 4K79 and 4K5U, respectively.
RESULTS
Thermodynamic Analysis of Glycan Binding to VLRB.aGPA.23
We previously analyzed the binding selectivity of VLRB.aGPA.23 using glycan microarrays (11). The main reactive structures were: TFα-Ser, blood group H type 3 trisaccharide (BG-H3; Fucα1–2Galβ1–3GalNAcα), blood group H disaccharide (BG-H; Fucα1–2Galβ), and aGPA, which is decorated with TFα-Ser (23).
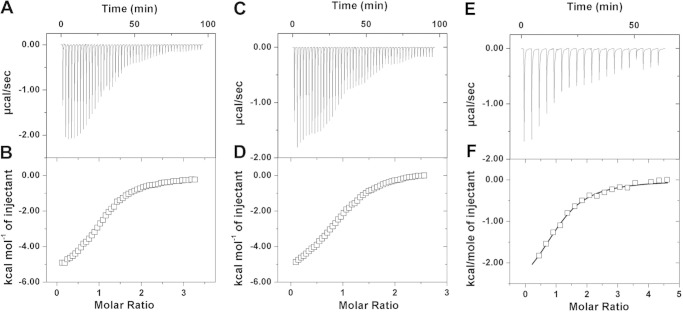
We used ITC to determine thermodynamic parameters for the binding of VLRB.aGPA.23 to TFα, TFα-Ser, TFα-Thr, BG-H, and galactose (Table 2). For this purpose, VLRB.aGPA.23 was expressed as a soluble monomeric protein by in vitro folding from bacterial inclusion bodies. As measured by ITC, VLRB.aGPA.23 bound the TFα disaccharide with an equilibrium binding constant (Kb) of 2.21 × 104 m−1 (Fig. 1, A and B). Importantly, this affinity is as high as has been reported for animal or plant lectin binding to disaccharide ligands (24). It is also nearly identical to the affinities determined for TFα-Ser (Kb = 2.04 × 104 m−1) (Fig. 1, C and D) and TFα-Thr (2.43 × 104 m−1) (see Table 2), showing that the amino acid moiety made no appreciable contribution to binding VLRB.aGPA.23, at least in solution. In terms of energetics, all three reactions were exothermic (negative ΔHb), indicating favorable contacts between the sugars and protein. Whereas the binding of TFα and TFα-Ser was enthalpically driven, that of TFα-Thr was characterized by a large favorable entropy (positive TΔSb), which compensated for a reduced enthalpy to achieve similar affinity.
TABLE 2.
Thermodynamic parameters for the binding of saccharides to VLRB.aGPA.23
The stoichiometry (n) values ranged from 0.989 to 1.09 with uncertainties from 0.4 to 10.0%. The values in parentheses represent uncertainties of fit. The temperature was 25 °C.
| Ligand | Kb | −ΔGb | −ΔHb | TΔSb | ΔSb |
|---|---|---|---|---|---|
| × 104m−1 | kcal mol−1 | kcal mol−1 | kcal mol−1 | cal mol−1 K−1 | |
| TFα | 2.21 (±0.06) | 5.63 | 5.91 (±0.08) | −0.28 | −0.98 |
| TFα-Ser | 2.04 (±0.06) | 5.58 | 5.36 (±0.08) | 0.22 | 0.78 |
| TFα-Thr | 2.43 (±0.2) | 5.67 | 2.30 (±0.06) | 3.37 | 11.9 |
| BG-H | 0.96 (± 0.1) | 5.08 | 2.8 (±0.2) | 2.28 | 8.05 |
FIGURE 1.
Calorimetric titrations of VLRB.aGPA.23 with glycan ligands at 10 °C. A, raw data were obtained from 48 automatic injections of 0.5-μl aliquots of 5.02 mm TFα solution into 0.370 mm VLRB.aGPA.23 solution. B, shown is the incremental heat per mole of added ligand (open squares) for the titration in A. The Kb obtained from this titration is 2.21 ± 0.06 × 104 m−1 with n = 1.09 ± 0.01. C, raw data were obtained from 48 automatic injections of 0.5-μl aliquots of 4.80 mm TFα-Ser solution into 0.370 mm VLRB.aGPA.23 solution. D, shown is the incremental heat per mole of added ligand (open squares) for the titration in C; Kb = 2.04 ± 0.06 × 104 m−1 with n = 0.97 ± 0.10. E, raw data were obtained from 19 automatic injections of 2-μl aliquots of 7.4 mm BG-H solution into 0.310 mm VLRB.aGPA.23 solution. F, shown is a non-linear least-squares fit (solid line) of the incremental heat per mole of added ligand (open squares) for the titration in E; Kb = 2.04 ± 0.06 × 104 m−1 with n = 1.06 ± 0.07.
VLRB.aGPA.23 bound the BG-H disaccharide ∼2-fold less tightly than TFα (Kb = 0.96 × 104 m−1) (Fig. 1, E and F) (Table 2). However, VLRB.aGPA.23 showed no detectable binding to galactose (not shown), which demonstrates the contribution of fucose at the non-reducing end of BG-H and of N-acetylgalactosamine at the reducing end of TFα to ligand recognition by this VLR.
