Abstract
Mitochondrial DNA (mtDNA) is unquestionably the remnant of an α-proteobacterial genome, yet only ∼10%–20% of mitochondrial proteins are demonstrably α-proteobacterial in origin (the “α-proteobacterial component,” or APC). The evolutionary ancestry of the non-α-proteobacterial component (NPC) is obscure and not adequately accounted for in current models of mitochondrial origin. I propose that in the host cell that accommodated an α-proteobacterial endosymbiont, much of the NPC was already present, in the form of a membrane-bound metabolic organelle (the premitochondrion) that compartmentalized many of the non-energy-generating functions of the contemporary mitochondrion. I suggest that this organelle also possessed a protein import system and various ion and small-molecule transporters. In such a scenario, an α-proteobacterial endosymbiont could have been converted relatively directly and rapidly into an energy-generating organelle that incorporated the extant metabolic functions of the premitochondrion. This model (the “pre-endosymbiont hypothesis”) effectively represents a synthesis of previous, contending mitochondrial origin hypotheses, with the bulk of the mitochondrial proteome (much of the NPC) having an endogenous origin and the minority component (the APC) having a xenogenous origin.
Only ∼10%–20% of mitochondrial proteins are α-proteobacterial in origin. The remainder may be from a premitochondrion, a metabolic but non-energy-generating organelle that existed before the α-proteobacterial symbiont arrived.
Considering the central role played in all eukaryotic cells by mitochondria or mitochondrion-related organelles (MROs, such as hydrogenosomes and mitosomes) (Hjort et al. 2010; Shiflett and Johnson 2010; Müller et al. 2012), the question of the origin and subsequent evolution of the mitochondrion has long captivated and challenged biologists. In a recent article in this series (Gray 2012), I discussed in detail several aspects of mitochondrial evolution, focusing particularly on how well the accumulating molecular data can be accommodated in current models of mitochondrial origin. In this context, the origin and evolution of the mitochondrial proteome, as opposed to the origin and evolution of the mitochondrial genome, were examined from the perspective of comparative mitochondrial proteomics. Somewhat disconcertingly, as more data have become available, we find ourselves considerably less certain about key aspects of how mitochondria originated than we were (or thought we were) several decades ago.
Here, I summarize key points discussed in more detail in the previous article before presenting a novel perspective on how the mitochondrion might have originated. The new model proposed here, which represents a synthesis of both endogenous (“origin from within”) and xenogenous (“origin from outside”) modes, is advanced in an attempt to account for the inability of a purely endosymbiotic model, whose strongest support has come from studies of the mitochondrial genome, to adequately accommodate data on the mitochondrial proteome.
A RUMSFELDIAN TAKE ON THE ORIGIN OF THE MITOCHONDRION
… there are known knowns; there are things we know we know.
We also know there are known unknowns; that is to say, we know there are some things we do not know.
But there are also unknown unknowns—the ones we don’t know we don’t know.
—U.S. Secretary of Defense Donald Rumsfeld Press briefing, February 12, 2002
This Rumsfeldian view provides a useful framework for outlining key aspects of what we know and what we don’t know about the origin and evolution of the mitochondrion.
Known Knowns
We know from comparative mitochondrial genomics—through the analysis of genes contained in mtDNA and the protein sequences they specify—that the mitochondrial genome is of (eu)bacterial origin. These sequences point us to members of a particular bacterial phylum, α-proteobacteria (also termed Alphaproteobacteria), as the closest extant free-living relatives of mitochondria, and therefore the bacterial group from within which the mitochondrial genome emerged (Gray and Doolittle 1982; Gray 1992).
We know that the most bacteria-like (least-derived) and gene-rich mitochondrial genomes are found within the core jakobids (Fig. 1) (Lang et al. 1997; Burger et al. 2013), a group of flagellated protozoa that are members of the proposed eukaryotic supergroup Excavata (Hampl et al. 2009). Jakobid mtDNAs specify strikingly bacteria-like large subunit (LSU), small subunit (SSU) and 5S ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs), as well as upward of 70 proteins, including subunits of an α2ββ′σ RNA polymerase, the enzyme used for transcription in Bacteria. Other notable features include genes encoding transfer-messenger RNA (tmRNA) and the RNA component of RNase P; bacteria-like operon organization; and putative Shine–Dalgarno-like motifs involved in translation initiation in Bacteria.
We know that certain components of the original, bacteria-like systems used for replication and transcription of mtDNA (e.g., the mitochondrial RNA polymerase) have been replaced in some or virtually all eukaryotes by bacteriophage-like counterparts (Shutt and Gray 2006).
We know that mitochondrial genome evolution has been characterized by extensive gene loss, with relocation of many of the genes in the protomitochondrial genome to the nucleus and import of their protein products back into the organelle. Such endosymbiotic gene transfer (EGT) usually involves complete genes, but EGT may also relocate only a portion of a mtDNA-encoded gene, with the rest of that gene remaining in and expressed from the mitochondrial genome (Burger et al. 2003).
We know that the mitochondrion originated only once, that is, mitochondria in all eukaryotic lineages descend from a common ancestor, the LMCA (last mitochondrial common ancestor). Commonality in key aspects of genetic and biochemical function, the presence of shared mitochondrion-specific deletions in bacteria-like operons present in some mtDNAs, and phylogenetic tree reconstruction strongly support this inference (Gray et al. 1999).
We know that despite an evident α-proteobacterial origin of the mitochondrial genome, only ∼10%–20% of the mitochondrial proteome—the collection of proteins making up the organelle—can confidently be traced to α-proteobacteria (this α-proteobacterial component is referred to here as the APC). The 80%–90% non-α-proteobacterial component, or NPC, appears from phylogenetic analysis either to be of diverse prokaryotic (bacterial or archaeal) evolutionary ancestry or to have arisen specifically within eukaryotes (Karlberg et al. 2000; Kurland and Andersson 2000; Gray et al. 2001; Gabaldón and Huynen 2003, 2004, 2007; Szklarczyk and Huynen 2010).
