Abstract
Photosynthetic eukaryotes house two photosystems with distinct light absorption spectra. Natural fluctuations in light quality and quantity can lead to unbalanced or excess excitation, compromising photosynthetic efficiency and causing photodamage. Consequently, these organisms have acquired several distinct adaptive mechanisms, collectively referred to as non-photochemical quenching (NPQ) of chlorophyll fluorescence, which modulates the organization and function of the photosynthetic apparatus. The ability to monitor NPQ processes fluorometrically has led to substantial progress in elucidating the underlying molecular mechanisms. However, the relative contribution of distinct NPQ mechanisms to variable light conditions in different photosynthetic eukaryotes remains unclear. Here, we present a mathematical model of the dynamic regulation of eukaryotic photosynthesis using ordinary differential equations. We demonstrate that, for Chlamydomonas, our model recapitulates the basic fluorescence features of short-term light acclimation known as state transitions and discuss how the model can be iteratively refined by comparison with physiological experiments to further our understanding of light acclimation in different species.
Keywords: photosynthesis, light acclimation, state transitions, non-photochemical quenching, Chlamydomonas reinhardtii, mathematical modelling
1. Introduction
The light-dependent electron transfer reactions of eukaryotic photosynthesis are catalysed by serially linked protein complexes within the thylakoid membranes of chloroplasts [1–4]. Linear photosynthetic electron flow (LEF) commences with the light-harvesting complex II (LHCII) proteins associated with photosystem II (PSII), which channel light energy to the PSII reaction centre that catalyses the oxidation of water to form molecular oxygen and protons in the thylakoid lumen. The electrons derived from PSII water-splitting are transferred through the thylakoid membrane by plastoquinone (PQ) to the cytochrome b6f complex (Cytb6f). The Cytb6f catalyses the translocation of protons from the thylakoid stroma to lumen while transferring the electrons to PSI via plastocyanin or cytochrome c6. Light-harvesting proteins associated with PSI drive the transfer of these electrons through PSI to ferredoxin, which can subsequently reduce nicotinamide adenine dinucleotide phosphate (NADP+) to NADPH via ferredoxin–NADP reductase. The proton gradient generated by the activities of PSII and Cytb6f powers ATP synthase-catalysed phosphorylation of ADP. Cyclic electron flow (CEF) involves the flow of electrons from ferredoxin to the PQ pool, resulting in an increased acidification of the thylakoid lumen and increased production of ATP at the expense of NADPH [5,6]. A remarkable feature of oxygenic photosynthesis is the extremely wide range of reaction kinetics both within the electron transfer chain and the light-independent reactions of carbon fixation [7]. This is further complicated by the distinct light absorption spectra of PSII and PSI (the latter being excited by longer wavelengths than the former), as unbalanced excitation of the two photosystems can lead to over-reduction (or over-oxidation) of the electron carriers connecting the two photosystems, leading to possible photo-oxidative damage to the cell. To achieve ‘photostasis’ amidst the broad range of light quality and quantity found in nature [7], photosynthetic organisms must balance the harvesting, dissipation and utilization of light energy [8].
The mechanisms of dynamic light acclimation occur across a wide range of time scales. Long-term responses (LTRs) include possible adjustments of photosystem stoichiometries [9] and thylakoid membrane folding [10]. These LTR mechanisms do not require photoreceptors and are thought to be primarily regulated by the thylakoid redox poise, which controls retrograde signalling from the chloroplast to the nucleus [11]. Photoreceptor-dependent LTR mechanisms can affect the repositioning of chloroplasts along light gradients [12,13]. Motile photosynthetic organisms, for example, Chlamydomonas, perceive light via rhodopsins to trigger phototactic responses [14,15]. On shorter timescales, excess energy can be dissipated through the transfer of electrons to O2 via the plastid terminal oxidase [16–18] or at the PSI acceptor side via the Mehler reaction [19]. However, short-term acclimation processes act primarily at the level of PSII and are collectively referred to as non-photochemical quenching (NPQ) of chlorophyll fluorescence [20,21]. NPQ is defined as the difference between the fluorescence maximum of dark-adapted cells and the fluorescence maximum observed during subsequent illumination [20]. The deconvolution of NPQ into three distinct components qE, qT and qI can be derived from the relaxation kinetics of NPQ in the dark [22]. The qE component of NPQ operates within seconds to minutes and is regulated by the lumenal pH [23]. Acidification of the thylakoid lumen stimulates xanthophyll de-epoxidation and/or conformational changes of the PSII antennae resulting in the dissipation of excess energy as heat [24,25]. Lumen acidification also directly regulates photosynthetic electron flow by decreasing the rate of plastoquinol oxidation by Cytb6f [26]. The qT component, also referred to as ‘state transition’, operates on the scale of minutes and involves the reversible association of LHCII proteins with PSII and PSI to regulate the relative light absorption cross section of the two photosystems [27]. The mobilization of LHCII proteins between the photosystems is regulated by antagonistic kinases and phosphatases [28]. The qI component has been ascribed to photoinhibition, wherein photodamaged PSII complexes are partially disassembled and repaired on the scale of hours [29].