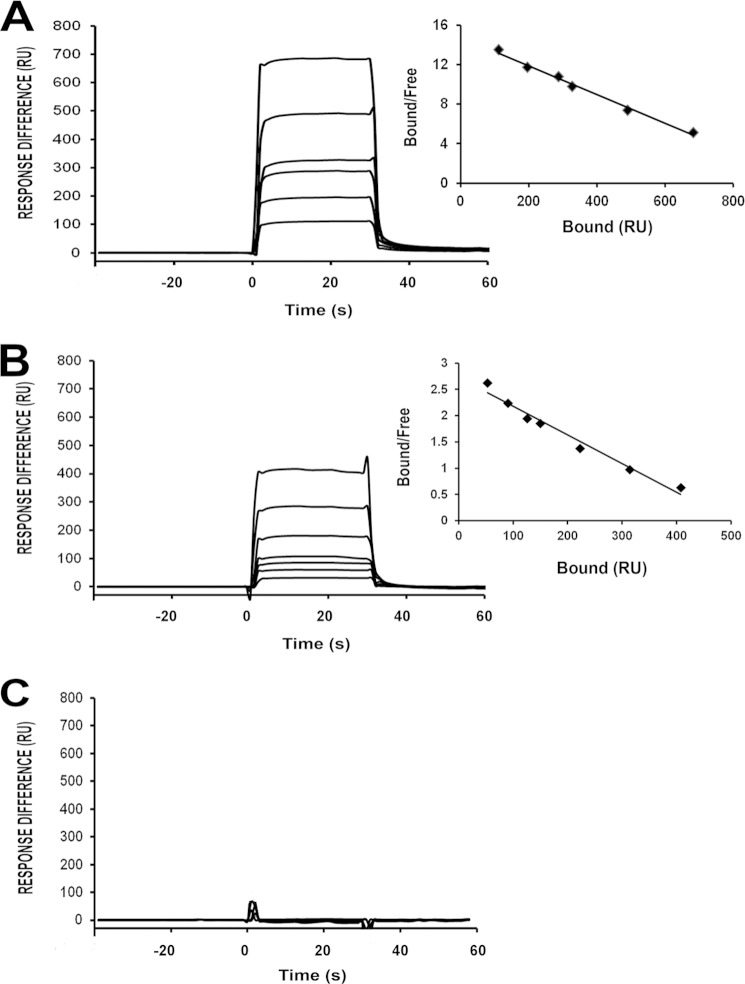
To independently confirm our Kb measurements from ITC, we used SPR. In these experiments a biotinylated synthetic polyacrylamide glycoconjugate of TFα (TFα-PAA) was immobilized on a streptavidin-coated biosensor surface, over which soluble monomeric VLRB.aGPA.23 was then injected (Fig. 2A). Under equilibrium binding conditions, a Kb of 1.5 × 104 m−1 was obtained, which is very similar to the Kb from ITC (2.21 × 104 m−1). The much higher effective affinity from SPR reported previously (Kb = 1.3 × 108 m−1) (11) may be explained by avidity effects. In that study we used a dimeric, not monomeric, form of VLR.aGPA.23 in which the VLR was fused to the Fc region of an antibody. In addition, the configuration of the SPR assay was reversed; multivalent TFα-PAA was flowed over immobilized dimeric (VLR.aGPA.23)2-Fc protein (11), which allowed multipoint binding that produced a large avidity effect.
FIGURE 2.
Equilibrium binding of VLRB.aGPA.23 to glycoconjugates. A, SPR sensograms for the interaction of VLRB.aGPA.23 (267.4, 133.7, 66.8, 33.4, 26.7, 16.7, 8.3 μm) with immobilized TFα-PAA (578 resonance units (RU)) are shown. Kb (1.5 × 104 m−1) was calculated from a Scatchard plot (inset) after correction for nonspecific binding by measuring the concentration of free reactants and complex at equilibrium. B, shown is the interaction of VLRB.aGPA.23 (267.4, 133.7, 66.8, 33.4, 26.7, 16.7, 8.3 μm) with immobilized aGPA (215 RU); Kb = 0.56 × 104 m−1. C, shown is the interaction of VLRB.aGPA.23 with immobilized GPA (217 RU).
We also compared the binding of VLRB.aGPA.23 to immobilized aGPA versus native GPA (Fig. 2, B and C). Whereas VLRB.aGPA.23 bound aGPA with Kb = 0.56 × 104 m−1, which is ∼3-fold lower than its affinity for TFα, no binding to native GPA was detected, in agreement with previous results showing high selectivity for the desialylated protein (11).
Overview of the VLRB.aGPA.23-TFα Complex
We determined the crystal structure of VLRB.aGPA.23 bound to TFα-Ser to 2.2 Å resolution by molecular replacement using VLRB.RBC36 (16) as a search model (Table 1; Fig. 3A). The root mean squared deviation in α-carbon positions for the four VLRB.aGPA.23 molecules in the asymmetric unit of the VLRB.aGPA.23-TFα crystal was in the range of 0.14–0.25 Å, indicating close similarity. Unambiguous electron density corresponding to the Galβ1–3GalNAcα moiety of TFα-Ser was found in the binding site, as evident from the 2Fo − Fc electron density map (Fig. 3B). However, no density was observed for the serine residue, suggesting flexibility due to lack of stable contacts with VLRB.aGPA.23. In agreement with this interpretation, TFα and TFα-Ser bound VLRB.aGPA.23 with very similar affinities and enthalpy changes (Table 2). The average temperature factor (B) for the Galβ1–3GalNAcα portion of TFα-Ser was 34 Å2, compared with 27 Å2 for main-chain atoms of the protein, indicating that the disaccharide is well ordered in the crystal.