We know that the core of the mitochondrial proteome was already in place in the LMCA, that is, the LMCA was likely as complex in its fundamental functions as a contemporary aerobic mitochondrion. We also know that subsequent retailoring of this proteome, involving both gain and loss of protein components and functions, occurred to different extents in diverse eukaryotic lineages, leading to the emergence of lineage-specific mitochondrial proteins characterized by a restricted phylogenetic distribution (Huynen et al. 2013).
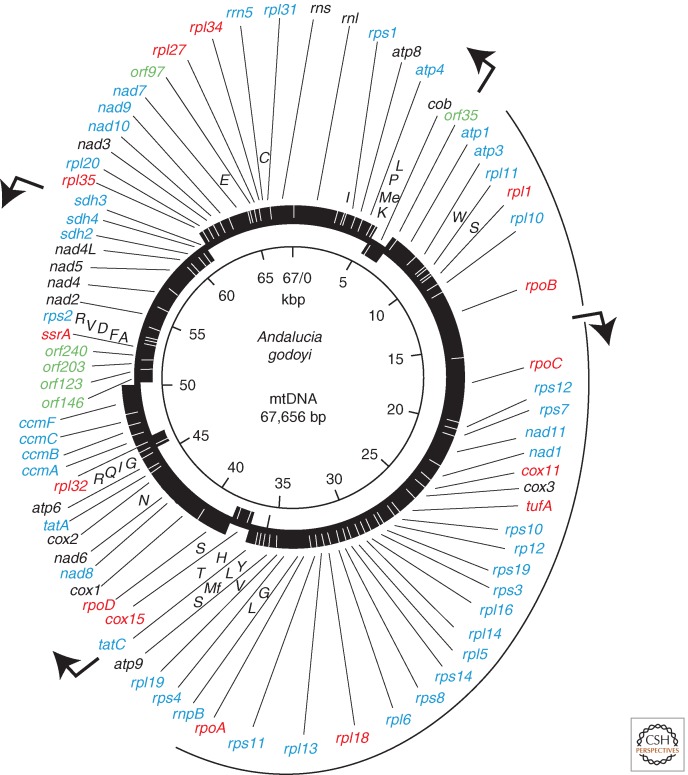
Figure 1.
Genetic map of the strikingly bacteria-like mitochondrial genome of the core jakobid, Andalucia godoyi: the most gene-rich mitochondrial genome known (Burger et al. 2013). (Solid rectangles) Genes, with those on the outer circle transcribed in a clockwise direction, and those on the inner circle transcribed counterclockwise. Colors denote taxonomic gene distribution: (black) common genes found in most animal, fungal, and protist mtDNAs; (blue) expanded gene set, mostly present in protist and plant mtDNAs; (red) genes predominantly found in jakobid mtDNAs; (green) hypothetical protein-coding genes (ORFs). A. godoyi mtDNA encodes 25 proteins involved in coupled electron transport–oxidative phosphorylation (nad, sdh, cob, cox, atp; complexes I–V, respectively); 12 SSU (rps) and 16 LSU (rpl) ribosomal proteins; one translation elongation factor (tuf); four subunits of a bacteria-like multisubunit RNA polymerase (rpoA,B,C,D); eight proteins involved in protein import (ccmA,B,C; tatA,C) and protein maturation (cox11,15; ccmF); LSU (rnl), SSU (rns), and 5S (rrn5) rRNAs; 25 tRNAs—single uppercase letters: (Mf) initiator methionine; (Me) elongator methionine; transfer-messenger RNA (tmRNA) (ssrA); and the RNA component of RNase P (rnpB). Five additional open reading frames (orf) encode putative proteins of unknown function. (Arrows) Positions of predicted promoters. (Figure courtesy of G. Burger.)
Known Unknowns
Despite what we are confident we know we know, several key aspects of mitochondrial origin and evolution remain unresolved:
We do not know precisely which extant α-proteobacterial lineage is most likely to be the descendant of the lineage that was the evolutionary source of the mitochondrial genome (for more detailed discussion of this point and references, see Gray 2012; Lang and Burger 2012). Various studies have consistently pointed to Rickettsiales, one of six or more orders within α-proteobacteria—and comprising obligate intracellular parasites such as Rickettsia, Anaplasma, and Ehrlichia—as the closest relatives of mitochondria. Rickettsiales comprises two distinct families, Rickettsiaceae and Anaplasmataceae, with several studies suggesting that mitochondria are more closely related to the former family, containing various Rickettsia species, than to the latter, which encompasses the genera Anaplasma, Ehrlichia, and Wolbachia. However, there is currently no consensus in the literature as to whether mitochondria branch within or as sister to either of the two Rickettsiales families, or outside of Rickettsiales altogether. Several newly identified free-living α-proteobacteria have recently been considered candidates for the title of “closest mitochondrial relative” (Brindefalk et al. 2011; Georgiades et al. 2011; Viklund et al. 2012), but methodological issues complicate phylogenetic tree reconstructions (Thrash et al. 2011; Rodríguez-Ezpeleta and Embley 2012), and thus the jury is still out on this point.
We do not know the evolutionary origin of most of the mitochondrial proteome (i.e., the NPC). The general assumption has been that these proteins were added to the α-proteobacterial symbiont during its transformation into an organelle, but where these proteins came from and how and when they were added to the evolving mitochondrion is not spelled out in models of mitochondrial origin. A substantial portion of the NPC appears to be eukaryote-specific (i.e., there are no evident homologs within either of the domains Bacteria or Archaea) and thus is presumed to have arisen within the domain Eucarya, although by what mechanism(s) is unclear.