Although the molecular mechanisms of light acclimation are becoming clear, the conditions required for their induction and their relative contribution to photostasis across variable light conditions remain poorly understood. Light acclimation processes also differ substantially among photosynthetic species and cell types. For example, plastoquinone terminal oxidase (PTOX)-mediated reduction of O2 is a minor pathway in land plants but can absorb up to 50% of the electrons derived from water-splitting in cyanobacteria and some eukaryotic algae [30,31]. Land plants and algae have distinct molecular mechanisms underlying qE. The PsbS protein is critical for qE in vascular land plants [32] but its role in green algae is unclear [33] and diatoms apparently do not encode this protein [34]. Conversely, the qE mechanism of eukaryotic algae, for example, Chlamydomonas, relies heavily on the light-harvesting complex stress-related (LHCSR) proteins, which are absent in vascular land plants [34]. State transitions also differ substantially between land plants and algae. Whereas land plants mobilize only 15–20% of the LHCII pool, state transitions in the alga Chlamydomonas reinhardtii have been reported to involve up to 80% of the LHCII antennae [35]. Some regulatory features of the LHCII kinase (Stt7/STN7) are shared between algae and land plants, such as its decreased activity under high light stress [36–38] and its activation by the binding of plastoquinol to Cytb6f [39–41]. It is currently unknown whether the LHCII phosphatase PPH1/TAP38 [42,43] is regulated, and specific orthologues have not been characterized outside the land plant lineage.
The analysis of light acclimation mutants has been useful in identifying distinct molecular mechanisms; however, these processes do not operate in isolation and they form an interdependent and complex regulatory network. Indeed, the fine-tuning of LEF and CEF also varies substantially across species and cell types, with CEF reaching high rates in bundle sheath cells of C4 leaves, whereas CEF rarely exceeds 10% in C3 leaves at steady state [44,45]. The relationship between the rate of CEF and NPQ also remains poorly understood. The processes are evidently linked based on the analysis of mutants, which are defective in both processes. For example, mutation of PGR5/PGRL1 in Arabidopsis, two proteins required for the ‘antimycin A-sensitive’ CEF pathway [46], also suppresses qE [47], most probably because of the diminished capacity of lumen acidification in the light. In Chlamydomonas, the analysis of npq4 stt7 double mutants indicates that qE and qT are functionally linked during high light stress [48]. The qT component has also been linked to the regulation of CEF by the analysis of stt7 mutants [49,50] and the enrichment of CEF supercomplexes in conditions favouring the association of mobile LHCII antennae with PSI [51,52]. However, this conclusion has been recently questioned by a study demonstrating that although the migration of LHCII antennae to PSI can enhance CEF in light-limiting conditions, Chlamydomonas cells may switch from low to high rates of CEF independently of the LHCII kinase Stt7 [53].
Mathematical modelling is a useful tool for studying the dynamic behaviour of biological networks. Discrepancies between the predictions of a mathematical model and observations from physiological and biochemical experiments represent gaps in our theoretical understanding. The predictive power of the mathematical model can be iteratively refined by exploring its parameter space and connectivity based on comparison with empirical data [54–56]. Here, we present a mathematical model describing short-term acclimation of the photosynthetic electron transfer chain to changing light. Most of the initial parameters of the model, including reaction kinetics and the variable connectivity of photosynthetic electron transport, were derived from the literature, and the dynamic processes are represented as ordinary differential equations. We demonstrate that our model can reproduce the basic fluorescence measurements during short-term acclimation under conditions promoting state transitions and discuss how the parameter space of the model can be further refined to improve our understanding of the mechanisms of adaptation to changing light.