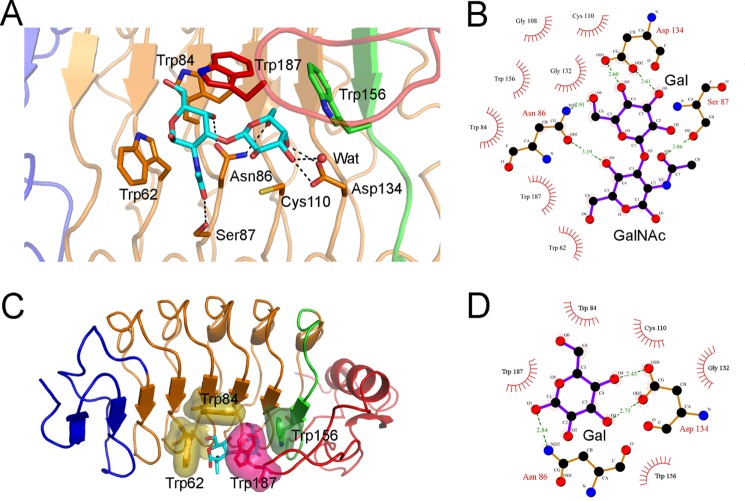
FIGURE 3.
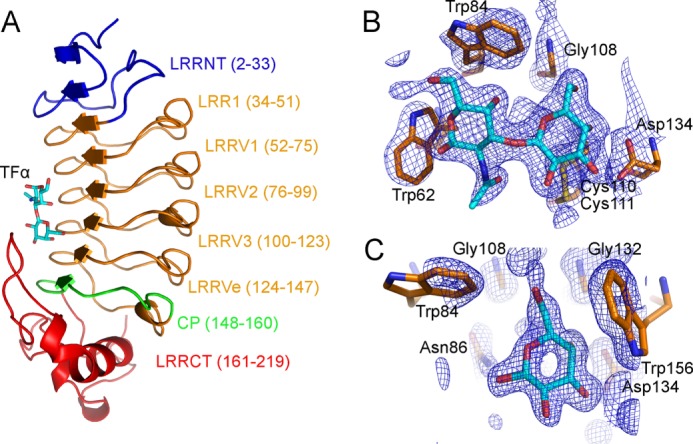
Structure of the VLRB. aGPA.23-TFα complex. A, a ribbon diagram of the VLRB.aGPA.23-TFα complex shows the concave antigen binding surface of the VLR solenoid. Blue, LRRNT; orange, LRR1, LRRV1–3, and LRRVe; green, CP; red, LRRCT. Carbons, nitrogens, and oxygens of TFα are cyan, blue, and red, respectively. B, electron density from the final 2Fo − Fc map (contoured at 1σ) of VLRB.aGPA.23-TFα at 2.2 Å resolution shows the Galβ1–3GalNAcα1 moiety of TFα. C, electron density from the final 2Fo − Fc map (contoured at 1σ) of the VLRB.aGPA.23-BG-H complex at 1.7 Å resolution show of the galactose moiety of BG-H. RU, relative units.
We also determined the structure of VLRB.aGPA.23 in complex with the BG-H disaccharide Fucα1–2Galβ to 1.9 Å resolution (Table 1). The galactose moiety of BG-H was clearly defined in the electron density (Fig. 3C). However, no density was observed for fucose, even though this moiety contributes significantly to binding (see above). The average B factor for the galactose was 35 Å2, compared with 20 Å2 for main-chain atoms of VLRB.aGPA.23.
VLRB.aGPA.23 adopts a horseshoe-shaped solenoid fold characteristic of other VLRs (Fig. 3A) (12, 16–19, 25). The structure comprises a 32-residue N-terminal LRR capping module (LRRNT), an 18-residue LRR1, four 24-residue LRRVs (LRRV1, LRRV2, LRRV3, and LRRVe), a 13-residue truncated LRR designated the connecting peptide (CP), and a 59-residue C-terminal LRR capping module (LRRCT). The inner, concave surface, through which VLRB.aGPA.23 binds TFα, is composed of eight β-strands (two from LRRNT, five from LRRs, and one from CP), that assemble into a continuous β-sheet (Fig. 2A). Residues Glu-13, Val-14, and Asn-15 of LRRNT initiate an anti-parallel β-strand that extends the continuous parallel β-sheet. The outer, convex surface of VLRB.aGPA.23 comprises an array of six loops and one α-helix. The region between the helices and strands constitutes the hydrophobic core of the VLR whose exposed ends are capped by LRRNT and LRRCT. In LRRNT, the characteristic four-cysteine motif of N-terminal LRR capping modules (CXnCXCXnC) forms two sets of disulfides (Cys-3--Cys-9 and Cys-7--Cys-16); a similar motif in LRRCT gives rise to disulfides Cys-164--Cys-198 and Cys-166--Cys-218. In their respective structures, the TFα and BG-H ligands are sandwiched between the concave surface of VLRB.aGPA.23 and a protruding loop of LRRCT (residues 184–190) (Fig. 3A), which corresponds to the highly variable insert of VLRs (12). This insert is absent from the LRRCTs of all other LRR-containing proteins, except platelet receptor glycoprotein Ibα (13).
