Abstract
Over-excitation of photosynthetic apparatus causing photoinhibition is counteracted by non-photochemical quenching (NPQ) of chlorophyll fluorescence, dissipating excess absorbed energy into heat. The PsbS protein plays a key role in this process, thus making the PsbS-less npq4 mutant unable to carry out qE, the major and most rapid component of NPQ. It was proposed that npq4 does perform qE-type quenching, although at lower rate than WT Arabidopsis. Here, we investigated the kinetics of NPQ in PsbS-depleted mutants of Arabidopsis. We show that red light was less effective than white light in decreasing maximal fluorescence in npq4 mutants. Also, the kinetics of fluorescence dark recovery included a decay component, qM, exhibiting the same amplitude and half-life in both WT and npq4 mutants. This component was uncoupler-sensitive and unaffected by photosystem II repair or mitochondrial ATP synthesis inhibitors. Targeted reverse genetic analysis showed that traits affecting composition of the photosynthetic apparatus, carotenoid biosynthesis and state transitions did not affect qM. This was depleted in the npq4phot2 mutant which is impaired in chloroplast photorelocation, implying that fluorescence decay, previously described as a quenching component in npq4 is, in fact, the result of decreased photon absorption caused by chloroplast relocation rather than a change in the activity of quenching reactions.
Keywords: energy dissipation, non-photochemical quenching, Lhcb, xanthophyll, photosynthesis, chloroplast avoidance
1. Introduction
Plants use light as an energy source for their metabolism: solar energy is absorbed, and excitons rapidly transferred to the photosynthetic reaction centres by the light-harvesting complexes (LHC) of each photosystem (PS). These bind a large array of chlorophylls (Chls) that are tightly connected to yield more than 80% quantum efficiency of photochemical reactions [1]. Under stable light conditions, plants optimize the efficiency of photosynthetic machinery and yet light intensity changes during the day and the rapid fluctuations imposed by shading in the canopy or by clouds, easily result in over-excitation. Thus, quantum efficiency needs to be rapidly downregulated to decrease the probability of Chl a triplet (3Chl*) and singlet oxygen formation [2,3]. This is performed by photoprotection mechanisms including leaf and chloroplast avoidance movement, reactive oxygen species (ROS) scavenging, and quenching of triplet and singlet Chl excited states [4–6]. The ability of plants to modulate light utilization efficiency in fluctuating light is crucial for plant fitness [7]. A major role for prevention of over-excitation is played by a set of inducible mechanisms referred to as non-photochemical quenching (NPQ) [8] that are triggered by a feedback loop in which excess light (EL) induces lumenal acidification, detected by the thylakoid protein PsbS, which in turn triggers qE, the most rapid component of NPQ, leading to dissipation of excess energy with a half-life of 1–2 min [5,9]. As PsbS is not a Chl-binding protein [10], its effect on Chl fluorescence must be achieved through interaction with the antenna system binding the xanthophylls zeaxanthin (Zea) and lutein (Lut) [11], in the absence of which quenching does not occur [12].
Besides qE, NPQ includes a slowly relaxing component (τ > 60 min) qI, which is independent of lumenal pH and has been attributed to inactive PSII centres produced by EL stress [13]. Additional quenching components with intermediate half-lives (τ = 10–15 min) were originally attributed to state 1–state 2 transitions [8], and more recently to Zea binding to the LHC proteins, hence the names qT or qZ [14,15].
Two types of mechanism have been proposed for activation of quenching by PsbS: the first proposes a direct interaction of PsbS with a neighbour antenna protein, either LHCII or a monomeric complex, which causes a conformational change activating quenching site(s) within the antenna subunit itself [16,17] or through the trapping of a Zea molecule at the PsbS–LHCII interface [11]. The first type of mechanism relies on the notion that LHC proteins exist in two conformations with different fluorescence lifetimes [18], whose interconversion is controlled by changes in protein–protein interactions in the membrane, which are promoted by activation of PsbS [19,20]. The recent report that npq4 plants lacking PsbS are competent in quenching, although longer exposure to EL is required than in WT plants [21], supports the first hypothesis.
In this work, we have studied the properties of light-induced fluorescence decrease in the npq4 mutant, which develops a slow fluorescence decay. A component, qM, with the same amplitude and half-life, could also be deconvoluted from the kinetics of WT plants, was uncoupler-sensitive and unaffected by treatments inhibiting PSII repair or mitochondrial ATP production. Also it was induced by white light but not by red light. A targeted reverse genetic analysis showed that the npq4phot2 double mutant which was impaired in chloroplast avoidance, was devoid of qM. On this basis, we propose that the fluorescence decay previously described as a quenching component in npq4 is, in fact, the result of decreased photon absorption caused by chloroplast relocation rather than by a change in the activity of quenching reactions. This finding supports a direct role of PsbS in triggering the quenching reactions.
2. Experimental procedures
(a). Plant material
Arabidopsis thaliana T-DNA insertion mutants (Col-0) npq1 (At1G08550) and npq2 (At5G67030) were a kind gift of K.K. Niyogi (University of California at Berkeley). Mutant lut2 (At5G57030) was obtained from the NASC collection, Salk line 005018. koLhcb4, koLhcb5 and koLhcb6 were obtained as described in [22,23]. Mutants npq4 and npq4ch1 were a kind gift of K. K. Niyogi, stn7npq4 was provided by E.-M. Aro (University of Turku, Finland), and phot2 by M. Wada (Kyushu University, Japan). Double mutants were obtained by crossing single mutant plants and selecting progeny either by pigment analysis, western blotting [22,23] or by the light-induced change in the green colour of leaf blades [24]. WT and mutant plants were grown on compost in a growth chamber for five weeks under controlled conditions (150 µmol photons m−2 s−1, 23°C, 8 L/16 D cycle, 70% relative humidity).
(b). In vivo fluorescence and non-photochemical quenching measurements
NPQ of Chl fluorescence was measured on leaves at room temperature (RT; 23°C) with a PAM 101 fluorometer (Walz, Germany). NPQ was calculated according to Van Kooten & Snel [25]. When red actinic light was used, the light intensities for these experiments were chosen in order to produce the same value of qL in all genotypes. When indicated, fluorescence was measured on detached leaves infiltrated with 150 mM sorbitol containing either 50 μM nigericin [26], 100 μM lincomycin [27] or 2 μM myxothiazol [28].
(c). Pigment analysis
Pigments were extracted from leaf discs using 85% acetone buffered with Na2CO3. Separation and quantification of pigments were performed by HPLC [29].
(d). Measurement of chloroplast movement
Chloroplast avoidance response was induced in leaves by EL treatment. Before measurements, plants were adapted in darkness for 1 h, then detached leaves on wet paper were exposed to 400 µmol photons m−2 s−1, white light at RT for 1 h. Distribution of chloroplasts in the mesophyll cells was determined by light microscopy. To take micrographs, we introduced the solution (150 mM sorbitol ± inhibitors) into intracellular spaces under weak negative pressure before EL treatment, and removed the upper epidermis from the leaves just before mounting the microscope slide.
3. Results
(a). Kinetics of non-photochemical quenching induction and relaxation in npq4 versus wild-type leaves
Plants lacking PsbS are completely devoid of the fast quenching phase qE when illuminated for 8 min at 1200 μmol photons m−2 s−1 (see electronic supplementary material, figure S1a), consistent with literature data [30]; the fluorescence quenching reached a maximum value four times lower than in WT plants. Dark relaxation of fluorescence quenching was clearly different between genotypes: npq4 showed almost no relaxation, while WT recovered the most fluorescence quenching. Upon exposure to longer periods of actinic light, the quenching behaviour of npq4 mutants showed a relative increase, reaching an NPQ value of 1.9, thus 45% with respect to WT plants (see electronic supplementary material, figure S1b). A major change was observed in the fluorescence recovery of npq4 mutants in the dark, which showed a higher rate, although still slower than in WT plants. Thus, although the rapidly relaxing phase of the dominant qE component was missing in npq4 mutants, a slowly reversible phase contributed to relaxation recovery, not only in WT plants but also in npq4 mutants. Yet, the dark recovery was slower than in WT plants, yielding a higher qI.