2. Methods
(a). Experimental
Chlamydomonas reinhardtii cells were grown in acetate medium (TAP) under dim light (10 µE m−2 s−1 from white fluorescent tubes) at 25°C to a density of 2 × 106 cells ml−1. They were collected by centrifugation and resuspended in minimal medium (HSM) at a density of 2 × 107 cells ml−1 and incubated under agitation in dim light for 1 h for anoxia-induced state transitions and in complete darkness for 1 h for light regime-induced state transitions. Fluorescence traces were recorded with a PAM Fluorescence Monitoring System (Hansatech, UK).
(b). Theoretical
The mathematical model as described below and detailed in the electronic supplementary material comprises a set of seven coupled ordinary differential equations. The equations are presented and explained in detail in the electronic supplementary material. The parameters were taken from the literature and subsequently adapted to fit the experimental curves. The numerical simulations of the equations were carried out with MATLAB.
3. Results and discussion
We have built a mathematical model with the goal to provide a theoretical framework in which short-term acclimation processes can be investigated by computer simulations. The purpose of the model is to verify whether current concepts are sufficient to explain experimental observations, to provide a platform to test mechanistic hypotheses and to make theoretical predictions about the dynamics of variable quantities, which cannot be monitored continuously. The model includes all relevant processes involved in the electron transport chain but is deliberately kept as simple as possible. We here focus on the investigation of state transitions in the green alga C. reinhardtii and thus see our work as a continuation of previous theoretical approaches to study NPQ, which addressed in particular the qE component [57,58].
Our experimental data were obtained for Chlamydomonas cells grown in dim light, which do not express LhcSR3 and have little capacity for qE-dependent quenching [59]. This allowed us in the first step to investigate exclusively the qT component and study its dynamics and regulation. A schematic of the model is illustrated in figure 1a. As the model aims at describing adaptation processes in the timescale of seconds to minutes, it does not include a detailed description of the internal processes of the photosynthetic complexes but rather uses a heuristic approach to represent their overall function. Thus, PSII, cytochrome b6f (Cytb6f) and photosystem I (PSI) are treated as oxidoreductases, of which the two photosystems are light-driven. In conjunction, these three complexes mediate linear electron flow from water to ferredoxin. The protons concomitantly translocated from stroma to lumen drive the ATP synthase, generating ATP in the stroma. Electrons from ferredoxin can either be used to produce redox equivalents in the form of NADPH by the enzyme ferredoxin NADP reductase (FNR) or re-injected into the PQ pool (CEF). A central aim of the model is to study state transitions. Therefore, the action of the kinase Stt7 and the phosphatase Pph1 are represented in the model. We chose to make the simplifying assumption that the action of Stt7 directly results in a translocation of antenna from PSII to PSI and that Pph1 mediates the reverse translocation. As a result, the cross sections of the photosystems change, leading to an altered differential activation. In the model, the Stt7 kinase is regulated by the redox state of the PQ pool, reflecting the existing experimental evidence [28]. In the current version of the model, we assume that the Pph1 phosphatase is constitutively active.
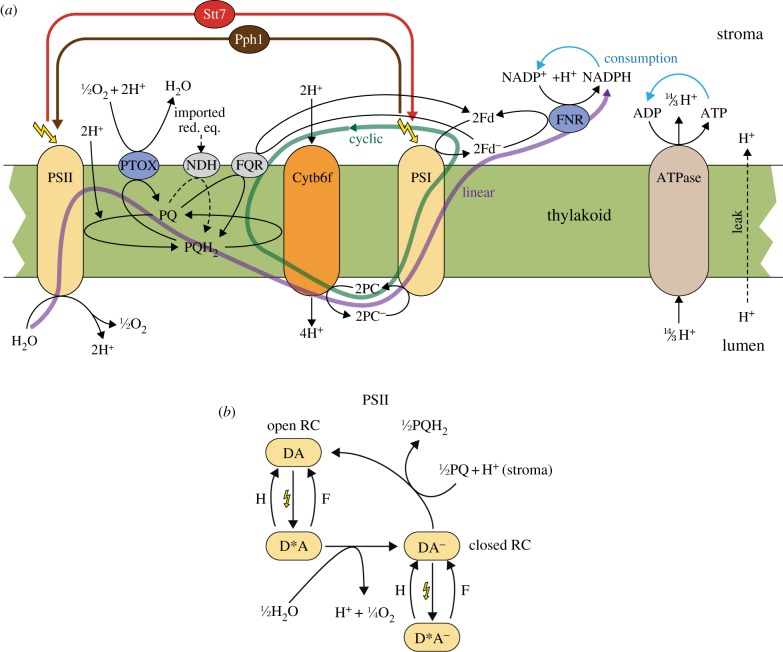
Figure 1.