The VLRB.aGPA.23-TFα Interface
The VLRB.aGPA.23-Galβ1–3GalNAcα complex buries a total solvent-accessible surface area of 270 Å2, to which the LRR (LRRV1–3 and LRRVe), CP, and LRRCT modules contribute 67, 6, and 26%, respectively. The specificity of the interaction is mediated mainly by five hydrogen bonds, three of which involve the galactose moiety: between Asn-86 Nδ2 and galactose O6′, between Asn-86 Oδ1 and N-acetylgalactosamine O4′, between Ser-87 Oγ and N-acetylgalactosamine O7′, between Asp-134 Oδ1 and galactose O4′, and between Asp-134 Oδ2 and galactose O3′ (Fig. 4A and B). Asn-86 and Ser-87 are located on LRRV2, and Asp-134 on LRRVe. Seven other residues (Trp-62, Trp-84, Gly-108, Cys-110, Gly-132, Trp-156, and Trp-187) stabilize the interaction with TFα via 53 van der Waals contacts. These residues, and in particular the four tryptophans, form a tight hydrophobic cage encasing the disaccharide that completely excludes buried water molecules (Fig. 4C). This cage together with the hydrogen bond network accounts for the exquisite glycan selectivity of VLRB.aGPA.23 (11). In addition, Trp-62 is positioned to sterically block the binding of oligosaccharides with extensions at the GalNAc end of the TFα disaccharide, further increasing selectivity (Fig. 4, A and C). Another key interaction with TFα involves Trp-187, whose indole is stacked parallel to the hydrophobic face of the galactose ring.
FIGURE 4.
The VLRB.aGPA.23-TFα binding interface. A, shown is a close-up view of VLRB.aGPA.23 residues involved in recognizing the TFα disaccharide. Colors (including residue labels) are as in Fig. 3. The side chains of contacting residues are drawn in stick representation with nitrogens in blue, oxygens in red, and sulfur in yellow. Hydrogen bonds are indicated by dotted lines. A water molecule bound to galactose O3′ is represented as a red sphere. B, shown is a schematic representation of interactions between VLRB.aGPA.23 and TFα. Residues making van der Waals contacts with TFα are indicated by arcs with spokes radiating toward the ligand moieties they contact. C, shown is tight packing of hydrophobic residues Trp-62, Trp-84, Trp-156, and Trp-187 around TFα, with their van der Waals radii drawn as surfaces. D, shown are schematic representations of interactions between VLRB.aGPA.23 and BG-H.
With the exception of Trp-187, which is in the LRRCT insert loop, all 10 TFα-contacting residues of VLRB.aGPA.23 are located on the β-strands of the LRR or CP modules: LRRV1 (Trp-62), LRRV2 (Trp-84, Asn-86, and Ser-87), LRRV3 (Gly-108 and Cys-110), LRRVe (Gly-132 and Asp-134), and CP (Trp-156). The antigen binding concave face of VLRB.aGPA.23, like that of other VLRs (12), is composed of three parallel ridges (designated R1, R2, and R3) associated with the beginning, middle, and end, respectively, of the β-strand of the LRR or CP modules (Fig. 5). Each β-strand contains a 6-residue motif, 1X(L/I)XLXX6, in which leucine or isoleucine at positions 2 and 4 form the hydrophobic core of the VLR, and the remaining four residues compose the three ridges: position 1 (R1), position 3 (R2), and positions 5 and 6 (R3). Sequence variability in VLRs is concentrated in R1–3 and in the LRRCT insert. In the VLRB.aGPA.23-TFα complex, only R2 and R3, in addition to the LRRCT insert, contact the disaccharide (Fig. 5A), whereas in VLR-protein complexes, contacts are typically distributed across all three binding site ridges due to the larger size of the ligand (Fig. 5B).
FIGURE 5.
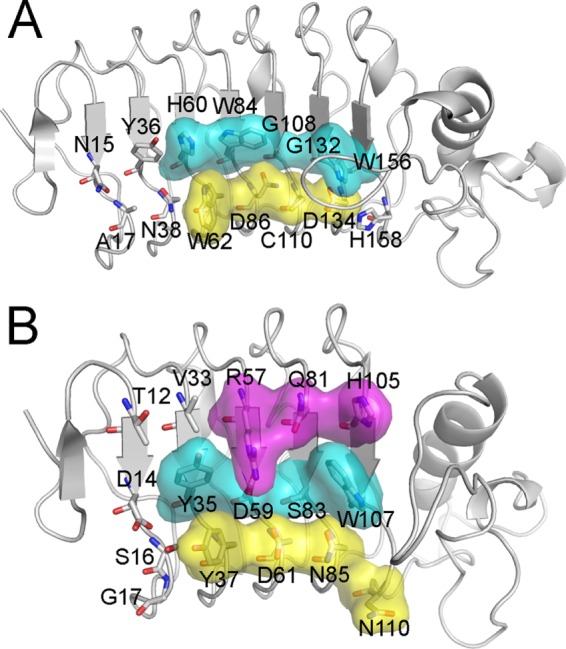
Molecular architecture of the glycan-binding site of VLRB.aGPA.23. A, shown is a surface representation of TFα-contacting residues in the R2 (cyan) and R3 (yellow) ridges of VLRB.aGPA.23. The disaccharide does not interact with R1. B, shown is a surface representation of hen egg white lysozyme-contacting residues in R1 (magenta), R2, and R3 ridges of VLRB.2D (Protein Data Bank accession code 3G3A) (17).
