Abstract
Background:
It is believed that the Helicobacter pylori (H. pylori) vacA gene, as a major virulence determinant (One of the major virulence determinant, not major), may be a risk factor for the development of gastroduodenal diseases. The frequency of vacA genotypes varies in different human populations. This study evaluated the prevalence of vacA alleles/genotypes among dyspeptic patients in Isfahan.
Materials and Methods:
One-hundred H. pylori-positive adult patients were examined in this study. After culture of gastric biopsies and DNA extraction from individual H. pylori isolates, the (all H. pylori strains harbor vacA alleles, please replace “presence” with “genotypes”) of the vacA s and m alleles were determined using polymerase chain reaction (PCR).
Results:
There were four vacA mosaicisms, including 28 for s1a/m1 (28%), 23 for s1b/m1 (23%), 26 for s1a/m2 (26%) and 23 for s1b/m2 (23%). The s2 allele was not found. The predominant vacA genotype in patients with non-ulcer dyspepsia and duodenal ulcer was s1a/m2, whereas in patients with adenocarcinoma was s1a/m1.
Conclusion:
The results showed there was no significant correlation between different genotypes of the vacA and the clinical outcomes and appears to vacA genotypes were not useful determinants for gastrointestinal diseases in our area.
Keywords: Adenocarsinoma, gastroduodenal diseases, Helicobacter pylori, Iran, Peptic ulcere, vacA gene
INTRODUCTION
Helicobacter pylori is a gram-negative, curved and microaerophilic bacterium. It infects the stomach mucosa of more than 50% of the world's human populations.[1,2,3] It may cause variety of clinical symptoms such as chronic gastric inflammation, peptic ulcer disease (PUD), MALT lymphoma and gastric adenocarcinoma.[4,5] The majority of H. pylori infected persons are asymptomatic, and only a fraction (10-20%) of carriers’ manifest clinical disease.[6,7] The prevalence of H. pylori infection is variable in different countries.[3] In Iran as a developing country, the rate of infection has been reported to be 60-90%.[8,9,10]
Some virulence-related genes such as vacA and cagA in the clinical isolates have been previously studied, however, the role of H. pylori isolates genotypes in different symptoms remains controversial.[11] The vacuolating-cytotoxin (VacA) is one of the major virulence factors, which encoded by the vacA gene.[12,13] This 87 kDa protein induces both vacuole formation and apoptosis in gastric epitheliums.[14,15] Although the vacA gene is present in all H. pylori strains, however, approximately 30-66% of clinical isolates produce the vacA cytotoxin.[4,16] The vacA gene contains two variable parts, the s region, and the m region, which encode the signal, and middle peptides, respectively.[17] The s region consisting of s1 (subtypes s1a, s1b and s1c) and s2 alleles, and m region also includes m1 (subtypes m1a, m1b and m1c) and m2 alleles, respectively.[14,18] The vacA s1 and m1 alleles have greater cytotoxic activity than s2 and m2 alleles.[19] A novel polymorphism in the intermediate (i) region (contain i1 and i2 alleles) between the s and m areas. They found that the i1 allele in Iran is significantly associated with gastric cancer.[20] The existence of different genotypes of vacA (s1/m1, s1/m2, s2/m1 and s2/m2) is due to various possible combinations of s and m alleles, that are virulence-associated.[18,21] Among these genotypes, the frequency of vacA s2/m1 is rare.[22] H. pylori s1/m1 genotypes offer the high levels of cytotoxic activity, and are associated to inflammation and ulcer[18,23] whereas, s1/m2 strains produce moderate amounts of toxin, and s2/m2 strains produce very little or no toxin.[14,15,18]
The correlation between the vacA genotypes and gastroduodenal manifestations are variable in terms of geographic areas. On the other hand, the distinct dominant vacA genotypes differ among clinical isolates from the various geographic regions,[13] because of the lack any report in the case of H. pylori vacA genotypes in Isfahan (center of Iran); therefore, to understand the clinical relevance of H. pylori genotyping in predicting infection outcome, we carried out this study in order to survey prevalence of vacA genotypes, and their relevance with different gastroduodenal diseases.