(a) Schematic of the model of the photosynthetic electron transport chain and state transitions. All processes included in the mathematical model are depicted. The model includes linear and cyclic electron flow (CEF), ATP generation and a dynamic adjustment of antenna cross sections. Linear electron flow (purple arrow) comprises photosystem II (PSII), the cytochrome b6f complex (Cytb6f), photosystem I (PSI) and the ferredoxin-NADPH reductase (FNR). PSII is treated as a light-activated oxidoreductase transferring electrons from water to plastoquinone (PQ), resulting in the reduced form PQH2. In this process, two protons are taken up from the stromal side of the thylakoid membrane and two protons, resulting from splitting water, are released to the lumen. Cytb6f is modelled as an oxidoreductase transferring electrons from PQH2 to plastocyanin (PC), which simultaneously uses a process called the Q-cycle to release four protons in the lumen. PSI is treated as the second light-activated oxidoreductase, transferring electrons from reduced PC (PC−) to ferredoxin (Fd). The final step in the linear electron transport chain is catalysed by FNR, which transfers electrons from reduced Fd (Fd−) to NADP+ resulting in the formation of NADPH. CEF (green arrow) is represented by a single pathway involving ferredoxin-PQ reductase (FQR), which transfers electrons from Fd− back to PQ. Additionally, the model includes processes of the chlororespiratory pathway. Under aerobic conditions, the plastoquinone terminal oxidase (PTOX) oxidizes the PQ pool. In anaerobic conditions, electrons, e.g. from glycolysis, are imported into the chloroplast and lead to a reduction in the PQ pool by NADH dehydrogenase (NDH). Linear and CEF lead to a net translocation of protons into the lumen. The resulting proton gradient drives the ATP synthase (ATPase). The net products of photosynthetic electron transport, ATP and NADPH, are consumed by external processes that are simulated as lumped reactions. State transitions are modelled by two processes, relocation of antenna complexes from PSII to PSI and vice versa. The former is triggered by the kinase Stt7, whereas the latter is dependent on the phosphatase Pph1. In the model, Stt7 is activated by a reduced PQ pool while Pph1 is constitutively active. For details on the kinetics, see the electronic supplementary material. (b) Schematic of the internal processes modelled in PSII. This simplified description of PSII is implemented to calculate the reaction rate of PSII and the fluorescence emitted from PSII (see electronic supplementary material for details). Open reaction centres (DA) are excited by light (yellow flash). The excited state (D*A) can either relax to the ground state DA by heat emission (H) or fluorescence (F), or it can perform charge separation and recharge the donor side through water-splitting, resulting in the closed state (DA−). The closed state is also excited by light, resulting in state D*A−, which can only relax back to the unexcited state DA− either by heat (H) or fluorescence (F) emission. Closed reaction centres are re-opened by electron transfer to the PQ pool.
A minimally invasive way to continuously monitor the state of the electron transport chain is through chlorophyll fluorescence measurements. In order to allow comparison of model predictions with such experimental observations, we need to calculate the fluorescence emitted from PSII. For this, we include in the model a simplified representation of the internal processes in PSII (see figure 1b). Again, to keep the model as simple as possible, we exploit the timescale separation (electron flow within the photosystems is much faster than electron transfer between the complexes of the transport chain) and treat the internal states of PSII as being approximately in steady state (standard quasi-steady-state assumption, see the electronic supplementary material for details).
State transitions can be induced by altering the redox state of the PQ pool either by changing the illumination state (dark versus light) or through switching between aerobic and anaerobic conditions. Under anaerobic conditions in the dark, mitochondrial respiration is arrested and the demand for ATP induces an increase in glycolysis and an accumulation of reducing equivalents (Pasteur effect) that lead to a reduction of the PQ pool and a transition to state 2 [60]. To calibrate and test the model, we compared the simulations with fluorescence traces for state transitions observed experimentally under these two schemes.