The VLRB.aGPA.23-BG-H Interface
The VLRB.aGPA.23-Fucα1–2Galβ complex buries a total solvent-accessible surface area of 141 Å2, to which the LRR (LRRV1–3 and LRRVe), CP, and LRRCT modules contribute 58, 10, and 31%, respectively. The specificity of the interaction is mediated mainly by three hydrogen bonds (Fig. 4D). Two of these hydrogen bonds, between Asp-134 Oδ1 and galactose O4′ and between Asp-134 Oδ2 and galactose O3′, are found in the VLRB.aGPA.23-TFα complex (Fig. 4B). The third hydrogen bond, between Asn-86 Nδ2 and galactose O1′, is unique to the VLRB.aGPA.23-BG-H complex. Six other residues (Trp-84, Asn-86, Cys-110, Gly-132, Trp-156, and Trp-187), which also contact TFα, stabilize the interaction with BG-H through 29 van der Waals contacts with the galactose (Fig. 4D).
Comparison with the VLRB.RBC36-BG-H2 Complex
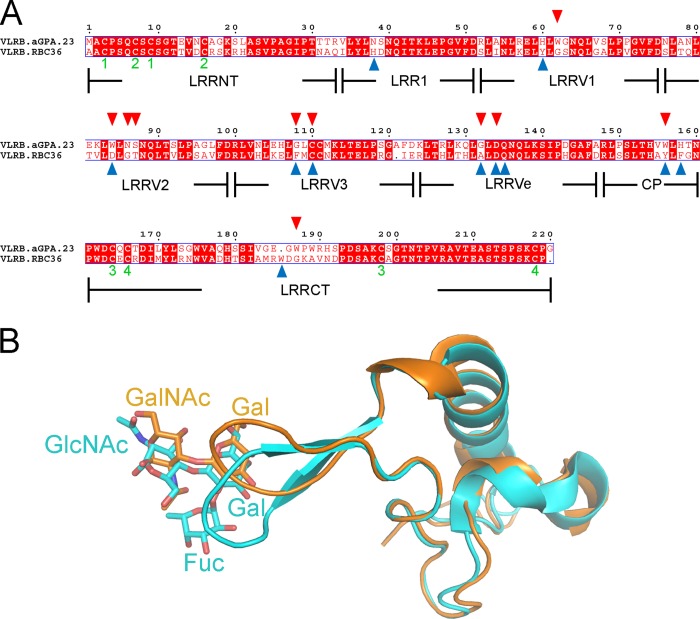
A comparison of VLRB.aGPA.23 with VLRB.RBC36, which recognizes blood group H type 2 trisaccharide (BG-H2; Fucα1–2Galβ1–4GlcNAcα) (16), showed high structural similarity (66% sequence identity; root mean squared difference = 0.95 for 194 α-carbon atoms) (Fig. 6A). This similarity is remarkable given the vast size and diversity of VLR repertoires in terms of both length and sequence (15) and the fact that VLRB.aGPA.23 and VLRB.RBC36 were isolated from different lampreys by different laboratories using different techniques (11, 16). However, VLRB.aGPA.23 differs from VLRB.RBC36 at 8 of 10 TFα-contacting positions (Fig. 6A). Conversely, VLRB.RBC36 differs from VLRB.aGPA.23 at 8 of 11 BG-H2-contacting positions. It is, therefore, not surprising that VLRB.aGPA.23 exhibited no reactivity toward BG-H2 in glycan array profiling (11). Indeed, the only conserved interactions in the two complexes are two hydrogen bonds between Asp-134, present in both VLRs, and the common galactose moiety of TFα and BG-H2. These interactions impose a similar location and orientation on this galactose in the VLRB.aGPA.23-TFα and VLRB.RBC36-BG-H2 structures (Fig. 6B).
FIGURE 6.
Comparison with the VLRB.RBC36-BG-H2 complex. A, structure-based sequence alignment of VLRB.aGPA.23 and VLRB.RBC36 (3E6J) (16) is shown. Boundaries of the LRRNT, LRR1, LRRV1–3, LRRVe, CP, and LRRCT modules are indicated. White characters on a red background show strictly conserved residues. Residues that are well conserved are drawn in red and framed in blue. The remaining residues are black. Residues that contact TFα in the VLRB.aGPA.23-TFα complex are marked with red triangles. Residues that contact BG-H2 in the VLRB.RBC36-BG-H2 complex are marked with blue triangles. The paired green numbers (1–4) indicate the bonded cysteine residues in the VLR structures. Sequence alignments were performed with the programs MultAlin and ESPript. B, the structure of the LRRCT inserts of VLRB.aGPA.23 (orange) and VLRB.RBC36 (cyan) shows interactions with TFα and BG-H2, respectively.
The main structural difference between VLRB.RBC36 and VLRB.aGPA.23 resides in their LRRCT inserts, which differ in both sequence and secondary structure. In VLRB.RBC36, the 10-residue insert forms a β-hairpin, whereas in VLRB.aGPA.23 the 9-residue insert is a loop whose overall shape differs markedly from that of the VLRB.RBC36 insert (Fig. 6B). In VLRB.RBC36, Trp-204 is located at the end of the first β-strand of the LRRCT insert, before the β-hairpin turn, and interacts only with the galactose of BG-H2. The corresponding residue of VLRB.aGPA.23, Trp-187, is located at the tip of the insert loop and interacts with both the galactose and N-acetylgalactosamine of TFα, thereby covering much more of the ligand (Fig. 4A).