Unknown Unknowns
We have no way of knowing what we might still discover about mitochondrial evolution, although we may be confident that further investigation of diverse mitochondrial genomes and proteomes will continue to surprise us. Unanticipated and increasingly bizarre forms of mitochondrial genome organization and expression continue to be reported, a recent example being from the euglenozoon Diplonema papillatum. Here, mitochondrial genes are fragmented, with the individual gene pieces distributed among several small, circular molecules and the corresponding transcripts processed at both ends before being joined together in the correct order to generate a translatable mRNA, a process that also involves U insertion RNA editing (Vlček et al. 2011; Kiethega et al. 2013). (Notably, D. papillatum, whose mitochondrial genetic system is arguably one of the most radically divergent yet characterized, is a member of Excavata, which as noted above contains the core jakobids, the eukaryotic group featuring the most primitive [least-derived] mitochondrial genomes known.) Investigation of the mitochondrial genetic system has uncovered a wealth of unexpected phenomena, including variant genetic codes, a bewildering array of RNA editing processes, self-splicing introns, fragmented rRNAs, and tRNA import (for review, see Gray et al. 1998; Gray 2003), as well as documenting clear examples of mitochondrion-to-nucleus gene transfer (Adams and Palmer 2003). Based on the experience of the last three decades, we may expect more of the same.
CONTENDING SYMBIOTIC MODELS FOR THE ORIGIN OF MITOCHONDRIA
Endosymbiotic models for the origin of mitochondria (for review, see Martin et al. 2001) can be grouped into two scenarios, depending on whether the organelle is seen to have originated after or at the same time as the rest of the eukaryotic cell (Gray et al. 1999). These two scenarios have been dubbed “archezoan” and “symbiogenesis,” respectively (Koonin 2010). The archezoan scenario hypothesizes that an amitochondriate eukaryotic host cell phagocytosed an α-proteobacterium, which was subsequently transformed into the mitochondrion. In contrast, the symbiogenesis scenario hypothesizes that a physical and metabolic fusion between an α-proteobacterium and an archaeal cell generated the ancestor of the eukaryotic cell, with the α-proteobacterial partner subsequently becoming the mitochondrion. The archezoan scenario most closely approximates the classical endosymbiont hypothesis of mitochondrial origin (Margulis 1970; Doolittle 1980), whereas the hydrogen hypothesis (Martin and Müller 1998) is an example of the symbiogenesis scenario (for more detailed discussion, see Koonin 2010; Gray 2012).
The archezoan scenario takes its name from the “archezoa hypothesis” (Cavalier-Smith 1987, 1989), which posited that a group of putatively amitochondrial eukaryotes was, in fact, primitively amitochondriate (i.e., never possessed mitochondria). As such, archezoans were offered as potential modern representatives of the sort of host cell that would have taken up the mitochondrial endosymbiont. The fact that archezoans branched early in SSU rRNA trees supported the idea that these eukaryotes were, indeed, primitive eukaryotes. However, the archezoa hypothesis was abandoned after the discovery that all putatively amitochondriate eukaryotes so far investigated contain a mitochondrion-related organelle (MRO) and that the early branching position of archezoans is a methodological artifact attributable to a relatively high rate of sequence divergence in archezoan SSU rRNA genes, such that these divergent sequences are incorrectly pulled toward the base of the eukaryotic tree (“long-branch attraction”). The demise of the archezoa hypothesis removes a base of support for the archezoan scenario of mitochondrial origin (admittedly, we cannot exclude the possibility that primitively amitochondriate eukaryotes still exist but have not yet been discovered—albeit not for lack of exhaustive searching—or that such lineages, if they once existed, have all become extinct). On the other hand, as critically discussed in more detail elsewhere (Koonin 2010; Forterre 2011; Gray 2012; Lang and Burger 2012), symbiogenesis models have their own problems both with respect to the origin of the mitochondrion and the origin of the eukaryotic cell per se. A particular difficulty is the fact that collectively more bacterial-type genes in eukaryotes appear to derive from diverse non-α-proteobacterial lineages or fail to affiliate robustly with any specific bacterial phylum (Pisani et al. 2007), whereas in a symbiogenesis scenario, one would anticipate that any bacterial genetic signal in the nuclear genome would be predominantly α-proteobacterial.
DECONSTRUCTING THE MITOCHONDRIAL PROTEOME
The mosaic nature of the mitochondrial proteome (Karlberg et al. 2000; Szklarczyk and Huynen 2010) is proving increasingly difficult to accommodate within current schemes of mitochondrial origin. A specific contribution from α-proteobacteria is strongly indicated, comprising the mitochondrial genome, the few proteins it encodes, and a limited number of other proteins whose genes we infer once resided in mtDNA but were subsequently relocated to the nucleus via EGT. This α-proteobacterial component (APC) forms a crucial part of the evidence that has been offered in support of an endosymbiotic origin of mitochondria; thus, it was a surprise to discover that the APC is a quantitatively minor (albeit qualitatively essential) element of the contemporary oxygen-using and ATP-producing organelle. The non-α-proteobacterial component (NPC), the evolutionary origin of which appears both phylogenetically diverse and largely obscure, dominates the mitochondrial proteome. Any mitochondrion origin scenario must take into account the accumulating evidence that says that only a minor portion of the mitochondrial proteome is clearly of α-proteobacterial ancestry, whereas the evolutionary origin of the major portion of mitochondrial proteins is obscure. Indeed, it is important to emphasize that, strictly speaking, the data used to “prove” the endosymbiotic origin of the mitochondrion in essence only prove the origin of the mitochondrial genome per se.
In both classical endosymbiotic (archezoan) and symbiogenesis (fusion) scenarios for the origin of the eukaryotic cell, an α-proteobacterial symbiont is assumed to have been extensively retailored to create the modern organelle, both by wholesale gene loss from the much larger endosymbiont genome and by the addition of new proteins. Comparatively few mitochondrial proteomes have been investigated directly (e.g., via mass spectrometry), yet the available data already show that key mitochondrial protein assemblages and pathways existed in essentially their modern form in the last eukaryotic common ancestor (LECA). Lineage-specific modification through both loss and addition of mitochondrial proteins has certainly occurred, and specific cases of mitochondrial protein acquisition through lateral gene transfer (LGT) can definitely be identified, but the fact remains that the basic structure and complex function of the mitochondrion were likely already well established in the LECA. Thus, between the events that created both the modern mitochondrion and the rest of the cell in which it resides, much reorganization of symbiont and host cell would have had to occur, whether the acquisition of the mitochondrion was through a symbiogenesis pathway or an archezoan one.