MATERIALS AND METHODS
Patients and clinical samples
H. pylori strains, isolated from 100 adult patients of both sexes with gastroduodenal manifestations at the Department of Gastroenterology of Hospital Al-Zahra in Isfahan, Iran, were included. All patients underwent upper gastrointestinal endoscopy for both visual examination and biopsy collection. Exclusion criteria were anti-H. Pylori therapy intake in the last 4 weeks. Based on endoscopic examination and clinical signs, patients were classified into the following groups: non-ulcer dyspepsia (n = 40), duodenal ulcer (n = 40), and adenocarcinoma (n = 20). From each patient, three antrum biopsy samples were taken for rapid urease test (RUT), histology and culture.
Culture and identification of H. pylori
For bacterial culture, biopsy specimens were processed according to the method of Lang and Garcia.[24] Plates were incubated in microaerobic conditions (5% O2, 10% CO2 and 85% N2) in anaerobic jars (MART, ANOXOMAT, Lichtenvoorde, the Netherlands) at 37°C for 3-5 days. Colonies were identified as H. pylori based on biochemical tests. All the isolates were stored at −80°C in aliquots of BHI broth (Merck, Germany) supplemented with 20% (v/v) glycerol.
DNA extraction
For each isolate, extraction of chromosomal DNA was performed using phenol-chloroform method.[25] Briefly, H. pylori cells were washed twice in 1000 μl TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0), then the pelletes were resuspended in 500 μl TE buffer and 100 μl lysozyme (10 mg/ml), and then lysed using SDS – proteinase K at 56°C for 1 h. The cell extract was then treated with phenol/chloroform isoamylalcohol, ethanol precipitation for DNA extraction.
Polymerase chain reaction
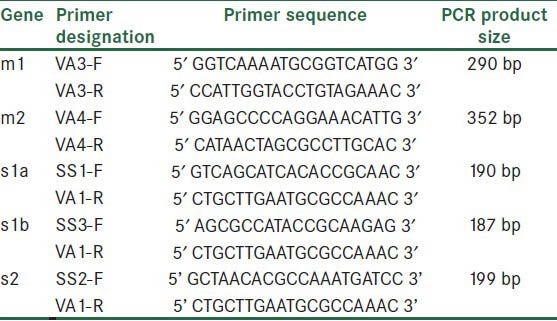
The primers was chosen for H. pylori vacA evaluation, are listed in Table 1. PCR was performed as described in previous references.[17]
Table 1.
Oligonucleotide primers used for H. pylori vacA alleles
Data analysis
The association between H. pylori genotypes and clinical relevance was analyzed by Pearson Chi-square or Fisher's exact test (when necessary). P value <0.05 was considered statistically significant.
RESULTS
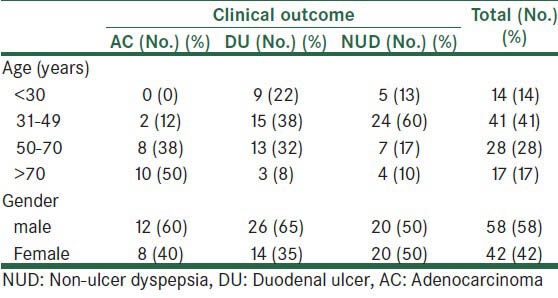
Our population consisted of 100 adult patients (55 males and 45 females) ranging in age from <30 to >70 years (mean 43 years). The distribution of patients according to age and sex is shown in Table 2.
Table 2.
Frequency of patients according to age and gender
H. pylori vacA alleles and genotypes
Correlation between vacA subtypes and clinical diseases is shown in Table 3.
Table 3.
Relationship between H. pylori vacA alleles/genotypes and clinical status
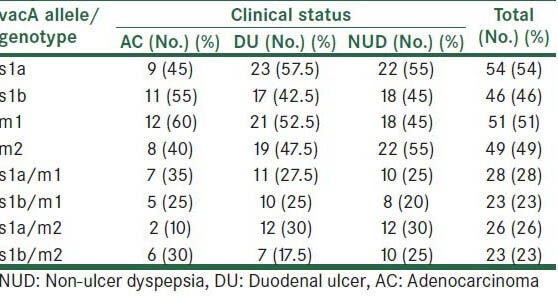
The most prevalent vacA allele in different samples was s1a (54%), m1 being present in 51%, followed by m2 and s1b with frequencies of 49 and 46%, respectively. Overall, 28% of isolates were grouped as vacA s1a/m1, 23% as s1b/m1, 26% as s1a/m2, and 23% as s1b/m2 genotypes. However, vacA s2 allele in none of patients was observed. The dominant vacA genotype in patients with NUD and DU was s1a/m2, whereas in patients with AC was s1a/m1. In our study, the prevalence of H. pylori vacA genotypes was not significantly associated with different clinical manifestations (P > 0.005).