DISCUSSION
Carbohydrate recognition by proteins is a topic of considerable interest with practical implications for basic research and clinical applications once the specifics of the molecular recognition process are understood. For carbohydrate binding proteins to achieve high selectivity, they must meet the unique challenge of discriminating among a vast array of sugar structures arising from the stereochemistry of hydroxyl groups in monosaccharides and different sugar linkage possibilities. A substantial amount of thermodynamic and structural information is available on glycan binding by plant and animal lectins (24, 26) and by anti-carbohydrate antibodies (27–31). These studies have shown that lectins and antibodies recognize sugars through the stacking of aromatic residues against the sugar ring and hydrogen bonding of sugar hydroxyls to polar amino acid side chains.
VLRs, such as VLRB.aGPA.23, represent a new type of binding proteins with exceptional selectivity for carbohydrates, as demonstrated previously by glycan array profiling (11). Moreover, the affinity of VLRB.aGPA.23 for TFα measured here by ITC (Kb = 2.21 × 104 m−1) was at the upper end of the spectrum for plant or animal lectins binding to disaccharide ligands (24). For example, the anti-HIV algal lectin griffithsin binds dimannoses with Kb values up to 2 × 104 m−1 (32). More typically, lectins display ∼10-fold lower affinity for disaccharides (24). Affinities in excess of 104 m−1 generally require longer oligosaccharides, which can interact with a more extended surface on the lectin (24, 26).
For anti-carbohydrate monoclonal antibodies, affinities in the range of 104-106 m−1 have been reported for tri-, tetra-, and pentasaccharides derived from Salmonella and Shigella lipopolysaccharide antigens (27–30). It should be noted, however, that these antibodies were obtained from mice immunized with the corresponding bacteria, whereas VLRB.aGPA.23 was isolated from lampreys that were not challenged with TFα (11). In addition, it should be possible to increase the affinity of glycan-specific VLRs such as VLRB.aGPA.23 by in vitro directed evolution, as in the case of VLRs reactive with the protein lysozyme, for which binding improvements of up to 1300-fold were achieved (33).
In common with other protein-carbohydrate interactions (24, 26), the binding of VLRB.aGPA.23 to all four glycans tested was characterized by a favorable enthalpy term, presumably due to the formation of hydrogen bonds and van der Waals contacts with the ligands (Table 2). In the case of TFα and TFα-Ser, the entropic contribution to the binding energy is slightly favorable (TFα-Ser) or slightly unfavorable (TFα), whereas for TFα-Thr and BG-H the entropy term makes a similar (BG-H) or greater (TFα-Thr) contribution to driving binding as the enthalpy term. This is somewhat atypical for sugar binding to proteins, where entropy is usually unfavorable due to loss of ligand conformational flexibility, and binding is enthalpically driven (24, 26). In this regard, structural studies of small glycopeptides have shown that GalNAcα attached to serine and threonine prefer very different rotomer populations, both in solution (34) and in silico (35), which could affect binding thermodynamics.
The absence of electron density for fucose in the VLRB.aGPA.23-Fucα1–2Galβ structure is surprising, as fucose contributes significantly to binding. Although we do not have a satisfactory explanation for this result, the lack of density does not necessarily mean there is no interaction at all between fucose and VLR.aGPA.23 but only that the interaction may not be sufficiently stable to be visualized in the crystal. It is also conceivable that the conformational entropy of the Fucα1–2Galβ ligand increases upon binding VLR.aGPA.23, thereby contributing favorably to the binding affinity. Although this may appear counterintuitive, NMR relaxation studies of galectin-3 have demonstrated that ligand binding increases the conformational entropy of the protein, which contributes favorably to the free energy of binding (36).
The architecture of the glycan-binding site of VLRB.aGPA.23 differs markedly from those of lectins or antibodies, which typically consist of long grooves on the protein surface for accommodating oligosaccharide chains (24, 26, 28–30, 32). In the VLRB.aGPA.23-TFα complex, the disaccharide is sandwiched between the LRRCT insert loop and the concave surface of the VLR solenoid formed by the short β-strands of the LRR and CP modules. This parallel β-sheet does not lend itself to the construction of carbohydrate binding grooves of the type seen in lectins and antibodies (37), and none is present in VLRB.aGPA.23 or VLRB.RBC36 (16). Based on the VLRB.aGPA.23-TFα structure, longer oligosaccharides with extensions at the reducing end of the TFα disaccharide (GalNAcα) would project perpendicularly across the β-strands, contacting additional LRR modules beyond LRRV1 (Fig. 6A). Although VLRB.aGPA.23 contains only two additional N-terminal modules (LRRNT and LRR1), which would limit the potential contacting surface to perhaps a tetrasaccharide, other VLRs contain as many as six additional modules (12, 15). However, the curvature of the binding surface would increase progressively (37), requiring the oligosaccharide to bend to conform to the concavity of the rigid VLR solenoid to maintain contacts with the protein. By contrast, the carbohydrate binding surfaces of lectins and antibodies are generally flat or convex (24, 26, 28–30, 32). Despite these striking differences in binding site architecture, our structural and thermodynamic study of VLRB.aGPA.23 has revealed how VLRs utilize the LRR scaffold to recognize glycans with an affinity and selectivity rivaling that of lectins and antibodies, making VLRs a highly promising class of natural glycan-binding proteins for basic research and clinical applications.
Acknowledgments
We thank Yili Li and Lu Deng for assistance with x-ray data collection and Mingming Gao for advice on data scaling and refinement.