Symbiont-to-mitochondrion transformation is generally considered to have occurred in a stepwise fashion, involving both loss and addition of proteins and functions. In comparing a reconstructed protomitochondrial proteome with the proteomes of contemporary α-proteobacteria and mitochondria (yeast and human), Gabaldón and Huynen (2007) inferred that the transformation of an α-proteobacterial symbiont to an organelle involved loss of some protomitochondrial metabolic pathways or their movement to other parts of the cell, as well as acquisition of new proteins leading to the gain of novel pathways, such as the protein import machinery and mitochondrial carriers. In addition, new proteins have been recruited to existing mitochondrial complexes resulting, for example, in the expansion of respiratory complex I from 17 subunits in α-proteobacteria to more than 40 subunits in eukaryotes (Cardol 2011). Such restructuring over a long period of time would presumably have generated a series of intermediate organelles differing in the range of functions they possessed. A key question that such a scenario poses is how the hundreds (perhaps thousands) of proteins making up the mitochondrial proteome were introduced. In particular, what was the evolutionary source of the NPC? I argue here that neither the archezoan nor the symbiogenesis scenario accommodates, in a compelling way, the diverse functional and phylogenetic character of the NPC.
At this point, a word of caution is in order with respect to how we view the NPC. In particular, it is possible (perhaps even likely) that a portion of the NPC consists of originally α-proteobacterial proteins that are too divergent in sequence to be recognized as such, so that the 10%–20% estimated α-proteobacterial contribution to the mitochondrial proteome is probably an underestimate, and instead represents a minimal contribution from the α-proteobacterial endosymbiont. Marked differences in phylogenetic resolution are to be expected in a protein collection as complex as the mitochondrial proteome, even though it is notable that the α-proteobacterial signal in the recognizable APC is particularly strong (Lang and Burger 2012). Nevertheless, it does not seem probable that the “bacterial” portion of the NPC could all be accounted for by divergent proteins of originally α-proteobacterial origin.
Another suggestion is that the α-proteobacterial symbiont was itself chimeric as a consequence of prior and extensive LGT. In this case, the symbiont could have contributed genes encoding proteins that appear to come from other bacterial lineages. A problem with this suggestion is that this non-α-proteobacterial contribution would have had to have been extensive enough to quantitatively swamp the α-proteobacterial component in the contemporary mitochondrion, which would imply a highly chimeric symbiont genome and/or differential loss of α-proteobacterial genes in the subsequent symbiont-to-organelle transformation. Moreover, it is not clear from this scenario why extant α-proteobacterial genomes generally lack orthologs corresponding to this putative LGT component. Thiegart et al. (2012) attribute this phylogenetic diversity to “a single origin of mitochondria followed by subsequent LGT among free-living prokaryotes.” However, this explanation would seem to demand that for mitochondrial homologs transferred in this way, the transferred genes would have to have evolved more slowly than donor (α-proteobacterial) ones, or would have to have been selectively lost from α-proteobactrerial lineages. Otherwise, they would still score in phylogenetic reconstructions as “α-proteobacterial.”
In yeast, about half of the prokaryote-like sequences in the mitochondrial proteome are common to Bacteria and Archaea as well as eukaryotes, and thus may well have been present in the last universal common ancestor (LUCA) (Karlberg et al. 2000). Of the remainder, few affiliate convincingly with specific non-α-proteobacterial bacterial lineages, whereas the α-proteobacterial signal is particularly robust (Lang and Burger 2012). Moreover, we still have to account for the large proportion (up to 50%) of the mitochondrial proteome that is unique to eukaryotes and that has been assumed to have evolved within the eukaryotic lineage. Thus, even if we are liberal in assigning a percentage to the APC (e.g., doubling it to 20%–40%), we are still left with upward of 60%–80% of the mitochondrial proteome that does not appear to have come from an α-proteobacterial source.
In this context, a final possibility is that at least some of the NPC proteins predated the endosymbiotic event, preexisting in whatever the host cell was that accommodated the symbiont. For example, in a recent review on plastid endosymbiosis, Keeling (2013a) has suggested a “targeting rachet” model in which protein targeting is established in the host before the endosymbiont is permanently integrated. The idea is that this process would target preexisting transporters to the membrane of transient symbionts, accelerating and facilitating the symbiont-to-organelle transformation.
The possibility that some portion of what would become the mitochondrial NPC preexisted in the host is attractive because it would effectively “precondition” the host–symbiont interaction. Rather than the NPC appearing on the scene after the acquisition of the endosymbiont (e.g., via LGT) or actually being part of the endosymbiont, it might be that much of the NPC was already present in the host cell that accommodated the α-proteobacterial symbiont. If such were the case, raw material for initiating the conversion of a bacterium to an organelle would be readily available.
The above proposal demands a rather different view of the eukaryotic cell and how it arose than is suggested in current proposals. In particular, the idea that the eukaryotic cell is fundamentally a fusion between bacterial and archaeal partners, with a large number of novel proteins arising subsequently and specifically within the eukaryotic lineage, is increasingly being questioned (e.g., Koonin 2010; Forterre 2011; Lang and Burger 2012). In proposing yet another symbiogenesis scenario (Planctomycetes, Verrucomicrobia, Chlamydiae [PVC] Bacteria—Thaumarchaeota—Virus,” or PTV), Forterre (2011) states that “the transformation of a FECA (first eukaryotic common ancestor) born from a fusion event into a reasonable ancestor of modern eukarya remains difficult to imagine in a convincing way in the PTV scenario as in all other fusion scenarios.” In fact, Forterre is led to conclude that eukaryotes “probably evolved from a specific lineage, according to the three domains scenario originally proposed by Carl Woese.”