The amplitude and kinetics of NPQ induction and relaxation were previously shown to be related to the activity of the xanthophyll cycle in EL [31]. We thus determined for Zea the extent of both EL-formation and dark-reconversion under the same experimental condition used to follow NPQ kinetics. Results in table S1 available in the electronic supplementary material show that the Zea content was the same in WT and npq4 leaves for each condition. Both genotypes retained approximately 50% of the Zea produced upon in EL, upon 1 h dark recovery.
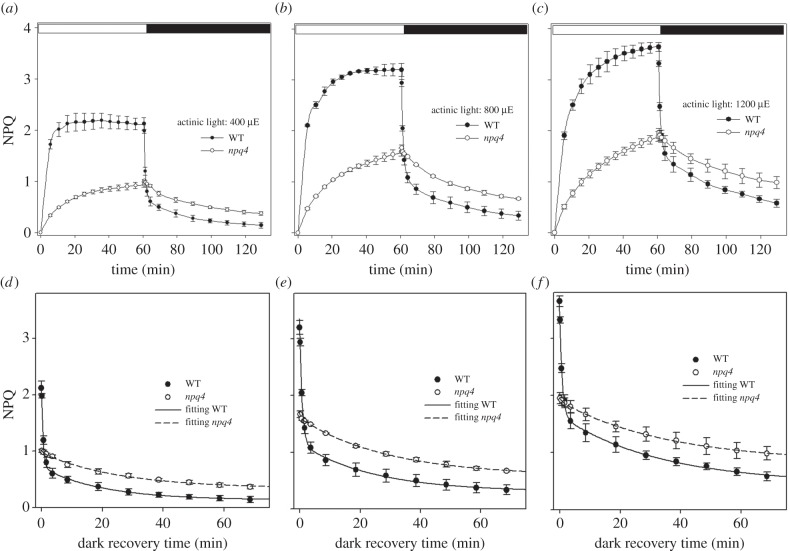
The NPQ kinetics of WT and npq4 leaves were measured at different actinic intensities (figure 1a–c), showing similar behaviour of npq4 leaves at all the light intensities used, including the rise in the light to half the amplitude of the WT leaves followed by a slow dark recovery.
Figure 1.
Kinetic deconvolution of NPQ dark relaxation of WT and npq4 leaves. The dark recovery of NPQ was measured in intact leaves of Arabidopsis WT and npq4 mutants upon illumination for 60 min with different intensities of white actinic light: (a) 400, (b) 800, (c) 1200 µmol photons m−2 s−1. Symbols and error bars show mean ± s.d. (n = 3). Each dataset was fitted with a bi-exponential function NPQ = AqI + AqE e(−t/τqE) + AqM e(−t/τqM) (d–f) and the kinetics of qE and qM relaxation were assessed in the different genotypes (table 1).
The kinetic analysis of NPQ dark relaxation is frequently used as a tool for the characterization of quenching dynamics [15,32]. Thus, NPQ relaxation kinetics (figure 1d–f) were fitted with a bi-exponential decay function: NPQ = A1 e(−t/τ1) + A2 e(−t/τ2) + A3. In WT plants, a fast decay component (τ1, half-life of 35–50 s) represented the dominant component of NPQ under all light conditions (around 60% of total quenching), and could reasonably be attributed to qE. The residual quenching at the end of dark recovery (A3) was due to processes that were essentially irreversible within the time range of the experiment (τ > 60 min), fitting with the characteristics of the photo-inhibitory quenching, qI [33]. In npq4 plants, the kinetics of NPQ relaxation were slowed down with respect to WT plants: a satisfactory fitting of NPQ dark relaxation could only be obtained with the introduction of an intermediate component with a decay rate intermediate between those of qE and qI (τ2, half-life of 20–35 min). This component also improved the fitting of WT curves where it contributes to about 30% of total NPQ (figure 1d–f and table 1). From now on this component will be referred to as qM. The rapid (qE) kinetic component was missing in npq4 mutants under all experimental conditions explored in this work. According to the former attributions, exponential parameters used for fitting of NPQ dark relaxation will be hereafter called AqE and τqE (qE amplitude and half-life, respectively), AqM and τqM (qM amplitude and half-life), AqI (qI amplitude).
Table 1.
Kinetics of NPQ dark relaxation in WT and npq4 leaves. The dark-recovery of NPQ was measured in intact leaves of Arabidopsis WT and npq4 mutants upon 60 min illumination with different actinic intensities (400, 800 and 1200 µmol photons m−2 s−1, white light, RT). Each dataset was fitted with a bi-exponential function NPQ = AqI + AqE e(−t/τqE) + AqM e(−t/τqM) and the kinetics of qE and qM relaxation were assessed in the different genotypes by comparing amplitudes of parameter A, which describes the slope of exponential functions. Significantly different values (Student's t-test) with respect to WT are marked with asterisks.
| actinic intensity (µmol photons m−2 s−1) | genotype | τqE (min) | AqE | τqM (min) | AqM | AqI |
|---|---|---|---|---|---|---|
| 400 | WT | 0.56 ± 0.02 | 1.26 ± 0.05 | 19.9 ± 4.4 | 0.58 ± 0.04 | 0.14 ± 0.13 |
| npq4 | — | — | 25.7 ± 3.9 | 0.63 ± 0.03 | 0.39 ± 0.07* | |
| 800 | WT | 0.83 ± 0.04 | 1.73 ± 0.08 | 24.3 ± 6.8 | 0.85 ± 0.07 | 0.33 ± 0.09 |
| npq4 | 29.3 ± 2.2 | 1.02 ± 0.03* | 0.67 ± 0.02* | |||
| 1200 | WT | 0.81 ± 0.04 | 1.65 ± 0.08 | 33.3 ± 8.4 | 1.20 ± 0.10 | 0.58 ± 0.08 |
| npq4 | 35.8 ± 12.8 | 1.07 ± 0.13 | 1.01 ± 0.13* |
Interestingly, the amplitude and half-life of the intermediate phase qM did not significantly differ between WT and npq4 plants at 400 and 1200 μmol photons m−2 s−1, while AqM was only slightly higher in npq4 than WT plants at 800 μmol photons m−2 s−1 (table 1); therefore, these data suggest that the component qM accounts for a fluorescence decay process, common to WT and npq4 plants, activated upon prolonged illumination and distinct from qE and qI.
Increasing the actinic light intensity caused a linear increase of the slowly relaxing component qI (AqI, table 1), that can reasonably be attributed to PSII photoinhibitory processes in both genotypes. In npq4, the parameter Fv/Fm (PSII maximal quantum efficiencies) reached lower values than WT upon illumination at intensities higher than 400 μmol photons m−2 s−1 (table S2 in the electronic supplementary material), thus showing that induction of PSII damage is more pronounced in npq4 plants. However, the extent of PSII photoinhibition at 400 μmol photons m−2 s−1 was nearly identical to WT, which indicates that this light treatment did not cause differential PSII damage in the two genotypes; moreover, qM amplitude and kinetics were similar in WT and npq4 plants using this light regime (table 1), thus showing that the middle decay phase is not influenced by qE and is not related to PSII photoinhibition. Therefore, the molecular basis of the component qM was further investigated under the optimal NPQ induction conditions of 400 μmol photons m−2 s−1, 60 min.