DISCUSSION
H. pylori is strongly associated with gastritis and peptic ulcer.[12] The variable prevalence of infection caused by this bacterium in various countries is due to its variable clinical isolates.[16] Considering the necessity of H. pylori genotype analysis in different populations and various areas of the world, we conducted this study to evaluate the prevalence of vacA genotypes and its relationship to different clinical outcomes in of Isfahan. Our study shows there is no s2 allele in H. pylori genome in the area of Isfahan, whereas m1 and m2 alleles were found in the mid region and s1a and s1b alleles were present in the signal sequence of H. pylori genome. A study conducted by Van Doorn et al.[26] in 1998 reported the frequency of s1a/m1, s1a/m2, s1b/m1, s1b/m2 and s2/m2 to be 36, 23, 2, 5 and 20%, respectively. In our study these values were found to be 28, 26, 23 and 23%, respectively, which indicates the various genotype frequencies according to different geographical regions. A study performed by Strobel et al.[27] in Germany also indicates that 96% of the strains isolated in this country contain s1 gene while s2 and m1 genes account for 4% and 51% of the strains, respectively. In a study carried out in Turkey it was shown that 87.7% of the strains isolated from gastrointestinal patients (NUD, DU, GC) harboured s1 gene, 12.3% contained s2 gene and 43% contained m1 gene[28] indicating the similarity of the studies carried out in Turkey and Germany. A meta-analysis of 1646 patients in the Middle East reported the frequency of the genotypes s1/m1 and s1/m2 in the northern and southern countries of this region to be 34%, 14.8%, 44.8% and 30.1%, respectively(29). These data are significantly different from the results of our study. In a study performed in Thailand the frequency of the genotypes s1 and s2 was shown to be 100% and 0%. The allelic variant vacA s1/m1 was more prevalent than s1/m2. Different genes either single or in combination had no significant relationship with clinical diseases.[11] This is in accordance with the study carried out in Isfahan with the exception that there was no significant difference in the prevalence of the above mentioned genotypes in our study. In four studies carried out in Iran the prevalence of s1 allele as the most common allele of VacA cytotoxin subtypes was found to be 71%, 69%, 69% and 70.5%, respectively and is in accordance with our study.[30,31,32,33] But contrary to our study, s2 allele was present in all these studies. The dominant genotype in Jafari, Dabiri and Molaei studies was vacA s1/m2, whereas in Isfahan this prevalence was 51% for vacA s1/m1 and 49% for s1/m2. In a study carried out in Pakistan[15] the most prevalent genotype was found to be s1b/m2 followed by s1a/m1, whereas in our study s1b/m2 and s1b/m1 had the lowest frequencies. In the mentioned study the frequency of s1a/m1 and cagA was lower than those reported by other neighbouring countries. In a study performed in Costa Rica the frequency of the genotypes s1b and m1 was reported 75.2% and 74.2%, respectively. In this study a significant association was observed between m1 genotype and gastritis.[34] The frequency of genotypes s1b and m1 was 45% each in our work, which is a lower percentage compared to the above study, although no significant correlation was observed between genotype m1 and gastritis. In a study performed in Thailand 58 gastritis patients (Ga), 28 gastric ulcer (GU), 45 duodenal ulcer (DU) and 4 gastric cancer (GC) patients were analysed, which the frequency of s1a and s1a/m1 genotypes were reported 81% and 21%, respectively.[35] The frequency of s1a allele is lower in our study while the frequency of s1a/m1 genotype is similar (28%) to the above study. In a study conducted in Pakistan,[16] the frequency of s1a/m1 was found to be 69% which was much higher than our findings. In the mentioned study in Turkey has shown the frequency of s1/m1 and s1/m2 genotypes was 41% and 48%, respectively. They also reported the frequency of s1a allele in NUD, DU and GC cases as 67, 97 and 88%, respectively. In a survey carried out in Bahrain, all patients with ulcer and 82.4% of patients with NUD contained the s1 strains.[36] In the present study all patients with DU and NUD had strains containing the s1 genotype. In a study conducted by Boyanova et al.[6] patients with ulcer had a higher number of virulent strains compared to other patients. In our study patients with AC and DU had a higher number of strains containing the s1/m1 genotype compared to subjects with NUD (64.7% compared to 35.3%). In addition, in the mentioned study similar to ours, s2 allele was not found in patients with ulcer but was only found in 13% of other patients. No vacA s2 genotype was reported in the study carried out by Sheu et al.[13] In a study conducted by Ohno et al.[37] among several Asian and European countries the vacA genotypes was shown to be different in these countries and the s2/m2 genotype was not found in Asian patients similar to our findings, whereas this genotype was found in western patients. In a study by Chomvarin et al.[11] no significant relationship was detected between vacA genotypes and other virulence genes such as iceA, cagA, babA2 and the complication produced in the patient, which confirms the findings in this report. These results were also reported in the previous studies carried out in Iran, Taiwan and Mexico. In the studies conducted by Yakoob, et al.,[16] Molaei, et al.,[33] Chiarini, et al.,[38] Bindayna and Mahmeed[36] significant association was found between vacA genotypes and clinical outcomes. Hussein et al.[19] also found such relationship between vacA genotypes and PUD in strains isolated from Iraqi patients, however, this relationship was not significant in the Iranian strains.