In contrast to the prevailing symbiogenesis view, a radically different proposal for the origin of the eukaryotic lineage has just been published by Harish et al. (2013), drawn from phylogenetic reconstructions based on genome content of compact protein domains, as defined in the SCOP (structural classification of proteins) database. These investigators identify Archaea and Bacteria as sister domains in a lineage (“akaryotes”) that descends in parallel, but independently, from an entity they term the “MRUCA” (most recent universal common ancestor), which “is not a bacterium.” According to their analysis, Harish et al. (2013) assert that MRUCA would have contained 75% of the unique SFs (SCOP superfamilies) encoded by extant genomes of the domains Bacteria, Archaea, and Eucarya, each specifying a proteome that partially overlaps each of the other two: “the consequence of descent from a universal common ancestor, MRUCA.” The authors state: “we cannot draw the conclusion that MRUCA was morphologically similar to a eukaryote,” but they do conclude that “elements and cohorts of SFs from its proteome are recognizable in the proteomes of modern eukaryotes. Thus, SFs homologous to those of modern mitochondria, chloroplasts, nuclei, spliceosomes, cytoskeleton, and the like are evident in the proteome of MRUCA.”
A notable implication of the proposal by Harish et al. (2013) is that the LECA could well have harbored a complex proteome that already included much of what was destined to become the mitochondrial NPC. This inference would be consistent with a new model of mitochondrial evolution outlined below, which hypothesizes that essential elements of the NPC already existed in the host cell that formed an association with the APC-contributing α-proteobacterium.
THE “PREMITOCHONDRION”: A PUTATIVE ORGANELLE PREDATING THE α-PROTEOBACTERIAL ENDOSYMBIOSIS
If we accept the possibility that much of the mitochondrial proteome was already present in the eukaryotic host cell before the α-proteobacterial symbiont arrived, what might it have been doing? To address this question, I propose a new model (the “pre-endosymbiont hypothesis”) (Fig. 2) that envisages an endogenously evolved organelle that we might term the “pre-mitochondrion,” which I suggest preexisted in what was in many respects already a eukaryotic cell (the “pre-eukaryote”). According to this scheme, the premitochondrion was essentially a metabolic rather than an energy-generating organelle: a membrane-bound structure compartmentalizing many (perhaps most) of the pathways and reactions characteristic of extant mitochondria, such as amino acid, nucleotide, and lipid metabolism; coenzyme biosynthesis; iron–sulfur cluster formation, and so on. The premitochondrion would already have membrane structural proteins and a protein import system, as well as transporters for various ions and metabolites, including amino acids and nucleotides. However, because the premitochondrion would be energy-consuming rather than energy-generating, it would possess a nucleotide carrier system to import ATP and export ADP, the reverse of what occurs in the contemporary mitochondrion.
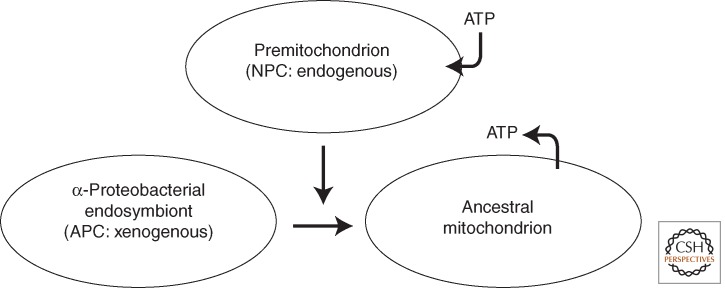
Figure 2.
A schematic view of the pre-endosymbiont hypothesis. The premitochondrion is seen as a membrane-bound entity endowed with a protein import system and various ion/small-molecule transporters, compartmentalizing many of the metabolic functions of the mitochondrion. The premitochondrion is assumed to have evolved endogenously within the pre-eukaryote cell from proteins that now constitute a major portion of the NPC (non-α-proteobacterial component) of the contemporary mitochondrial proteome. An α-proteobacteria-like endosymbiont is converted to the ancestral mitochondrion, effectively “capturing” the protein components and functions of the premitochondrion. The endosymbiont contributes the APC (α-proteobacterial component) of the mitochondrial proteome, which is largely directed toward specification of energy-generating capacity (in the form of coupled electron transfer/oxidative phosphorylation). The premitochondrion → ancestral mitochondrion conversion is greatly facilitated by the existence of the reservoir of premitochondrial proteins in the host cell. Endosymbiont-to-nucleus gene transfer (EGT) coupled with rationalization of redundant pathways results in the formation of evolutionarily chimeric enzymatic pathways and protein complexes in the ancestral mitochondrion, as well as functional relocation of the products of some transferred α-proteobacterial genes to other cellular compartments. The premitochondrion is assumed to be a non-energy-generating organelle that imports ATP; a key innovation in the transition to the contemporary mitochondrion is the generation/acquisition of an ADP/ATP transporter that reverses this flow, ultimately allowing the mitochondrion to become the primary site of ATP generation for cellular functions.
In this scenario, a large part of the mitochondrial proteome (much of the NPC) would be of endogenous origin, having evolved within the pre-eukaryotic cell, along with its other subcellular components and compartments. In some respects, the premitochondrion would resemble non-DNA-containing MROs such as hydrogenosomes and mitosomes, which perform a variable subset of normal mitochondrial metabolic functions but lack the capacity to generate ATP via coupled electron transport–oxidative phosphorylation. Such MROs could be regarded as representing a reversion to an ancestral form.
Transformation of an α-proteobacterial symbiont into an integrated organelle would occur as conventionally envisaged, but it would be substantially simplified if, in fact, the bulk of the proteins required for such a radical reorganization were already present, in the form of a functional entity. Because coupled oxidative phosphorylation would have existed and functioned in the symbiont to generate both a proton gradient and ATP, it is likely that it was the symbiont that was modified by premitochondrial components to become the mitochondrion, rather than the symbiont genome being introduced somehow into and becoming functional within the premitochondrion.
The pre-eukaryote cell would thus resemble the proposed host cell in the classical serial endosymbiont hypothesis (Taylor 1987) except that a progenitor of the mitochondrion (the pre-mitochondrion) was already present. Subsequent transformation of the α-proteobacterial symbiont, via incorporation of structural and functional elements of the premitochondrion, would effectively add energy-generating capacity, in the form of electron transport and coupled oxidative phosphorylation, to the varied metabolic capabilities already possessed by the premitochondrion. Indeed, it is important to emphasize that the essential function of the contemporary mitochondrial genome is the specification of critical protein subunits of the four membrane-bound complexes (I–IV) of the mitochondrial electron transport chain and the ATP synthase (complex V), whereas other APC proteins are largely involved in supporting mitochondrial genome expression. In essence, the pre-endosymbiont hypothesis incorporates aspects of both endogenous and xenogenous scenarios for the origin of the mitochondrion.