(b). Effect of ΔpH and D1 turnover on qM
Build-up of trans-thylakoid ΔpH is the key event that triggers NPQ; indeed, ΔpH collapses upon treatment with the ionophore nigericin and the activation of thermal energy dissipation is prevented. Npq4 leaves vacuum-infiltrated with the ionophore still maintained a rise of NPQ in the light, whose amplitude was comparable to that of untreated npq4 leaves (figure 2a); nevertheless, NPQ relaxation in the dark was missing, thus suggesting that the quenching was mainly because of photoinhibition of PSII reaction centres (table 2). A recent report [34] suggested that the slow phase of NPQ relaxation in the dark would reflect the consumption of ATP accumulated in the light phase. To test this possibility, amplitude and kinetics of qM in npq4 leaves were measured upon inhibition of the respiratory chain with myxothiazol: upon this treatment, cytoplasm is depleted in ATP in the dark, therefore acting as a sink for chloroplastic ATP, and accelerating NPQ relaxation [34]. However, myxothiazol treatment did not affect either qM amplitude or its half-life (figure 2b and table 2), thus indicating that the middle phase of NPQ decay was not related to slow ΔpH relaxation.
Figure 2.
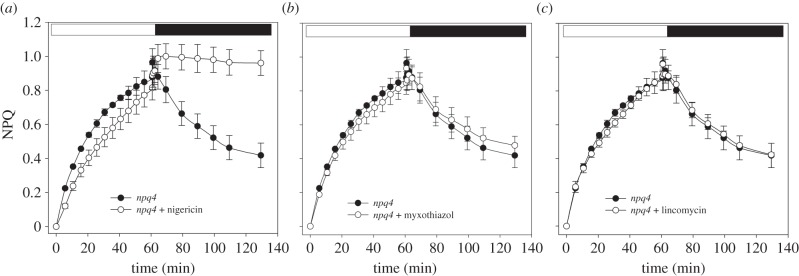
NPQ analysis of npq4 leaves upon inhibition of trans-thylakoid ΔpH, PSII repair mechanism or mitochondrial ATP production. Kinetics of NPQ induction and relaxation were measured in dark-adapted leaves, upon 60 min illumination at 400 µmol photons m−2 s−1, followed by a further 60 min of dark relaxation, in the absence or presence of 50 µM nigericin (a), 2 µM myxothiazol (b) or 100 µM lincomycin (c). Symbols and error bars show mean ± s.d. (n = 3).
Table 2.
Kinetics of qM dark relaxation in npq4 leaves. The kinetic components qM and qI were deconvolved from dark recovery of NPQ in npq4 leaves. Dark-adapted leaves were exposed to white actinic light for 60 min at 400 µmol photons m−2 s−1, RT, following 60 min of dark recovery. (upper section) Before NPQ induction, leaves were vacuum-infiltrated with 150 mM sorbitol and either 50 µM nigericin (uncoupler, collapsed the ΔpH across the thylakoid membranes), 2 µM myxothiazol (respiratory chain inhibitor) or 100 µM lincomycin (chloroplast protein biosynthesis inhibitor). (middle section) The kinetic components qM and qI were measured in npq4 double mutants lacking zeaxanthin (npq4npq1) or lacking lutein (npq4lut2). (lower section) Components qM and qI were measured in npq4 double mutants depleted of Lhcb subunits CP26 and CP24 (npq4koLhcb5/6), CP29 and CP24 (npq4koLhcb4/6), lack the entire LHC (npq4ch1), or unable to activate state transition (npq4stn7) or chloroplast avoidance movement (npq4phot2). Each dataset was fitted with an exponential function NPQ = AqI + AqM e(−t/τqM) and the kinetics of qM relaxation were assessed in the different samples by comparing amplitudes of parameters A. Significantly different values (Student's t-test) with respect to WT are marked with asterisks.
| τqM (min) | AqM | AqI | |
|---|---|---|---|
| npq4 | 28.4 ± 4.5 | 0.63 ± 0.03 | 0.35 ± 0.03 |
| npq4 + nigericin | — | — | 0.90 ± 0.03* |
| npq4 + myxothiazol | 34.7 ± 13.9 | 0.52 ± 0.08 | 0.40 ± 0.09 |
| npq4 + lincomycin | 37.3 ± 6.3 | 0.58 ± 0.04 | 0.39 ± 0.04 |
| npq4npq1 | 15.7 ± 1.3* | 0.56 ± 0.02 | 0.34 ± 0.01 |
| npq4lut2 | 27.7 ± 3.1 | 0.80 ± 0.03* | 0.33 ± 0.03 |
| npq4koLhcb5/6 | 22.8 ± 3.4 | 0.49 ± 0.03 | 0.28 ± 0.03 |
| npq4koLhcb4/6 | 20.7 ± 3.5 | 0.49 ± 0.03 | 0.29 ± 0.03 |
| npq4ch1 | 14.3 ± 1.2* | 0.71 ± 0.03* | 0.11 ± 0.03* |
| npq4stn7 | 14.2 ± 1.9* | 0.55 ± 0.04 | 0.37 ± 0.06 |
| npq4phot2 | 15.3 ± 5.8* | 0.28 ± 0.05* | 0.27 ± 0.09* |
We further tested the possibility that turnover of D1 could account for qM; thus NPQ kinetics were measured on npq4 leaves upon treatment with lincomycin, a plastid protein synthesis inhibitor. Results (figure 2c and table 2) show that lincomycin treatments failed to affect NPQ rise and decay in npq4 leaves, indicating that turnover of the PSII reaction centre did not significantly contribute to qM.
To further investigate the molecular basis of qM, we used a targeted reverse genetic approach: we constructed a series of double and triple mutants combining npq4 with mutations affecting mechanisms that are known to influence Chl fluorescence yield in vivo.
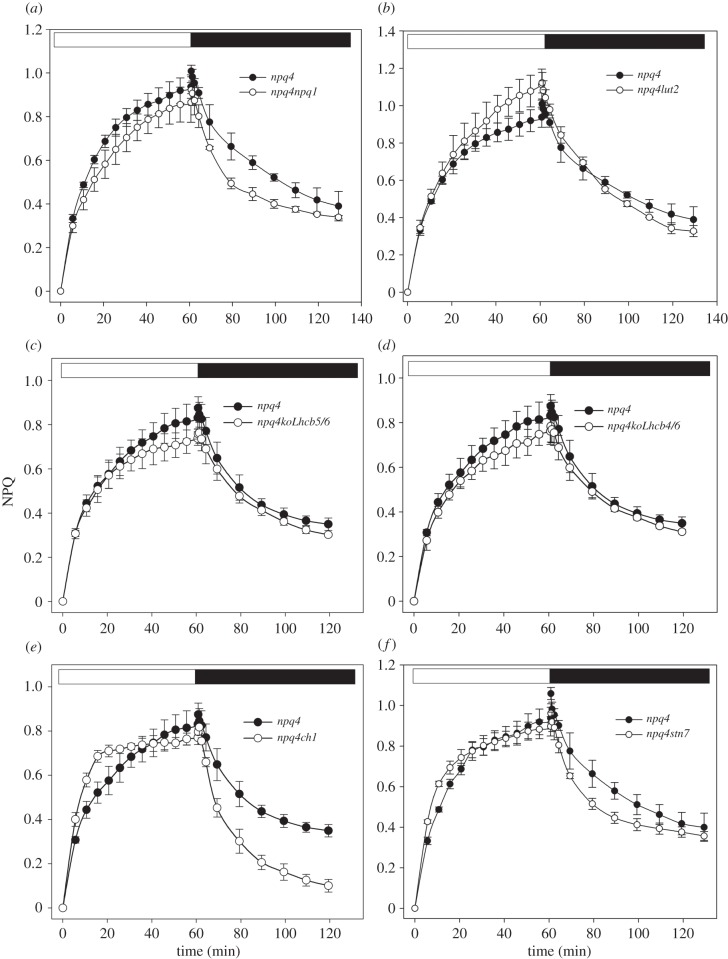
(c). Role of xanthophyll composition
Induction and relaxation of NPQ were measured on Arabidopsis npq4 mutants with altered xanthophyll composition (figure 3). The npq1 mutant lacks violaxanthin de-epoxidase activity and is thus unable to convert Viola into Zea; qE in npq1 has approximately 40% amplitude with respect to WT [12], showing that Zea synthesis is needed for full expression of qE. A comparison of quenching dynamics showed that qM kinetics are very similar in npq4 and npq4npq1 plants (figure 3a): although npq4 showed a somewhat more rapid NPQ rise within the first minutes of illumination, prolonged treatment lead to overlapping amplitude in both genotypes (table 2); likewise, npq4npq1 plants showed a dark recovery which was initially faster than in npq4, while the extent of reversible NPQ was similar in the two genotypes at the end of dark period. The half-life of fluorescence dark recovery is lower in npq4npq1 than in npq4, suggesting that residual Zea in the dark could account for the different kinetics in dark recovery.