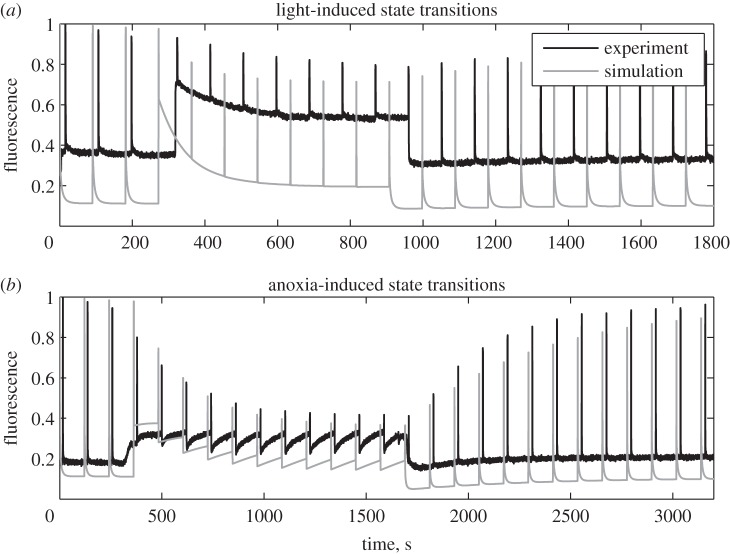
Figure 2a presents experimental data (black) and simulation (grey) for dark-adapted wild-type cells that were illuminated by weak light (100 µE m−2 s−1) for 10 min. It can be observed that the key features of the experimental fluorescence signal are correctly reproduced by the simulations. The dynamics of the saturating peaks (FM’) reflect the change in antenna cross section of PSII. Illumination causes a reduction in the PQ pool (see electronic supplementary material, figure S1), activating the kinase Stt7, and thus a partial transition to state 2. Antenna migration to PSI leads to a weaker activation of PSII and thus a slower reduction of the PQ pool. This negative feedback of the state transitions on PQ reduction leads to the establishment of a stationary and stable redox poise. In darkness, the process slowly reverses towards state 1. The steady-state fluorescence (FS) in the dark is also very well captured, including the slow fluorescence decline between consecutive peaks caused by reoxidation of the PQ pool after its transient reduction through the actinic effect of the saturating flashes. The relative FS levels in the light compared with darkness is not precisely reproduced. Why this is the case needs further exploration and it is probable that a more detailed model of the internal processes in PSII will be necessary to refine the calculation of the fluorescence signal. However, the important feature, i.e. the tendency of FS to increase shortly after light exposure and then to decline on a longer timescale as shown in the first studies of state transitions [61], is also clearly visible in the simulation results.
Figure 2.
Experimental (black) and simulated (grey) fluorescence traces. (a) Dark-adapted Chlamydomonas cells were further incubated in the dark for 5 min, exposed for 10 min to weak light (100 μE m−2 s−1) and then returned to darkness. Saturating light flashes were applied at regular intervals and the fluorescence signal (black) was recorded. The same protocol was simulated with the mathematical model and the calculated fluorescence signal (grey) is shown. The dynamics of FM reflect the transition to state 2 in the light and the reverse transition to state 1 in darkness. (b) Anoxia was induced by sealing the culture and allowing respiration to consume the available oxygen. After 15 min of anoxia air was re-applied by bubbling it in the sample. Saturating flashes were applied at regular intervals. The experimental (black) and simulated (grey) fluorescence are shown. Again, the FM dynamics represent transitions to state 2 in anoxic and to state 1 in aerobic conditions. The Fs dynamics between flashes reflect the redox poise of the PQ pool.
Figure 2b shows experimental data and simulations for state transitions induced by shifting low-light-adapted wild-type cells from aerobic to anaerobic conditions and back in darkness. Importantly, the simulations have been performed with exactly the same parameters as were used to fit the curves in figure 2a. Again, the important features of the fluorescence signals are well reproduced by the model. The dynamics of FM’ follow the change in antenna cross section of PSII, whereas FS reflects, at least qualitatively, the change of redox state of the PQ pool. Interestingly, the interflash dynamics of FS again are reproduced with remarkable accuracy. While in the beginning of the anoxic period FS remains approximately constant between two consecutive flashes, later in that phase it exhibits a slight increase. Investigating the variables that are not directly observable explains this behaviour (see electronic supplementary material, figure S2). While the PQ pool gets reduced by electrons from glycolysis, the cross section of PSII simultaneously decreases. Early during anoxia, these processes proceed with approximately the same rate and the opposite effects on fluorescence cancel each other. Later, state transition is mostly completed and the reduction in cross section is slowed, leading to a fluorescence rise resulting from PQ reduction.