SOME IMPLICATIONS AND CONSIDERATIONS
Internal membrane-bound compartments such as the endoplasmic reticulum, Golgi body, and other components of the endomembrane system are assumed to have emerged endogenously within the evolving eukaryotic cell. Thus, it is not inconceivable that an organelle such as the premitochondrion could likewise have evolved endogenously. Indeed, the premitochondrion is conceptually similar to the peroxisome, a membrane-bound, non-DNA-containing eukaryotic organelle that is generally considered to have had an endogenous origin (Gabaldón 2013). One might imagine that the selective advantage for such compartmentation would be the concentration of enzymes and reactants in various metabolic pathways, in the face of a steady increase in cell size, one of the hallmarks of the eukaryotic cell.
Symbiotic models of mitochondrial origin usually invoke the introduction of aerobic (oxygen-dependent) metabolism into an essentially anaerobic host cell (Margulis 1970), the initial selective advantage being either more efficient ATP generation in an increasingly oxygen-rich environment or oxygen detoxification (the “ox-tox” hypothesis) (Andersson and Kurland 1999). However, there are good arguments to consider that the ancestral eukaryotic cell was, in fact, already capable of aerobic metabolism (Raff and Mahler 1972). In that case, assuming that an aerobic α-proteobacterium was taken up by an aerobic pre-eukaryote via phagocytosis (another eukaryotic hallmark), there need have been no selective advantage initially that depended on the symbiont being an aerobe. Moreover, the relationship of the ingestee to ingester might be less one of food (as generally assumed) and more one of a coexisting long-term inhabitant, as is the situation in contemporary eukaryotes that harbor other organisms, such as the amoebozoan protist Acanthamoeba castellanii. Although this phagotroph preys on other microbes for food, it also hosts a wide variety of Bacteria (including various types of α-proteobacteria) in stable intracellular associations (Marciano-Cabral 2004). This type of relationship would seem much more conducive to promoting a symbiont-to-organelle transformation in the long term than one in which the symbiont happens to be a rare escapee from digestion. As Lang and Burger (2012) have commented, “While ancient symbiotic events are often viewed as driven by selective advantage for both partners, it is equally possible that they have started out by pure accident. …”
If we accept that the host cell was capable of aerobic metabolism, what type of energy-generating system might it have used? Given the ubiquity and antiquity of aerobic metabolism based on cytochrome oxidase (Castresana et al. 1994), it is possible that both host cell and symbiont had parallel systems for energy generation. Here again, no selective advantage would accrue to the host from a symbiont bringing such a system into it. Initially, the energy requirements of host and symbiont would be met by their respective energy-generating systems. However, the equation would shift dramatically once ADP import into the symbiont and ATP export from it evolved. That change might have occurred through a “reverse flow” mutation of the ATP-importing/ADP-exporting translocase postulated here to have been present in the premitochondrion, or through elaboration of a completely new transporter. Evidently, the required transporter was not contributed by the symbiont (Karlberg et al. 2000).
The emergence of an ADP-importing/ATP-exporting translocase is a fundamental element in all models of mitochondrial evolution. Subsequent intracellular proliferation of the evolving mitochondrion would lead to a pronounced increase in bioenergetic membranes supporting coupled electron transport and oxidative phosphorylation, greatly increasing the production of ATP available to the cell as a whole. Indeed, Lane and Martin (2010) argue persuasively that the increased energy provided by specialization of the endosymbiont into an ATP-generating organelle and increasing organelle copy number was “a prerequisite to eukaryote complexity: the key innovation to multicellular life.”
Even before the elaboration of an ADP-importing/ATP-exporting translocase, acquisition by the symbiont of the protein import system and other transporters of the premitochondrion would set the stage for relocation to the symbiont of many of the metabolic pathways of the premitochondrion. The advantage of performing these activities in the symbiont rather than in the premitochondrion would be that the symbiont would have generated the ATP in situ for biosynthetic pathways requiring it, whereas the premitochondrion would have had to import ATP from the cytosol for these same reactions. Once ATP export from evolving organelle to cytosol was established, the selective advantage would be greatly enhanced, because the evolving mitochondria could proliferate and thereby sequester an increasingly larger proportion of the structural and enzymatic components required to make the functional premitochondrion. Intracellular competition of these sorts would inevitably lead to the disappearance of the premitochondrion as a separate entity and the fixation of its successor, the modern mitochondrion.
Finally, because host and symbiont genomes would initially encode a number of the same or very similar metabolic pathways, a process of genome reduction would ensue whereby duplicate genes were lost from one or the other genome. Most of this reduction appears to have involved the endosymbiont genome, with many genes being lost or transferred to the nucleus (Gabaldón and Huynen 2007). In the latter case, relocated genes would have had to acquire signals for nuclear expression and mitochondrial targeting, although some α-proteobacterial symbiont genes have evidently been retargeted to other cellular locations, where they currently function (Gabaldón and Huynen 2007). Conversely, host genes have evidently substituted for symbiont genes in specifying components of many mitochondrial pathways.
A particularly notable example in this regard is the tricarboxylic acid (TCA) cycle, genes for which were presumably contained originally in the symbiont genome but moved to the nuclear genome. However, not all of the mitochondrial TCA cycle enzymes are decidedly α-proteobacterial in origin (e.g., Kurland and Andersson 2000; Schnarrenberger and Martin 2002). Current models of mitochondrial evolution generally assume that LGT from another bacterial source led to replacement of the originally α-proteobacterial gene: an assumption that requires many separate LGT events to account for the large number of non-α-proteobacterial constituents of the mitochondrial proteome. The scenario presented here suggests a simplified alternative: that the non-α-proteobacterial components of the TCA cycle (and other mitochondrial pathways) are encoded by genes that were originally present in the ancestral eukaryotic host before the arrival of the mitochondrial endosymbiont, and that they replaced the homologous endosymbiont genes at an early stage in the symbiont-to-organelle transition.