Figure 3.
NPQ analysis of npq4 leaves with altered xanthophyll compositions or depleted of either PSII LHCs or state transition. NPQ induction and relaxation were measured in dark-adapted npq4 plants lacking zeaxanthin (npq4npq1 (a)) or lutein (npq4lut2 (b)), or devoid of either minor Lhcb CP26 and CP24 (koLhcb5/6 (c)), CP29 and CP24 (koLhcb4/6 (d)) or the entire LHC owing to mutation (ch1 (e)), or blocked in state transitions (npq4stn7 (f)). NPQ kinetics were measured in dark-adapted leaves, upon 60 min illumination at 400 µmol photons m−2 s−1, followed by further 60 min of dark relaxation. Symbols and error bars show mean ± s.d. (n = 3).
Long-term NPQ measurements were performed on lut2, an Arabidopsis mutant devoid of lutein (figure 3b). Lut, together with Zea, affects quenching dynamics by modulating qE [35]. Prolonged illumination leads to overlapping NPQ traces in npq4 and npq4lut2, and qM half-life and amplitude were very similar in both genotypes (table 2), implying that qM is not affected by Lut depletion. In conclusion, the analysis of NPQ dark relaxation kinetics in npq4 double mutants indicates that qM dynamics were only slightly affected, if at all, by mutations in xanthophyll composition.
(d). Role of light-harvesting complexes and state 1–state 2 transitions
As NPQ depends on the antenna proteins [14,22,23,36], we evaluated the capacity of npq4 mutants, devoid of specific LHC gene products, to modulate qM.
In mutants devoid of both Lhcb5 and Lhcb6 subunits (figure 3c and table 2), the amplitude and relaxation of NPQ were essentially the same as observed in npq4 mutants. Similar results were obtained upon removal of both Lhcb4 and Lhcb6 (figure 3d and table 2), thus ruling out the possibility that minor antennae modulate qM amplitude and kinetics. These data are consistent with the behaviour of npq4ch1 that lacks Chlb, and is thus devoid of all LHCs [37,38]; the slow phase of NPQ relaxation was found to be independent of LHC composition, indeed qM amplitude was similar in npq4 and npq4ch1 mutants (figure 3e and table 2). State transitions lead to quenching of LHCII fluorescence by PSI [39] upon phosphorylation of LHCII, by STN7 kinase, driving its migration from PSII to PSI [40]; by using the mutant npq4stn7, we checked the possibility that state transition were involved in qM. The maximal amplitude of NPQ in npq4stn7 was essentially the same as in npq4 plants (figure 3f). The only difference was found in the kinetics of qM dark-recovery, which was faster in the double mutant than in the npq4 mutant (τ about 15 min in npq4stn7 versus 29 min in npq4; table 2).
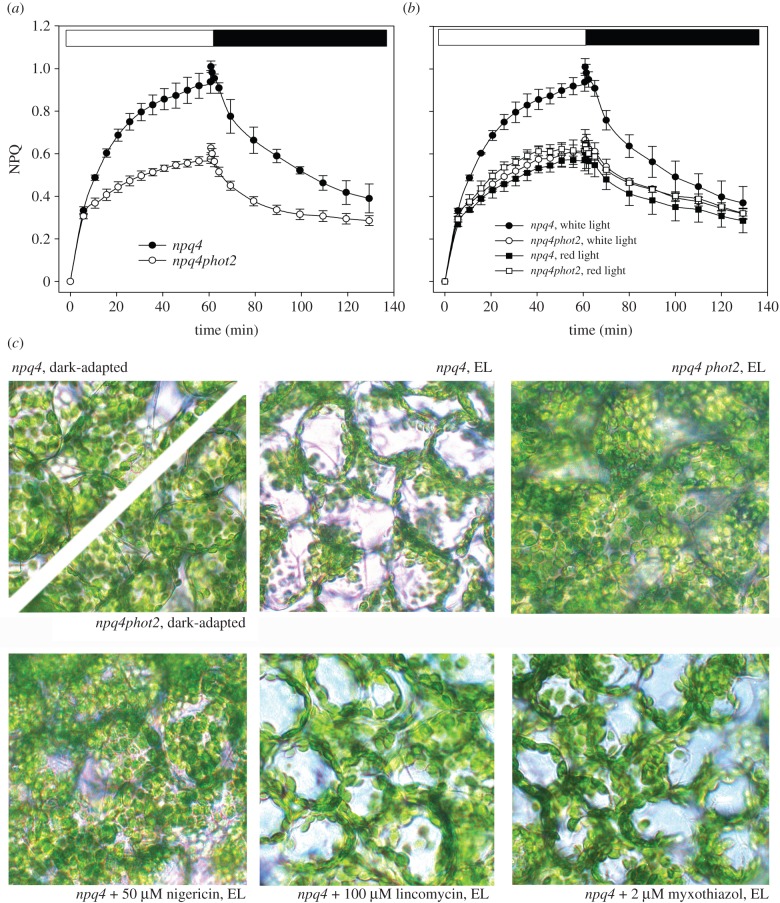
(e). Role of chloroplast photorelocation
Previous reports showed that light-induced chloroplast movements could affect Chl fluorescence emission [41,42]. We thus determined the qM relaxation kinetics in a double mutant npq4phot2, lacking the phototropin PHOT2 which activates the blue-light-dependent chloroplast avoidance movement [24].
Although two different components of NPQ dark-relaxation (qM and qI) were still detected in npq4phot2, the kinetics of the qM component were faster than in npq4 (figure 4a and table 2), and the amplitude of the middle phase was decreased by 50% compared with npq4.
Figure 4.
NPQ analysis of npq4 leaves depleted of chloroplast avoidance movement. (a) PsbS-minus plants were crossed with Arabidopsis knock-out lines lacking the photoprotective mechanism of chloroplast avoidance movement (phot2). Kinetics of NPQ induction and relaxation of npq4 and npq4phot2 were measured in dark-adapted leaves, as described for figure 3. (b) NPQ kinetics were measured on npq4 and npq4phot2 leaves upon illumination with either white actinic light (400 μmol photons m−2 s−1) or red light (350 μmol photons m−2 s−1, 600 < λ < 750 nm). Symbols and error bars show mean ± s.d. (n = 3). (c) Distribution of chloroplasts in the mesophyll cells of npq4 and npq4phot2 was determined by light microscopy. Leaves were dark-adapted for 1 h and then irradiated with white light at 400 µmol m−2 s−1 for 1 h. Prior to light treatment, detached leaves were infiltrated with 150 mM sorbitol containing either 50 μM nigericin, 100 μM lincomycin or 2 μM myxothiazol. (Online version in colour.)
To further verify the hypothesis that phototropins were involved in the modulation of qM, we repeated NPQ measurements by using red light (600 < λ < 750 nm, 350 μmol photons m−2 s−1, 23°C) as actinic source. Results are shown in figure 4b: both npq4 and npq4phot2 matched the kinetics of npq4phot2 in white light, implying that a specific fluorescence decay component is activated by actinic light λ < 600 nm but not by illumination with red light, and affects the amplitude of qM specifically. This point was further studied through the analysis of Chl fluorescence parameters 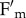 and
and 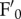 during photosynthesis: we found that, upon illumination with white light at 400 µmol photons m−2 s−1, npq4 leaves underwent a decrease in both
during photosynthesis: we found that, upon illumination with white light at 400 µmol photons m−2 s−1, npq4 leaves underwent a decrease in both 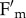 and
and 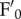 as the result of change in both quantum yield and leaf transmittance, respectively, while in npq4phot2 only
as the result of change in both quantum yield and leaf transmittance, respectively, while in npq4phot2 only 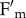 decreased upon irradiation (see electronic supplementary material, figure S2).
decreased upon irradiation (see electronic supplementary material, figure S2).