The fact that the simulations are in good agreement with the experimental data allows us to conclude that our current knowledge and assumptions about the molecular mechanisms of state transitions in Chlamydomonas are sufficient to reproduce and explain most of the fluorescence changes, which are related to the dynamic allocation of antenna to the two photosystems. However, mathematical models are useful not only to reproduce and explain observed behaviour but also to make further predictions and attempt to answer more fundamental questions. For example, to address the question why state transitions are more pronounced in low light and seem to be repressed under high light conditions, we employed the model with the parameters used above to systematically analyse the steady-state behaviour for different light intensities and different relative antenna cross sections. For this, the rate constants for the kinase Stt7 and the phosphatase Pph1 were set to zero and the relative cross section was fixed to different values. Subsequently, the system was simulated until it reached steady state. Figure 3 displays the computed steady-state redox state of the PQ pool as a function of the relative cross section and the total light intensity. A remarkable observation is a sharp transition of the PQ redox state for low light intensities. If the cross section of PSII is too small, the pool is almost completely oxidized, whereas for cross sections that are too large it is extremely reduced. This illustrates the need to finely adapt the relative antenna cross section under low-light conditions to maintain a redox poise. By contrast, for high light conditions the transition from oxidized to reduced PQ is much smoother, indicating greater flexibility with respect to relative cross sections to maintain a redox poise.
Figure 3.
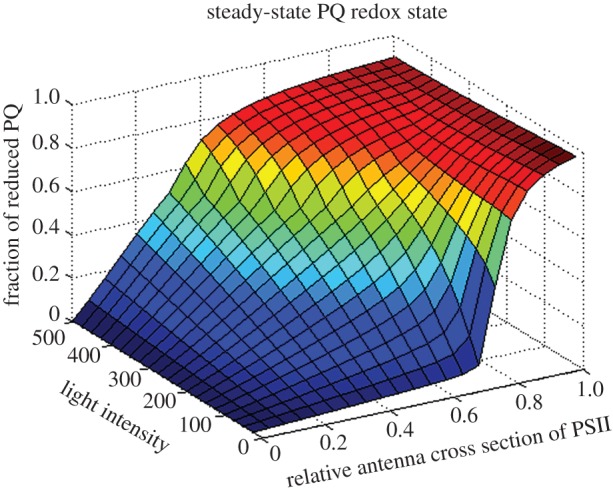
Predicted stationary states of the reduction level of the PQ pool for different light intensities and different states. For these predictions, state transitions were modelled by imposing values for the relative antenna cross sections (x-axis) between zero, representing all antennae on PSI, and unity, representing all antennae on PSII and keeping them fixed throughout the simulations. Light intensity (y-axis) was varied and for each combination of light intensity and antenna cross sections, and the system was simulated until steady state was reached. On the z-axis, the reduced fraction of the PQ pool in stationary state is plotted. For low light intensities, a sharp transition between a strongly oxidized and a highly reduced PQ pool is observed, demonstrating that only tightly controlled cross sections can ensure a balanced redox poise and thus explaining the importance of state transitions in this low light regime. For higher light intensities, the transition is smoother, indicating a higher flexibility in cross section to achieve redox poise.
So far, we have deliberately excluded the qE component of NPQ, thus mimicking low-light-adapted Chlamydomonas cells. With the modelling framework developed here, it is straightforward to include equations representing energy-dependent quenching of chlorophyll fluorescence, which may reflect the situation in high-light-adapted Chlamydomonas or plants. In the first approach, we have included a simplified mechanism [57] in which one pH-induced quencher increases the rate of chlorophyll de-excitation. When repeating the steady-state calculation (see electronic supplementary material, figure S3 and the text thereupon), an explanation for the different behaviours in different light regimes can be found. While the presence of qE-dependent quenching does not alter the behaviour for low light, in high light it results in the formation of a plateau in which the redox state of the PQ pool remains relatively constant for a wide range of relative cross sections. These theoretical predictions lead us to hypothesize that the intrinsic dynamic properties of the electron transport chain demand the ability to adjust antenna cross sections, but that this requirement is weaker for high light conditions. Indeed, the phosphorylation of the LHCII antenna, which is largely mediated by the STN7/Stt7 kinase in low light, is inhibited at high light intensities [48,62]. This has been ascribed to the negative regulation of the kinase through the thioredoxin pathway in high light [36,63] or to a conformational change in the PSII antenna [64].