CONCLUDING REMARKS
The new model of mitochondrial origin and evolution outlined here (Fig. 2) is an attempt to integrate conflicting data that are not readily accommodated by current hypotheses. In particular, the pre-endosymbiont hypothesis accounts in a relatively straightforward way for the presence and characteristics of both α-proteobacterial and non-α-proteobacterial components (APC and NPC, respectively) of the mitochondrial proteome. The existence of an ancestral metabolic organelle (the premitochondrion) would effectively “precondition” the host cell for the successful conversion of a bacterial endosymbiont into an integrated organelle because much of the material necessary for this transformation would already be available in the host, in the form of much of the NPC. Furthermore, the hypothesis presented here provides an explanation for what the preexisting portion of the NPC would be doing before the endosymbiont arrived; moreover, it helps to explain not only the mosaic nature of the mitochondrial proteome overall, but also the mosaic character of the enzymatic pathways contained therein.
The pre-endosymbiont hypothesis elaborated here effectively represents a synthesis of previous, contending mitochondrial origin hypotheses, with much of the mitochondrial proteome (represented within the NPC) having an endogenous origin and the minority component (the APC) having a xenogenous origin. In some respects, this hypothesis hearkens back to the (nonsymbiotic) model for the origin of mitochondria published more than 40 years ago by Raff and Mahler (1972). That publication predated the molecular evidence that convincingly established the α-proteobacterial nature of the mitochondrial genome: data that had a profound influence on the demise of nonsymbiotic models and the ascendency of endosymbiotic models of mitochondrial origin (Gray and Doolittle 1982; Gray 1992). The molecular data, drawn in large part from analyses of mtDNA and the genes it contains, leave little doubt that bacterial endosymbiosis played a crucial role in the origin of the mitochondrion, although I would contend that that role is considerably more circumscribed than we initially imagined. Because the mitochondrion was originally viewed as a singular entity rather than as a mosaic, proof for the origin of the mitochondrial genome was generally accepted as constituting proof for the origin of the organelle as a whole. In retrospect, that extrapolation turned out to be overly simplistic. In the context of a discussion elsewhere in this series (Keeling 2013b), it is worth contemplating how different the historical record of our views about mitochondrial origin and evolution might have been had our detailed knowledge and understanding of the mitochondrial proteome predated that of the mitochondrial genome.
ACKNOWLEDGMENTS
This essay is dedicated to my PhD supervisor, Professor Byron G. Lane, on the occasion of his 80th birthday, in recognition and deep appreciation of 50 years of mentorship and friendship. I am grateful to P.J. Keeling, J.M. Archibald, and A.J. Roger for critical comment on an early draft of this article; to C.G. Kurland for stimulating discussion about mitochondrial evolution; and to G. Burger for provision of the A. godoyi mtDNA gene map shown in Figure 1.
Footnotes
Editors: Patrick J. Keeling and Eugene V. Koonin
Additional Perspectives on The Origin and Evolution of Eukaryotes available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Adams KL, Palmer JD 2003. Evolution of mitochondrial gene content: Gene loss and transfer to the nucleus. Mol Phylogenet Evol 29: 380–395 [DOI] [PubMed] [Google Scholar]
- Andersson SGE, Kurland CG 1999. Origins of mitochondria and hydrogenosomes. Curr Opin Microbiol 2: 535–541 [DOI] [PubMed] [Google Scholar]
- Brindefalk B, Ettema TJG, Viklund J, Thollesson M, Andersson SGE 2011. A phylometagenomic exploration of oceanic Alphaproteobacteria reveals mitochondrial relatives unrelated to the SAR11 clade. PLoS ONE 6: e24457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Gray MW, Lang BF 2003. Mitochondrial genomes: Anything goes. Trends Genet 19: 709–716 [DOI] [PubMed] [Google Scholar]
- Burger G, Gray MW, Forget L, Lang BF 2013. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol 5: 418–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardol P 2011. Mitochondrial NADH:ubiquinone oxidoreductase (complex I) in eukaryotes: A highly conserved subunit composition highlighted by mining of protein databases. Biochim Biophys Acta 1807: 1390–1397 [DOI] [PubMed] [Google Scholar]
- Castresana J, Lübben M, Saraste M, Higgins DG 1994. Evolution of cytochrome oxidase, an enzyme older than atmospheric oxygen. EMBO J 13: 2516–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T 1987. Eukaryotes with no mitochondria. Nature 326: 332–333 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 1989. Archaebacteria and archezoa. Nature 339: 100–101 [DOI] [PubMed] [Google Scholar]
- Doolittle WF 1980. Revolutionary concepts in evolutionary biology. Trends Biochem Sci 5: 146–149 [Google Scholar]
- Forterre P 2011. A new fusion hypothesis for the origin of Eukarya: Better than previous ones, but probably also wrong. Res Microbiol 162: 77–91 [DOI] [PubMed] [Google Scholar]
- Gabaldón T 2013. A metabolic scenario for the evolutionary origin of peroxisomes from the endomembrane system. Cell Mol Life Sci 10.1007/s00018-013-1424-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T, Huynen MA 2003. Reconstruction of the proto-mitochondrial metabolism. Science 302: 609. [DOI] [PubMed] [Google Scholar]
- Gabaldón T, Huynen MA 2004. Shaping the mitochondrial proteome. FEBS Lett 1659: 212–220 [DOI] [PubMed] [Google Scholar]
- Gabaldón T, Huynen MA 2007. From endosymbiont to host-controlled organelle: The hijacking of mitochondrial protein synthesis and metabolism. PLoS Comput Biol 3: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades K, Madoui M-A, Le P, Robert C, Raoult D 2011. Phylogenomic analysis of Odyssella thessalonicensis fortifies the common origin of Rickettsiales, Pelagibacter ubique and Reclimonas americana mitochondrion. PLoS ONE 6: e24857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW 1992. The endosymbiont hypothesis revisited. Int Rev Cytol 141: 233–357 [DOI] [PubMed] [Google Scholar]