This work was supported, in whole or in part, by National Institutes of Health (NIH) Grants AI036900 (to R. A. M.) and AI083892 (to Z. P.) and by the intramural research program of the NIH, NCI (to J. J. B. and J. C. G.). This work was also supported by National Science Foundation Grant MCB-0614672 (to Z. P.).
The atomic coordinates and structure factors (codes 4K79 and 4K5U) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- TFα
- Thomsen-Friedenreich antigen
- BG-H
- blood group H disaccharide
- BG-H3
- blood group H type 3 trisaccharide
- VLR
- variable lymphocyte receptor
- LRR
- leucine-rich repeat
- TFα-PAA
- TFα-polyacrylamide-biotin
- ITC
- isothermal titration calorimetry
- SPR
- surface plasmon resonance
- LRRNT
- N-terminal LRR capping module
- LRRV
- LRR variable module
- CP
- connecting peptide module
- LRRCT
- C-terminal LRR capping module
- GPA
- human glycophorin A
- aGPA
- asialo human glycophorin A.
REFERENCES
- 1. Varki A., Kannagi R., Toole B. P. (2009) in Essentials of Glycobiology (Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E., eds.) Chapter 44, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York: [PubMed] [Google Scholar]
- 2. Almogren A., Abdullah J., Ghapure K., Ferguson K., Glinsky V. V., Rittenhouse-Olson K. (2012) Anti-Thomsen-Friedenreich-Ag (anti-TF-Ag) potential for cancer therapy. Front. Biosci. Schol. Ed. 4, 840–863 [DOI] [PubMed] [Google Scholar]
- 3. Powlesland A. S., Hitchen P. G., Parry S., Graham S. A., Barrio M. M., Elola M. T., Mordoh J., Dell A., Drickamer K., Taylor M. E. (2009) Targeted glycoproteomic identification of cancer cell glycosylation. Glycobiology 19, 899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li Q., Anver M. R., Li Z., Butcher D. O., Gildersleeve J. C. (2010) GalNAcα1–3Gal, a new prognostic marker for cervical cancer. Int. J. Cancer 126, 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Manimala J. C., Roach T. A., Li Z., Gildersleeve J. C. (2006) High-throughput carbohydrate microarray analysis of 24 lectins. Angew. Chem. Int. Ed. Engl. 45, 3607–3610 [DOI] [PubMed] [Google Scholar]
- 6. Manimala J. C., Roach T. A., Li Z., Gildersleeve J. C. (2007) High-throughput carbohydrate microarray profiling of 27 antibodies demonstrates widespread specificity problems. Glycobiology 17, 17C[en]23C [DOI] [PubMed] [Google Scholar]
- 7. Sakai K., Yuasa N., Tsukamoto K., Takasaki-Matsumoto A., Yajima Y., Sato R., Kawakami H., Mizuno M., Takayanagi A., Shimizu N., Nakata M., Fujita-Yamaguchi Y. (2010) Isolation and characterization of antibodies against three consecutive Tn-antigen clusters from a phage display library displaying human single-chain variable fragments. J. Biochem. 147, 809–817 [DOI] [PubMed] [Google Scholar]
- 8. Hu D., Tateno H., Kuno A., Yabe R., Hirabayashi J. (2012) Directed evolution of lectins with a sugar-binding specificity for 6-sulfo-galactose. J. Biol. Chem. 287, 20313–20320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boltz K. W., Gonzalez-Moa M. J., Stafford P., Johnston S. A., Svarovsky S. A. (2009) Peptide microassays for carbohydrate recognition. Analyst 134, 650–652 [DOI] [PubMed] [Google Scholar]
- 10. Sun W., Du L., Li M. (2010) Aptamer-based carbohydrate recognition. Curr. Pharm. Des. 16, 2269–2278 [DOI] [PubMed] [Google Scholar]
- 11. Hong X., Ma M.Z., Gildersleeve J.C., Chowdhury S., Barchi J. J., Jr., Mariuzza R. A., Murphy M. B., Mao L., Pancer Z. (2013) Sugar-binding proteins from fish. Selection of high-affinity “lambodies” that recognize biomedically relevant glycans. ACS Chem. Biol. 8, 152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deng L., Luo M., Velikovsky A., Mariuzza R. A. (2013) Structural insights into the evolution of the adaptive immune system. Annu. Rev. Biophys. 42, 191–215 [DOI] [PubMed] [Google Scholar]
- 13. Rogozin I. B., Iyer L. M., Liang L., Glazko G. V., Liston V. G., Pavlov Y. I., Aravind L., Pancer Z. (2007) Evolution and diversification of lamprey antigen receptors. Evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat. Immunol. 8, 647–656 [DOI] [PubMed] [Google Scholar]
- 14. Boehm T. (2011) Design principles of adaptive immune systems. Nat. Rev. Immunol. 11, 307–317 [DOI] [PubMed] [Google Scholar]