CONCLUSION
Considering the mentioned studies, it is safe to say that the frequency of the s2 genotype is low in most studies which are confirmed by the results in our study. Other genotypes such as s1a, s1b, m1 and m2 are significantly different in different regions which indicate the various distributions of these genotypes. Overall, it is understood that there is no correlation between H. pylori vacA genotype and the resulting disease. Therefore each of the vacA genotypes causing the complication may be found in different patients.
ACKNOWLEDGMENTS
This research was supported by grant No. 82078 of Isfahan University of Medical Sciences.
Footnotes
Source of Support: This research was supported by grant No. 82078 of Isfahan University of Medical Sciences
Conflict of Interest: None declared.
REFERENCES
- 1.Blaser MJ. Ecology of Helicobacter pylori in the human stomach. J Clin Invest. 1997;100:759–62. doi: 10.1172/JCI119588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaser MJ. Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. J Infect Dis. 1990;161:626–33. doi: 10.1093/infdis/161.4.626. [DOI] [PubMed] [Google Scholar]
- 3.Dunn BE, Cohen H, Blaser MJ. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–41. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar S, Kumar A, Dixit VK. Direct detection and analysis of vacA genotypes and cagA gene of Helicobacter pylori from gastric biopsies by a novel multiplex polymerase chain reaction assay. Diagn Microbiol Infect Dis. 2008;62:366–73. doi: 10.1016/j.diagmicrobio.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 5.Forsyth MH, Atherton JC, Blaser MJ, Cover TL. Heterogeneity in levels of vacuolating cytotoxin gene (vacA) transcription among Helicobacter pylori strains. Infect Immun. 1998;66:3088–94. doi: 10.1128/iai.66.7.3088-3094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyanova L, Markovska R, Yordanov D, Marina M, Ivanova K, Panayotov S, et al. High prevalence of virulent Helicobacter pylori strains in symptomatic Bulgarian patients. Diagn Microbiol Infect Dis. 2009;64:374–80. doi: 10.1016/j.diagmicrobio.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Yamaota Y, Kato M, Asaka M. Geographic differences in gastric cancer incidence can be explained by differences between Helicobacter pylori strains. Intern Med. 2008;47:1077–83. doi: 10.2169/internalmedicine.47.0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massarrat S, Saberi-Firoozi M, Soleimani A, Himmelmann GW, Hitzges M, Keshavarz H. Peptic ulcer disease, irritable bowel syndrome and constipation in two populations in Iran. Eur J Gastroenterol Hepatol. 1995;7:427–33. [PubMed] [Google Scholar]
- 9.Siavoshi F, Malekzadeh R, Daneshmand M, Ashktorab H. Helicobacter pylori endemic and gastric disease. Dig Dis Sci. 2005;50:2075–80. doi: 10.1007/s10620-005-3010-1. [DOI] [PubMed] [Google Scholar]
- 10.Farshad S, Japoni A, Alborzi A, Hosseini M. Restriction fragment length polymorphism of virulence genes cagA, vacA and ureAB of Helicobacter pylori strains isolated from Iranian patients with gastric ulcer and nonulcer disease. Saudi Med J. 2007;28:529–34. [PubMed] [Google Scholar]
- 11.Chomvarin C, Namwat W, Chaicumpar K, Mairiang P, Sangchan A, Sripa B, et al. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA and babA2 genotypes in Thai dyspeptic patients. Int J Infect Dis. 2008;12:30–6. doi: 10.1016/j.ijid.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Paniagua GL, Monroy E, Rodríguez R, Arroniz S, Rodríguez C, Cortés JL, et al. Frequency of vacA, cagA and babA2 virulence markers in Helicobacter pylori strains isolated from Mexican patients with chronic gastritis. Ann Clin Microbiol Antimicrob. 