- Gray MW 2003. Diversity and evolution of mitochondrial RNA editing systems. IUBMB Life 55: 227–233 [DOI] [PubMed] [Google Scholar]
- Gray MW 2012. Mitochondrial evolution. Cold Spring Harb Perspect Biol 4: a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Doolittle WF 1982. Has the endosymbiont hypothesis been proven? Microbiol Rev 46: 1–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Brossard N, Delage F, Littlejohn TG, et al. 1998. Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res 26: 865–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF 1999. Mitochondrial evolution. Science 283: 1476–1481 [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF 2001. The origin and early evolution of mitochondria. Genome Biol 2: reviews1018–1018.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AGB, Roger AJ 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups.” Proc Natl Acad Sci 106: 3859–3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harish A, Tunlid A, Kurland CG 2013. Rooted phylogeny of the three superkingdoms. Biochimie 95: 1593–1604 [DOI] [PubMed] [Google Scholar]
- Hjort K, Goldberg AV, Tsaousis AD, Hirt RP, Embley TM 2010. Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos Trans R Soc Lond B Biol Sci 365: 713–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen MA, Duarte I, Szklarczyk R 2013. Loss, replacement and gain of proteins at the origin of the mitochondria. Biochim Biophys Acta 1827: 224–231 [DOI] [PubMed] [Google Scholar]
- Karlberg O, Canbäck B, Kurland CG, Andersson SGE 2000. The dual origin of the yeast mitochondrial proteome. Yeast 17: 170–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ 2013a. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu Rev Plant Biol 64: 583–607 [DOI] [PubMed] [Google Scholar]
- *.Keeling P 2013b. The impact of history on our perception of evolutionary events: Endosymbiosis and the origin of eukaryotic complexity. Cold Spring Harb Perspect Biol 6: a016196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiethega G, Yan Y, Turcotte M, Burger G 2013. RNA-level unscrambling of fragmented genes in Diplonema mitochondria. RNA Biol 10: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV 2010. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol 11: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Andersson SGE 2000. Origin and evolution of the mitochondrial proteome. Microbiol Mol Biol Rev 64: 786–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Martin W 2010. The energetics of genome complexity. Nature 467: 929–934 [DOI] [PubMed] [Google Scholar]
- Lang BF, Burger G 2012. Mitochondrial and eukaryotic origins: A critical review. In Mitochondrial genome evolution (ed. Maréchal-Drouard L), Advances in Botanical Research, Vol. 63, pp. 1–20 Elsevier, Amsterdam [Google Scholar]
- Lang BF, Burger G, O’Kelly CJ, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Gray MW 1997. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 387: 493–497 [DOI] [PubMed] [Google Scholar]
- Marciano-Cabral F 2004. Introductory remarks: Bacterial endosymbionts or pathogens of free-living amebae. J Eukaryot Microbiol 51: 497–501 [DOI] [PubMed] [Google Scholar]
- Margulis L 1970. Origin of eukaryotic cells. Yale University Press, New Haven, CT [Google Scholar]
- Martin W, Müller M 1998. The hydrogen hypothesis for the first eukaryote. Nature 392: 37–41 [DOI] [PubMed] [Google Scholar]
- Martin W, Hoffmeister M, Rotte C, Henze K 2001. An overview of endosymbiotic models for the origins of eukaryotes, their ATP-producing organelles (mitochondria and hydrogenosomes), and their heterotrophic lifestyle. Biol Chem 382: 1521–1539 [DOI] [PubMed] [Google Scholar]
- Müller M, Mentel M, van Hellemond JJ, Henze K, Woehle C, Gould SB, Yu RY, van der Giezen M, Tielens AGM, Martin WF 2012. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev 76: 444–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani D, Cotton JA, McInerney JO 2007. Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol Biol Evol 24: 1752–1760 [DOI] [PubMed] [Google Scholar]
- Raff RA, Mahler HR 1972. The non symbiotic origin of mitochondria. Science 177: 575–582 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Embley TM 2012. The SAR11 group of α-proteobacteria is not related to the origin of mitochondria. PLoS ONE 7: e30520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnarrenberger C, Martin W 2002. Evolution of the enzymes of the citric acid cycle and the glyoxylate cycle of higher plants. A case study of endosymbiotic gene transfer. Eur J Biochem 269: 868–883 [DOI] [PubMed] [Google Scholar]
- Shiflett AM, Johnson PJ 2010. Mitochondrion-related organelles in eukaryotic protists. Annu Rev Microbiol 64: 409–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutt TE, Gray MW 2006. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet 22: 90–95 [DOI] [PubMed] [Google Scholar]
- Szklarczyk R, Huynen MA 2010. Mosaic origin of the mitochondrial proteome. Proteomics 10: 4012–4024 [DOI] [PubMed] [Google Scholar]
- Taylor FJR 1987. An overview of the status of evolutionary cell symbiosis theories. In Endocytobiology III (ed. Lee JJ, Fredrick JF), Vol. 503, pp. 1–16 The New York Academy of Sciences, New York: [DOI] [PubMed] [Google Scholar]
- Thiegart T, Landan G, Schenk M, Dagan T, Martin WF 2012. An evolutionary network of genes present in the eukaryote common ancestor polls genomes on eukaryotic and mitochondrial origin. Genome Biol Evol 4: 466–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash JC, Boyd A, Huggett MJ, Grote J, Carini P, Yoder RJ, Robbertse B, Spatafora JW, Rappe MS, Giovannoni SJ 2011. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci Rep 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viklund J, Ettema TJG, Andersson SGE 2012. Independent genome reduction and phylogenetic reclassification of the oceanic SAR11 clade. Mol Biol Evol 29: 599–615 [DOI] [PubMed] [Google Scholar]
- Vlček C, Marande W, Teijeiro S, Lukeš J, Burger G 2011. Systematically fragmented genes in a multipartite mitochondrial genome. Nucleic Acids Res 39: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]