- 15. Boehm T., McCurley N., Sutoh Y., Schorpp M., Kasahara M., Cooper M. D. (2012) VLR-based adaptive immunity. Annu. Rev. Immunol. 30, 203–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han B. W., Herrin B. R., Cooper M. D., Wilson I. A. (2008) Antigen recognition by variable lymphocyte receptors. Science 321, 1834–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Velikovsky C. A., Deng L., Tasumi S., Iyer L. M., Kerzic M. C., Aravind L., Pancer Z., Mariuzza R. A. (2009) Structure of a lamprey variable lymphocyte receptor in complex with a protein antigen. Nat. Struct. Mol. Biol. 16, 725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deng L., Velikovsky C. A., Xu G., Iyer L. M., Tasumi S., Kerzic M. C., Flajnik M. F., Aravind L., Pancer Z., Mariuzza R. A. (2010) A structural basis for antigen recognition by the T cell-like lymphocytes of sea lamprey. Proc. Natl. Acad. Sci. U.S.A. 107, 13408–13413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kirchdoerfer R. N., Herrin B. R., Han B. W., Turnbough C. L., Jr., Cooper M. D., Wilson I. A. (2012) Variable lymphocyte receptor recognition of the immunodominant glycoprotein of Bacillus anthracis spores. Structure 20, 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Storoni L. C., McCoy A. J., Read R. J. (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 21. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 22. Emsley P., Lohkamp B., Scott W.G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pisano A., Redmond J. W., Williams K. L., Gooley A. A. (1993) Glycosylation sites identified by solid-phase Edman degradation. O-Linked glycosylation motifs on human glycophorin A. Glycobiology 3, 429–435 [DOI] [PubMed] [Google Scholar]
- 24. Dam T. K., Brewer C. F. (2002) Thermodynamic study of lectin-carbohydrate interactions by isothermal titration calorimetry. Chem. Rev. 102, 387–429 [DOI] [PubMed] [Google Scholar]
- 25. Kim H. M., Oh S. C., Lim K. J., Kasamatsu J., Heo J. Y., Park B. S., Lee H., Yoo O. J., Kasahara M., Lee J. O. (2007) Structural diversity of the hagfish variable lymphocyte receptors. J. Biol. Chem. 282, 6726–6732 [DOI] [PubMed] [Google Scholar]
- 26. Imberty A., Mitchell E. P., Wimmerová M. (2005) Structural basis of high-affinity glycan recognition by bacterial and fungal lectins. Curr. Opin. Struct. Biol. 15, 525–534 [DOI] [PubMed] [Google Scholar]
- 27. Brummell D. A., Sharma V. P., Anand N. N., Bilous D., Dubuc G., Michniewicz J., MacKenzie C. R., Sadowska J., Sigurskjold B. W., Sinnott B. (1993) Probing the combining site of an anti-carbohydrate antibody by saturation-mutagenesis. Role of the heavy-chain CDR3 residues. Biochemistry 32, 1180–1187 [DOI] [PubMed] [Google Scholar]
- 28. Bundle D. R., Eichler E., Gidney M. A., Meldal M., Ragauskas A., Sigurskjold B. W., Sinnott B., Watson D. C., Yaguchi M., Young N. M. (1994) Molecular recognition of a Salmonella trisaccharide epitope by monoclonal antibody Se155-4. Biochemistry 33, 5172–5182 [DOI] [PubMed] [Google Scholar]
- 29. Sigurskjold B. W., Bundle D. R. (1992) Thermodynamics of oligosaccharide binding to a monoclonal antibody specific for a Salmonella O-antigen point to hydrophobic interactions in the binding site. J. Biol. Chem. 267, 8371–8376 [PubMed] [Google Scholar]
- 30. Vyas N. K., Vyas M. N., Chervenak M. C., Johnson M. A., Pinto B. M., Bundle D. R., Quiocho F. A. (2002) Molecular recognition of oligosaccharide epitopes by a monoclonal Fab specific for Shigella flexneri Y lipopolysaccharide. X-ray structures and thermodynamics. Biochemistry 41, 13575–13586 [DOI] [PubMed] [Google Scholar]
- 31. Julien J. P., Lee P. S., Wilson I. A. (2012) Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA. Immunol. Rev. 250, 180–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moulaei T., Shenoy S. R., Giomarelli B., Thomas C., McMahon J. B., Dauter Z., O'Keefe B. R., Wlodawer A. (2010) Monomerization of viral entry inhibitor griffithsin elucidates the relationship between multivalent binding to carbohydrates and anti-HIV activity. Structure 18, 1104–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tasumi S., Velikovsky C. A., Xu G., Gai S. A., Wittrup K. D., Flajnik M. F., Mariuzza R. A., Pancer Z. (2009) High-affinity lamprey VLRA and VLRB monoclonal antibodies. Proc. Natl. Acad. Sci. U.S.A. 106, 12891–12896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Corzana F., Busto J. H., Jiménez-Osés G., García de Luis M., Asensio J. L., Jiménez-Barbero J., Peregrina J. M., Avenoza A. (2007) Serine versus threonine glycosylation: the methyl group causes a drastic alteration on the carbohydrate orientation and on the surrounding water shell. J. Am. Chem. Soc. 129, 9458–9467 [DOI] [PubMed] [Google Scholar]
- 35. Mallajosyula S. S., MacKerell A. D., Jr. (2011) Influence of solvent and intramolecular hydrogen bonding on the conformational properties of O-linked glycopeptides. J. Phys. Chem. B 115, 11215–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Akke M. (2012) Conformational dynamics and thermodynamics of protein-ligand binding studied by NMR relaxation. Biochem. Soc. Trans. 40, 419–423 [DOI] [PubMed] [Google Scholar]
- 37. Bella J., Hindle K. L., McEwan P. A., Lovell S. C. (2008) The leucine-rich repeat structure. Cell. Mol. Life Sci. 65, 2307–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]