Thus, the chloroplast avoidance response is, among all the mechanisms examined, the only significant modulator of the concentration of Chl excited states independent of PsbS. To confirm the effect of the phot2 mutation on the npq4 background, we examined leaf cells by light microscopy (figure 4c). Upon irradiation with white light at 400 µmol photons m−2 s−1, npq4 leaves showed chloroplasts located at the anticlinal cell walls, thus indicating that they had undergone avoidance movement. Instead, the chloroplasts of the mutant npq4phot2 retained their preferential association with the periclinal walls, as in leaves adapted to darkness for 1 h. Treatment with nigericin blocks the chloroplast avoidance response (figure 4c), consistent with the complete depletion of qM after treatment with the ionophore (figure 2a).
In conclusion, the analyses of quenching relaxation dynamics on a number of mutants in the npq4 genetic background, identified a kinetically intermediate component of fluorescence decay, distinct from either qE or qI, called qM, the triggering of which requires uniquely formation of a transmembrane proton gradient, but which is not related to xanthophyll or LHC composition, PSII turnover, consumption of ATP accumulated in the light phase or state transitions. Based on its relaxation kinetics, the mechanism of chloroplast photorelocation accounts for nearly 50% of qM amplitude.
4. Discussion
Here, we have investigated the light-induced decline of Chl fluorescence and its relaxation dynamics in the npq4 mutant of Arabidopsis, lacking the PsbS subunit essential for qE activity, in order to assess the basis for its residual light-induced fluorescence decline activity. Kinetic analysis of fluorescence dark recovery in vivo allowed an NPQ component to be identified which relaxes in the dark within the time range 16–25 min, intermediate between the fast qE component (1–2 min) and inhibitory quenching qI (more than 1 h). This component, qM, showed similar amplitude and half-life in WT and npq4 plants (figure 1 and table 1) and is uncoupler-sensitive (figure 2a). To search for the molecular basis of this process, the npq4 genotype was crossed with others, which affected photosynthetic components and mechanisms known to alter the characteristics of light-induced Chl fluorescence changes, and analysed the fluorescence quenching in each.
(a). The kinetic components of non-photochemical quenching dark relaxation in npq4
Analysis of NPQ dark relaxation of WT and npq4 leaves identified three distinct kinetic components (figure 1 and table 1). The rapid phase (half-life 35–55 s) detected in WT but not in npq4 leaves, can be safely assigned to qE [8]. The long-term relaxing component, whose half-life is longer than 60 min, can be assigned to photoinhibitory processes based on its amplitude dependence of photon fluency and increased incidence in npq4 [13]. The third, intermediate kinetic component (half-life 20–35 min), can be detected with similar amplitude in both WT and npq4 relaxation kinetics. Although the existence of a middle-phase kinetic component of NPQ has been reported previously [8,43], its physiological origin is still debated.
The middle phase component, qM, is saturated at moderate light intensity, maintaining the same amplitude at 800 and 1200 µmol photons m−2 s−1 (table 1). This suggests that qM is not related to PSII photoinhibition, as the amplitude of a photoinhibitory component is expected to increase with irradiance; indeed, the component here defined as qI increases from 0.67 at 800 µmol photons m−2 s−1 to 1.01 at 1200 µmol photons m−2 s−1 in npq4 (figure 1 and table 1). At all light regimes tested, the npq4 leaves showed higher qI than WT leaves, consistent with the photoprotective role of qE in short-term exposure to EL [44].
We used an actinic intensity of 400 µmol photons m−2 s−1 as at this irradiance qM is almost saturated and photoinhibition in npq4 is as low as in the WT leaves (table S2 available in the electronic supplementary material). Lincomycin treatment failed to affect NPQ decay in npq4 leaves, implying that at 400 µmol photons m−2 s−1 turnover of D1 did not significantly contribute to qM.
A slow phase of NPQ dark relaxation was reported to depend on the hydrolysis of ATP accumulated during a light phase [34]. However, myxothiazol treatment did not affect qM in npq4 (figure 2b and table 2), thus ruling out the possibility that the qM decay component was related to slow ΔpH relaxation.
It is interesting to note that qM is sensitive to uncouplers: indeed, dark relaxation of fluorescence decline is prevented in leaves infiltrated with the ionophore nigericin. Besides the loss of qM, nigericin led to a strong increase in qI amplitude, implying that both qE and qM are crucial for PSII photoprotection in EL conditions (figure 2a). The intermediate component has been previously defined as qT or qZ for its possible dependence on state transitions or Zea biosynthesis, respectively [8,15,43]. However, data reported here show that blocking these processes with specific mutations does not interfere with qM as determined in the npq4 strain. This mutant was chosen to avoid overlapping contribution of qE to the dark relaxation dynamics. In npq4, we could not detect qE type quenching (τ < 5 min) under any conditions, even upon 60 min of illumination (table 1).
(b). Targeted reverse genetic analysis to identify the molecular basis for qM
NPQ kinetic of nigericin-treated leaves demonstrated that triggering of qM requires transmembrane proton gradient formation during the light phase. Among the effects of EL treatment is thylakoid lumen acidification and Zea synthesis, which are needed for full expression of qE in Arabidopsis [31]. Instead, the double mutant npq4npq1, which is depleted in both qE and Zea, showed the same kinetics and amplitude of qM as npq4 (figure 3a). Therefore, unlike qE, Zea depletion did not prevent full expression of qM, which indeed reaches maximum value (although much lower than in WT) in both npq4 and npq4npq1 plants (table 2). The kinetics of Zea epoxidation in npq4 do not fit with those of NPQ dark recovery, the former being far slower than the kinetic relaxation of qM [15]. We conclude that the xanthophyll cycle, one of the most efficient modulators of qE [14,31], does not affect the amplitude of qM but only slowed the dark relaxation rate (figure 3a). Similar considerations can be applied to Lut; indeed, the amplitude and kinetics of qM are essentially the same in both npq4 and npq4lut2 plants (figure 3b). qE is located in the antenna system, and an important role in NPQ was attributed to Lhcb4 and Lhcb5 [36,45]. Nevertheless, light-induced fluorescence decline was essentially the same in npq4, npq4koLhcb4/6 and npq4koLhcb5/6 leaves. Moreover, no significant differences were found when comparing npq4 versus npq4ch1 leaves depleted in all LHC proteins, including LHCII (table 2) [37], suggesting that these changes were not the result of qE type quenching. Finally, we examined the hypothesis of the involvement of state 1–state 2 transitions in PsbS-independent fluorescence decline; however, the similar behaviour of npq4 and npq4stn7 leaves (figure 3f) excludes this possibility.