2009;8:14. doi: 10.1186/1476-0711-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheu SM, Hung KH, Sheu BS, Yang HB, Wu JJ. Association of nonsynonymous substitutions in the intermediate region of the vacA gene of Helicobacter pylori with gastric diseases in Taiwan. J Clin Microbiol. 2009;47:249–51. doi: 10.1128/JCM.01651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugimoto M, Yamaoka Y. The association of vacA genotype and Helicobacter pylori-related disease in Latin American and African populations. Clin Microbiol Infect. 2009;15:835–42. doi: 10.1111/j.1469-0691.2009.02769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmad T, Sohail K, Rizwan M, Mukhtar M, Bilal R, Khanum A. Prevalence of Helicobacter pylori pathogenicity-associated cagA and vacA genotypes among Pakistani dyspeptic patients. FEMS Immunol Med Microbiol. 2009;55:34–8. doi: 10.1111/j.1574-695X.2008.00492.x. [DOI] [PubMed] [Google Scholar]
- 16.Yakoob J, Abid S, Abbas Z, Jafri W, Ahmad Z, Ahmed R, et al. Distribution of Helicobacter pylori virulence markers in patients with gastroduodenal diseases in Pakistan. BMC Gastroenterol. 2009;9:87. doi: 10.1186/1471-230X-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atherton JC, Cao P, Peek RM, Jr, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–7. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 18.Sicinschi LA, Correa P, Peek RM, Jr, Camargo MC, Delgado A, Piazuelo MB, et al. Helicobacter pylori genotyping and sequencing using paraffin-embedded biopsies from residents of colombian areas with contrasting gastric cancer risks. Helicobacter. 2008;13:135–45. doi: 10.1111/j.1523-5378.2008.00554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussein NR, Mohammadi M, Talebkhan Y, Doraghi M, Letley DP, Muhammad MK, et al. Differences in virulence markers between Helicobacter pylori strains from Iraq and those from Iran: Potential importance of regional differences in H. pylori-associated disease. J Clin Microbiol. 2008;46:1774–9. doi: 10.1128/JCM.01737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhead JL, Letley DP, Mohammadi M, Hussein N, Mohagheghi MA, Eshagh Hosseini M, et al. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology. 2007;133:926–36. doi: 10.1053/j.gastro.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 21.Maciorkowska E, Roszko I, Kowalczuk O, Kaczmarski M, Chyczewski L, Kemona A. The evaluation of vacA gene allele's frequency in Helicobacter pylori strains in children and adults in Podlaskie region. Folia Histochem Cytobiol. 2007;45:215–9. [PubMed] [Google Scholar]
- 22.Mohammadi M, Oghalaie A, Mohajerani N, Massarrat S, Nasiri M, Bennedsen M, et al. Prevalence of Helicobacter pylori vacuolating cytotoxin and its allelic mosaicism as a predictive marker for Iranian dyspeptic patients. Bull Soc Pathol Exot. 2003;96:3–5. [PubMed] [Google Scholar]
- 23.Sgouras DN, Panayotopoulou EG, Papadakos K, Martinez-Gonzalez B, Roumbani A, Panayiotou J, et al. This CagA and VacA polymorphisms do not correlate with severity of histopathological lesions in Helicobacter pylori-infected Greek children. J Clin Microbiol. 2009;47:2426–34. doi: 10.1128/JCM.00159-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lang L, García F. Comparison of E-test and disk diffusion assay to evaluate resistance of Helicobacter pylori isolates to amoxicillin, clarithromycin, metronidazole and tetracycline in Costa Rica. Int J Antimicrob Agents. 2004;24:572–7. doi: 10.1016/j.ijantimicag.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 25.Hua J, Ng HC, Yeoh KG, Ho B. Predominance of a single strain of Helicobacter pylori in gastric antrum. Helicobacter. 1999;4:28–32. doi: 10.1046/j.1523-5378.1999.09043.x. [DOI] [PubMed] [Google Scholar]
- 26.van Doorn LJ, Figueiredo C, Sanna R, Plaisier A, Schneeberger P, de Boer W, et al. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998;115:58–66. doi: 10.1016/s0016-5085(98)70365-8. [DOI] [PubMed] [Google Scholar]
- 27.Strobel S, Bereswill S, Balig P, Allgaier P, Sonntag HG, Kist M. Identification and analysis of a new vacA genotype variant of Helicobacter pylori in different patient groups in Germany. J Clin Microbiol. 1998;36:1285–9. doi: 10.1128/jcm.36.5.1285-1289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erzin Y, Koksal V, Altun S, Dobrucali A, Aslan M, Erdamar S, et al. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA, babA2 genotypes and correlation with clinical outcome in Turkish patients with dyspepsia. Helicobacter. 2006;11:574–80. doi: 10.1111/j.1523-5378.2006.00461.x. [DOI] [PubMed] [Google Scholar]
- 29.Sugimoto M, Zali MR, Yamaoka Y. The association of vacA genotypes and Helicobacter pylori-related gastroduodenal diseases in the Middle East. Eur J Clin Microbiol Infect Dis. 2009;28:1227–36. doi: 10.1007/s10096-009-0772-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salehi Z, Abadi AS, Ismail PB, Kqueen CY, Jelodar MH, Kamalidehghan B. Evaluation of Helicobacter pylori vacA genotypes in Iranian patients with peptic ulcer disease. Dig Dis Sci. 2009;54:2399–403. doi: 10.1007/s10620-008-0633-z. [DOI] [PubMed] [Google Scholar]
- 31.Jafari F, Shokrzadeh L, Dabiri H, Baghaei K, Yamaoka Y, Zojaji H, et al. vacA genotypes of Helicobacter pylori in relation to cagA status and clinical outcomes in Iranian populations. Jpn J Infect Dis. 2008;61:290–3. [PMC free article] [PubMed] [Google Scholar]
- 32.Dabiri H, Maleknejad P, Yamaoka Y, Feizabadi MM, Jafari F, Rezadehbashi M, et al. Distribution of Helicobacter pylori cagA, cagE, oipA and vacA in different major ethnic groups in Tehran, Iran. J Gastroenterol Hepatol. 2009;24:1380–6. doi: 10.1111/j.1440-1746.2009.05876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molaei M, Foroughi F, Mashayekhi R, Haghazali M, Zojaji H, Jafari F, et al. CagA status and VacA subtypes of Helicobacter pylori in relation to histopathologic findings in Iranian population. Indian J Pathol Microbiol. 2010;53:24–7. doi: 10.4103/0377-4929.59178. [DOI] [PubMed] [Google Scholar]
- 34.Conzzz SA, Takeuchi H, Valerín AL, Con-Wong R, Con-Chin GR, Con-Chin VG, et al. Diversity of Helicobacter pylori cagA and vacA genes in Costa Rica: Its relationship with atrophic gastritis and gastric cancer. Helicobacter. 2007;12:547–52. doi: 10.1111/j.1523-5378.2007.00534.x. [DOI] [PubMed] [Google Scholar]
- 35.Linpisarn S, Suwan W, Lertprasertsuk N, Koosirirat C, Steger HF, Prommuangyong K, et al. Helicobacter pylori cagA, vacA and iceA genotypes in northern Thai patients with gastric disease. Southeast Asian J Trop Med Public Health. 2007;38:356–62. [PubMed] [Google Scholar]
- 36.Bindayna KM, Al Mahmeed A. VacA genotypes in Helicobacter pylori strains isolated from patients with and without duodenal ulcer in Bahrain. Indian J Gastroenterol. 2009;28:175–9. doi: 10.1007/s12664-009-0069-1. [DOI] [PubMed] [Google Scholar]
- 37.Ohno T, Sugimoto M, Nagashima A, Ogiwara H, Vilaichone RK, Mahachai V, et al. Relationship between Helicobacter pylori hopQ genotype and clinical outcome in Asian and Western populations. J Gastroenterol Hepatol. 2009;24:462–8. doi: 10.1111/j.1440-1746.2008.05762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiarini A, Calà C, Bonura C, Gullo A, Giuliana G, Peralta S, et al. Prevalence of virulence-associated genotypes of Helicobacter pylori and correlation with severity of gastric pathology in patients from western Sicily, Italy. Eur J Clin Microbiol Infect Dis. 2009;28:437–46. doi: 10.1007/s10096-008-0644-x. [DOI] [PubMed] [Google Scholar]