(c). Chloroplast avoidance response and qM
Among all the mutations introduced into the npq4 genetic background, phot2 was the only one that affected qM (figure 4a). Differences in npq4 versus npq4phot2 NPQ kinetics showed that the fluorescence recovery component is affected by chloroplast photorelocation. This process is mediated by the blue light receptors, phototropins. Consistently, we verified that the same effect on NPQ kinetics was obtained by using red light rather than white actinic light (figure 4b). Moreover, upon illumination with white light at 400 µmol photons m−2 s−1, npq4 leaves underwent a decrease in both 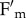 and
and 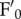 , as expected for concomitant changes in PSII quantum yield and leaf absorption, respectively, while in npq4phot2 leaves only
, as expected for concomitant changes in PSII quantum yield and leaf absorption, respectively, while in npq4phot2 leaves only 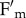 was decreased upon irradiation (see the electronic supplementary material, figure S2). Thus, an avoidance response, causing chloroplast movement towards cell walls parallel to incident light, affects the apparent kinetics of NPQ, particularly the slower components. This is consistent with reports showing that light-induced chloroplast movements could affect Chl fluorescence emission [41], and with recent results which highlighted, in phot2, the lack of a fluorescence decay kinetic component, qM, intermediate between qE and qI [46]. The similar amplitude and half-life of the qM component in WT and npq4 leaves (table 1), and its reduction in a mutant devoid of chloroplast photorelocation (table 2; see also [46]), strongly support the view that chloroplast relocation significantly influences the apparent kinetics of NPQ by decreasing the photon absorption of leaves, rather than changing the activity of quenching reactions.
was decreased upon irradiation (see the electronic supplementary material, figure S2). Thus, an avoidance response, causing chloroplast movement towards cell walls parallel to incident light, affects the apparent kinetics of NPQ, particularly the slower components. This is consistent with reports showing that light-induced chloroplast movements could affect Chl fluorescence emission [41], and with recent results which highlighted, in phot2, the lack of a fluorescence decay kinetic component, qM, intermediate between qE and qI [46]. The similar amplitude and half-life of the qM component in WT and npq4 leaves (table 1), and its reduction in a mutant devoid of chloroplast photorelocation (table 2; see also [46]), strongly support the view that chloroplast relocation significantly influences the apparent kinetics of NPQ by decreasing the photon absorption of leaves, rather than changing the activity of quenching reactions.
It should be noted, however, that this effect arises from decreased photon absorption which gives a lower fluorescence yield, rather than from a genuine quenching process. In fact, during illumination, chloroplast relocation induces a change in the distribution of pigments within the cell, with the formation of localized chloroplast stacks along anticlinal cell walls. This effect reduces the overall optical density of the cell because of a ‘sieve effect’ resulting from the formation of highly transmitting paths across the periclinal cell surfaces while increasing the optical density beyond linearity in the vicinity of the anticlinal cell walls [47]. Thus, the fraction of excited, fluorescence emitting chloroplasts is decreased because of shading by neighbouring, ones. It is worth noting that the kinetics and timescale of qM formation and relaxation at 400 μmol photons m−2 s−1 of white light (table 1) fits with that described for the chloroplast avoidance response under similar irradiance [48]. Light microscopy analysis (figure 4c) confirmed that movement of chloroplasts was inhibited in the presence of nigericin, consistent with the depletion in qM. The chloroplast avoidance response probably relies on the cytosolic Ca2+ signal for its activation [49]. Maintenance of a low cytosolic Ca2+ level requires an electrogenic pump which exploits, protonmotive force to actively extrude Ca2+ [50]. Phototropin signal transduction involves transient depolarization of the plasma membrane which, in turn, triggers cytosolic Ca2+ intake. Nigericin wrecks all the transmembrane electrochemical gradients, thus blocking several signal transduction events. The double effect of nigericin in collapsing the thylakoid pH gradient and in blocking chloroplast relocation (figure 4c) can easily lead to misinterpretation of qM as a slow qE response in the absence of PsbS.
Although chloroplast relocation is the major factor affecting the amplitude of qM in npq4 (figure 4a,b), the fluorescence recovery kinetics of npq4phot2 are not completely devoid of qM. The residual component accounts for about 18% of total reversible  quenching in WT (tables 1 and 2) and reflects mechanism(s) sensitive to uncouplers (figure 2a) and yet distinct from the avoidance response, as it is still active in npq4phot2. Previous work led to different proposals for mechanisms leading to fluorescence recovery components with intermediate half-life between qE and qI. First, it was attributed to state 1–state 2 transitions [43]; second to PSII photoinhibition [51]; third to a slowly developing component of qE dependent on Zea [52]; fourth to light-induced dissociation of the complex Lhcb4-Lhcb6-LHCII-M [19]. Here, we show that qM did not correlate with Zea accumulation nor was it related to qI. Although thylakoid membrane reorganization could well explain changes in chloroplast fluorescence yield since protein–protein interactions are responsible for nearly 50% of quenching [14,19], the need for PsbS to trigger domain reorganization [19,20] suggests this is not the source of residual qM. An interesting observation is that a substantial fraction of qM is retained in the absence of Lhcb in the npq4ch1 mutant (figure 3e), although with a somewhat shorter half-life (table 2). This is consistent with the characteristics of Zea-independent NPQ localized in the PSII core complex [53].
quenching in WT (tables 1 and 2) and reflects mechanism(s) sensitive to uncouplers (figure 2a) and yet distinct from the avoidance response, as it is still active in npq4phot2. Previous work led to different proposals for mechanisms leading to fluorescence recovery components with intermediate half-life between qE and qI. First, it was attributed to state 1–state 2 transitions [43]; second to PSII photoinhibition [51]; third to a slowly developing component of qE dependent on Zea [52]; fourth to light-induced dissociation of the complex Lhcb4-Lhcb6-LHCII-M [19]. Here, we show that qM did not correlate with Zea accumulation nor was it related to qI. Although thylakoid membrane reorganization could well explain changes in chloroplast fluorescence yield since protein–protein interactions are responsible for nearly 50% of quenching [14,19], the need for PsbS to trigger domain reorganization [19,20] suggests this is not the source of residual qM. An interesting observation is that a substantial fraction of qM is retained in the absence of Lhcb in the npq4ch1 mutant (figure 3e), although with a somewhat shorter half-life (table 2). This is consistent with the characteristics of Zea-independent NPQ localized in the PSII core complex [53].
Our results support the view that no qE occurs in npq4 leaves within a wide range of actinic light intensities. Moreover light-induced fluorescence decline in npq4 was always far lower than in WT plants, even upon 1 h of EL exposure (figure 1). Finally, the residual fluorescence decline in npq4 leaves is due to avoidance of photon absorption, while quenching mechanisms can only be responsible for a minor component associated with the PSII core (figure 3e), rather than to reactions within the antenna system. Our results significantly differ from those of Johnson & Ruban [21], who reported that qE could be catalysed, although at a slower rate, in npq4 plants. First, we found that the amplitude of fluorescence decline in npq4 leaves did not match that observed in WT under any conditions, the fraction of reversible  quenching in the absence of PsbS and triggered by lumen acidification being small (about 18%) (table 1). The photoprotective effect of fluorescence decline was consistently low, as shown from the higher amplitude of qI in npq4 leaves under EL conditions (table 1). Second, we found that uncouple sensitivity is the result of the disruption of chloroplast relocation, also involving a proton gradient for signal transduction upon blue light activation of phototropins [49]. Overall, these results point to a crucial role of PsbS in the modulation of NPQ and show that sensing of trans-thylakoid ΔpH by protonable residues in the LHC is not enough to induce WT levels of NPQ in the absence of PsbS. This conclusion is consistent with recent results showing that less than 0.5% of purified minor antennae underwent charge transfer quenching in vitro, whereas the fraction engaged in this process was more than 80 times higher in intact thylakoids with PsbS [36]. Thus, PsbS is indispensable for qE, within the trans-thylakoid ΔpH that can be obtained by light treatment of leaves, rather than being only a modulator of the proton–antenna association constant, pK, of qE activation [54].
quenching in the absence of PsbS and triggered by lumen acidification being small (about 18%) (table 1). The photoprotective effect of fluorescence decline was consistently low, as shown from the higher amplitude of qI in npq4 leaves under EL conditions (table 1). Second, we found that uncouple sensitivity is the result of the disruption of chloroplast relocation, also involving a proton gradient for signal transduction upon blue light activation of phototropins [49]. Overall, these results point to a crucial role of PsbS in the modulation of NPQ and show that sensing of trans-thylakoid ΔpH by protonable residues in the LHC is not enough to induce WT levels of NPQ in the absence of PsbS. This conclusion is consistent with recent results showing that less than 0.5% of purified minor antennae underwent charge transfer quenching in vitro, whereas the fraction engaged in this process was more than 80 times higher in intact thylakoids with PsbS [36]. Thus, PsbS is indispensable for qE, within the trans-thylakoid ΔpH that can be obtained by light treatment of leaves, rather than being only a modulator of the proton–antenna association constant, pK, of qE activation [54].
References
- 1.Croce R, Van Amerongen H. 2011. Light-harvesting and structural organization of photosystem II: from individual complexes to thylakoid membrane. J. Photochem. Photobiol. B 104, 142–153. ( 10.1016/j.jphotobiol.2011.02.015) [DOI] [PubMed] [Google Scholar]
- 2.Krieger-Liszkay A. 2005. Singlet oxygen production in photosynthesis. J. Exp. Bot. 56, 337–346. ( 10.1093/jxb/erh237) [DOI] [PubMed] [Google Scholar]
- 3.Alboresi A, Dall'Osto L, Aprile A, Carillo P, Roncaglia E, Cattivelli L, Bassi R. 2011. Reactive oxygen species and transcript analysis upon excess light treatment in wild-type Arabidopsis thaliana vs a photosensitive mutant lacking zeaxanthin and lutein. BMC. Plant Biol. 11, 62 ( 10.1186/1471-2229-11-62) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wada M, Kagawa T, Sato Y. 2003. Chloroplast movement. Ann. Rev. Plant Biol. 54, 455–468. ( 10.1146/annurev.arplant.54.031902.135023) [DOI] [PubMed] [Google Scholar]
- 5.Niyogi KK. 2000. Safety valves for photosynthesis. Curr. Opin. Plant Biol. 3, 455–460. ( 10.1016/S1369-5266(00)00113-8) [DOI] [PubMed] [Google Scholar]
- 6.Dall'Osto L, Holt NE, Kaligotla S, Fuciman M, Cazzaniga S, Carbonera D, Frank HA, Alric J, Bassi R. 2012. Zeaxanthin protects plant photosynthesis by modulating chlorophyll triplet yield in specific light-harvesting antenna subunits. J. Biol. Chem. 287, 41 820–41 834. ( 10.1074/jbc.M112.405498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulheim C, Agren J, Jansson S. 2002. Rapid regulation of light harvesting and plant fitness in the field. Science 297, 91–93. ( 10.1126/science.1072359) [DOI] [PubMed] [Google Scholar]
- 8.Walters RG, Horton P. 1991. Resolution of components of non-photochemical chlorophyll fluorescence quenching in barley leaves. Photosynth. Res. 27, 121–133. ( 10.1007/BF00033251) [DOI] [PubMed] [Google Scholar]
- 9.Briantais J-M, Vernotte C, Picaud M, Krause GH. 1980. Chlorophyll fluorescence as a probe for the determination of the photoinduced proton gradient in isolated chloroplasts. Biochim. Biophys. Acta 591, 198–202. ( 10.1016/0005-2728(80)90233-9) [DOI] [PubMed] [Google Scholar]
- 10.Dominici P, Caffarri S, Armenante F, Ceoldo S, Crimi M, Bassi R. 2002. Biochemical properties of the PsbS subunit of photosystem II either purified from chloroplast or recombinant. J. Biol. Chem. 277, 22 750–22 758. ( 10.1074/jbc.M200604200) [DOI] [PubMed] [Google Scholar]
- 11.Wilk L, Grunwald M, Liao PN, Walla PJ, Kuhlbrandt W. 2013. Direct interaction of the major light-harvesting complex II and PsbS in nonphotochemical quenching. Proc. Natl Acad. Sci. USA 110, 5452–5456. ( 10.1073/pnas.1205561110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niyogi KK, Shih C, Chow WS, Pogson BJ, DellaPenna D, Björkman O. 2001. Photoprotection in a zeaxanthin- and lutein-deficient double mutant of Arabidopsis. Photosynth. Res. 67, 139–145. ( 10.1023/A:1010661102365) [DOI] [PubMed] [Google Scholar]
- 13.Anderson JM, Park YI, Chow WS. 1997. Photoinactivation and photoprotection of photosystem II in nature. Physiol. Plant. 100, 214–223. ( 10.1111/j.1399-3054.1997.tb04777.x) [DOI] [Google Scholar]
- 14.Dall'Osto L, Caffarri S, Bassi R. 2005. A mechanism of nonphotochemical energy dissipation, independent from PsbS, revealed by a conformational change in the antenna protein CP26. Plant Cell 17, 1217–1232. ( 10.1105/tpc.104.030601) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nilkens M, Kress E, Lambrev P, Miloslavina Y, Muller M, Holzwarth AR, Jahns P. 2010. Identification of a slowly inducible zeaxanthin-dependent component of non-photochemical quenching of chlorophyll fluorescence generated under steady-state conditions in Arabidopsis. Biochim. Biophys. Acta 1797, 466–475. ( 10.1016/j.bbabio.2010.01.001) [DOI] [PubMed] [Google Scholar]
- 16.Ahn TK, Avenson TJ, Ballottari M, Cheng YC, Niyogi KK, Bassi R, Fleming GR. 2008. Architecture of a charge-transfer state regulating light harvesting in a plant antenna protein. Science 320, 794–797. ( 10.1126/science.1154800) [DOI] [PubMed] [Google Scholar]
- 17.Ruban AV, et al. 2007. Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature 450, 575–578. ( 10.1038/nature06262) [DOI] [PubMed] [Google Scholar]
- 18.Kruger TPJ, Novoderezhkin VI, Ilioaia C, van Grondelle R. 2010. Fluorescence spectral dynamics of single LHCII trimers. Biophys. J. 98, 3093–3101. ( 10.1016/j.bpj.2010.03.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betterle N, Ballottari M, Zorzan S, de Bianchi S, Cazzaniga S, Dall'Osto L, Morosinotto T, Bassi R. 2009. Light-induced dissociation of an antenna hetero-oligomer is needed for non-photochemical quenching induction. J. Biol. Chem. 284, 15 255–15 266. ( 10.1074/jbc.M808625200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson MP, Goral TK, Duffy CD, Brain AP, Mullineaux CW, Ruban AV. 2011. Photoprotective energy dissipation involves the reorganization of photosystem II light-harvesting complexes in the grana membranes of spinach chloroplasts. Plant Cell 23, 1468–1479. ( 10.1105/tpc.110.081646) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson MP, Ruban AV. 2010. Arabidopsis plants lacking PsbS protein possess photoprotective energy dissipation. Plant J. 61, 283–289. ( 10.1111/j.1365-313X.2009.04051.x) [DOI] [PubMed] [Google Scholar]
- 22.de Bianchi S, Dall'Osto L, Tognon G, Morosinotto T, Bassi R. 2008. Minor antenna proteins CP24 and CP26 affect the interactions between photosystem II subunits and the electron transport rate in grana membranes of Arabidopsis. Plant Cell 20, 1012–1028. ( 10.1105/tpc.107.055749) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Bianchi S, Betterle N, Kouril R, Cazzaniga S, Boekema E, Bassi R, Dall'Osto L. 2011. Arabidopsis mutants deleted in the light-harvesting protein Lhcb4 have a disrupted photosystem II macrostructure and are defective in photoprotection. Plant Cell 23, 2659–2679. ( 10.1105/tpc.111.087320) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M. 2001. Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science 291, 2138–2141. ( 10.1126/science.291.5511.2138) [DOI] [PubMed] [Google Scholar]
- 25.Van Kooten O, Snel JFH. 1990. The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynt. Res. 25, 147–150. ( 10.1007/BF00033156) [DOI] [PubMed] [Google Scholar]
- 26.Gilmore AM, Shinkarev VP, Hazlett TL, Govindjee G. 1998. Quantitative analysis of the effects of intrathylakoid pH and xanathophyll cycle pigments on chlorophyll a fluorescence lifetime distributions and intensity in thylakoids. Biochemistry 37, 13 582–13 593. ( 10.1021/bi981384x) [DOI] [PubMed] [Google Scholar]
- 27.Aro E-M, Virgin I, Andersson B. 1993. Photoinhibition of photosystem II - inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143, 113–134. ( 10.1016/0005-2728(93)90134-2) [DOI] [PubMed] [Google Scholar]
- 28.Garmier M, Carroll AJ, Delannoy E, Vallet C, Day DA, Small ID, Millar AH. 2008. Complex I dysfunction redirects cellular and mitochondrial metabolism in Arabidopsis. Plant Physiol. 148, 1324–1341. ( 10.1104/pp.108.125880) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilmore AM, Yamamoto HY. 1991. Zeaxanthin formation and energy-dependent fluorescence quenching in pea chloroplasts under artificially mediated linear and cyclic electron transport. Plant Physiol. 96, 635–643. ( 10.1104/pp.96.2.635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li XP, Bjorkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK. 2000. A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403, 391–395. ( 10.1038/35000131) [DOI] [PubMed] [Google Scholar]
- 31.Niyogi KK, Grossman AR, Björkman O. 1998. Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10, 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson MP, Davison PA, Ruban AV, Horton P. 2008. The xanthophyll cycle pool size controls the kinetics of non-photochemical quenching in Arabidopsis thaliana. FEBS Lett. 582, 262–266. ( 10.1016/j.febslet.2007.12.016) [DOI] [PubMed] [Google Scholar]
- 33.Krause GH. 1988. Photoinhibition of photosynthesis. An evaluation of damaging and protective mechanisms. Physiol. Plant. 74, 566–574. ( 10.1111/j.1399-3054.1988.tb02020.x) [DOI] [Google Scholar]
- 34.Joliot PA, Finazzi G. 2010. Proton equilibration in the chloroplast modulates multiphasic kinetics of nonphotochemical quenching of fluorescence in plants. Proc. Natl Acad. Sci. USA 107, 12 728–12 733. ( 10.1073/pnas.1006399107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, et al. 2009. Lutein accumulation in the absence of zeaxanthin restores nonphotochemical quenching in the Arabidopsis thaliana npq1 mutant. Plant Cell 21, 1798–1812. ( 10.1105/tpc.109.066571) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Avenson TJ, Ahn TK, Zigmantas D, Niyogi KK, Li Z, Ballottari M, Bassi R, Fleming GR. 2008. Zeaxanthin radical cation formation in minor light-harvesting complexes of higher plant antenna. J. Biol. Chem. 283, 3550–3558. ( 10.1074/jbc.M705645200) [DOI] [PubMed] [Google Scholar]
- 37.Havaux M, Dall'Osto L, Bassi R. 2007. Zeaxanthin has enhanced antioxidant capacity with respect to all other xanthophylls in Arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiol. 145, 1506–1520. ( 10.1104/pp.107.108480) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dall'Osto L, Cazzaniga S, Havaux M, Bassi R. 2010. Enhanced photoprotection by protein-bound vs free xanthophyll pools: a comparative analysis of chlorophyll b and xanthophyll biosynthesis mutants. Mol. Plant 3, 576–593. ( 10.1093/mp/ssp117) [DOI] [PubMed] [Google Scholar]
- 39.Galka P, Santabarbara S, Khuong TTH, Degand H, Morsomme P, Jennings RC, Boekema EJ, Caffarri S. 2012. Functional analyses of the plant photosystem I–light-harvesting complex II supercomplex reveal that light-harvesting complex II loosely bound to photosystem II is a very efficient antenna for photosystem I in state II. Plant Cell 24, 2963–2978. ( 10.1105/tpc.112.100339) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bellafiore S, Barneche F, Peltier G, Rochaix JD. 2005. State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433, 892–895. ( 10.1038/nature03286) [DOI] [PubMed] [Google Scholar]
- 41.Brugnoli E, Bjorkman O. 1992. Chloroplast movements in leaves—influence on chlorophyll fluorescence and measurements of light-induced absorbance changes related to DeltapH and zeaxanthin formation. Photosynth. Res. 32, 23–35. ( 10.1007/BF00028795) [DOI] [PubMed] [Google Scholar]
- 42.Davis PA, Hangarter RP. 2012. Chloroplast movement provides photoprotection to plants by redistributing PSII damage within leaves. Photosynt. Res. 112, 153–161. ( 10.1007/s11120-012-9755-4) [DOI] [PubMed] [Google Scholar]
- 43.Quick WP, Stitt M. 1989. An examination of factors contributing to non-photochemical quenching of chlorophyll fluorescence in barley leaves. Biochim. Biophys. Acta 977, 287–296. ( 10.1016/S0005-2728(89)80082-9) [DOI] [Google Scholar]
- 44.Li XP, Muller-Moule P, Gilmore AM, Niyogi KK. 2002. PsbS-dependent enhancement of feedback de-excitation protects photosystem II from photoinhibition. Proc. Natl Acad. Sci. USA 99, 15 222–15 227. ( 10.1073/pnas.232447699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miloslavina Y, de Bianchi S, Dall'Osto L, Bassi R, Holzwarth AR. 2011. Quenching in Arabidopsis thaliana mutants lacking monomeric antenna proteins of photosystem II. J. Biol. Chem. 286, 36 830–36 840. ( 10.1074/jbc.M111.273227) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cazzaniga S, Dall'Osto L, Kong SG, Wada M, Bassi R. 2013. Interaction between avoidance of photon absorption, excess energy dissipation and zeaxanthin synthesis against photoxidative stress in Arabidopsis. Plant J. 76, 568–579. ( 10.1111/tpj.12314) [DOI] [PubMed] [Google Scholar]
- 47.Davis PA, Caylor S, Whippo CW, Hangarter RP. 2011. Changes in leaf optical properties associated with light-dependent chloroplast movements. Plant Cell Environ. 34, 2047–2059. ( 10.1111/j.1365-3040.2011.02402.x) [DOI] [PubMed] [Google Scholar]
- 48.Kasahara M, Kagawa T, Oikawa K, Suetsugu N, Miyao M, Wada M. 2002. Chloroplast avoidance movement reduces photodamage in plants. Nature 420, 829–832. ( 10.1038/nature01213) [DOI] [PubMed] [Google Scholar]
- 49.Suetsugu N, Wada M. 2007. Chloroplast photorelocation movement mediated by phototropin family proteins in green plants. Biol. Chem. 388, 927–935. ( 10.1515/BC.2007.118) [DOI] [PubMed] [Google Scholar]
- 50.Ettinger WF, Clear AM, Fanning KJ, Peck ML. 1999. Identification of a Ca2+/H+ antiport in the plant chloroplast thylakoid membrane. Plant Physiol. 119, 1379–1386. ( 10.1104/pp.119.4.1379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruban AV, Horton P. 1995. An investigation of the sustained component of nonphotochemical quenching of chlorophyll fluorescence in isolated chloroplasts and leaves of spinach. Plant Physiol. 108, 721–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horton P. 1996. Nonphotochemical quenching of chlorophyll fluorescence. In Light as an energy source and information carrier in plant physiology (ed. Jennings RC.), pp. 99–111. New York, NY: Plenum Press. [Google Scholar]
- 53.Finazzi G, Johnson GN, Dall'Osto L, Joliot P, Wollman FA, Bassi R. 2004. A zeaxanthin-independent nonphotochemical quenching mechanism localized in the photosystem II core complex. Proc. Natl Acad. Sci. USA 101, 12 375–12 380. ( 10.1073/pnas.0404798101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson MP, Ruban AV. 2011. Restoration of rapidly-reversible photoprotective energy dissipation in the absence of PsbS protein by enhanced ΔpH. J. Biol. Chem. 286, 19 973–19 981. ( 10.1074/jbc.M111.237255) [DOI] [PMC free article] [PubMed] [Google Scholar]