4. Conclusion and perspectives
This work further extends previous studies on modelling high light acclimation in plants [57]. Here, our aim is to recapitulate the basic fluorescence features of short-term light acclimation in Chlamydomonas known as state transitions, which take place in low light (or in the dark under particular environmental conditions), and which are prominent in C. reinhardtii. To do so, we have implemented the representation of the photosynthetic electron transport chain in the model (figure 1a). In particular, we have included a heuristic description of CEF, which enhances the proton gradient and thereby triggers qE. We have also taken into account the chlororespiratory pathway, which modulates the redox state of the PQ pool in the dark, thereby activating the Stt7 kinase under anaerobic conditions (figure 2b). We have also implemented a mechanistic representation of the PSII catalytic cycle (figure 1b) to reproduce the changes of fluorescence yield induced by the saturating pulses which are used to monitor changes in the PSII absorption cross section in vivo (figure 2).
Overall, the fact that key experimental features of fluorescence traces are reproduced by the model with good agreement indicates that our model provides a reliable representation of state transitions in the light, and in the dark upon changes from aerobic to anaerobic conditions, despite the quantitative differences that simulations and experimental data show. This implies that this description of photosynthesis as a global process (from light capture to carbon assimilation) is sufficiently accurate to simulate redox changes in the electron transport chain, which take place either in the light through PSII and PSI photochemistry or in the dark through metabolic exchanges with the cell cytosol surrounding the chloroplast.
We can foresee several applications for this model, in particular, mechanistic issues related to photosynthetic activity. This is exemplified by the finding that, while the overall changes in the PSII absorption cross section in the light are well reproduced by the model, the large reduction in the PQ pool in the light cannot be properly simulated. This could suggest that the model is unable to describe the in vivo changes of the redox state of the PQ pool. However, this could also indicate that the ‘classic’ picture of state transitions in Chlamydomonas is not entirely correct. This view assumes that all the light-harvesting complexes migrate from PSII to PSI during the transition to state 2. By equilibrating their light-harvesting capacity, qT leads to a balance between PSII and PSI, which in our simulations restores the redox poise of the PQ pool between states 1 and 2 more extensively than in our experiments. On the other hand, previous studies have suggested that a substantial fraction of LHCII detaches from PSII during the transition to state 2, but does not associate with PSI and remains in a quenched state in the thylakoids [51]. In this case, state transitions are not expected to induce a complete redox balance of the PQ pool, because the decreased activity of PSII is not compensated by a concomitant enhancement of PSI activity. Obviously, the model can be used to test this hypothesis. Moreover, our model can bring essential information concerning the physiology of light acclimation by simulating the relative weight of qE and qT under changing light conditions, and therefore allows an exploration of the interplay between the two processes (e.g. [48]) under a wide range of simulated environmental conditions. Eventually, the model opens the possibility of investigating evolutionary issues. For example, it can be used to explore the different qE and qT capacities of plants and algae by modifying the appropriate parameters. It can also be used to understand why other ecologically relevant organisms (for example, diatoms) have evolved a strong qE response, without any need for qT development in their marine environment (e.g. [65,66]).
Acknowledgements
We thank Nicolas Roggli for preparing the figures.
Funding statement
M.G.-C. was supported by the SystemsX.ch RTD ‘Plant Growth in a Changing Environment’ and by the Swiss National Foundation (31003A_146300). O.E., G.F. and M.G.-C. benefited from the Marie Curie ITN ‘AccliPhot’ (GA 316 427). J.-D.R. acknowledges a grant from the Swiss National Foundation (3100A0_117712). G.F. was supported by an EMBO Post-Doctoral Fellowship. Mutual visits between Geneva and Aberdeen were funded by the Royal Society through the International Exchanges Grant (ref. IE110263). G.F. acknowledges funding by the French National Foundation Agency (ANR grant phytadapt ANR-NT09_567009) and the Labex GRAL (Grenoble Alliance for Integrated Structural Cell Biology) grants.
References
- 1.Arnon DI, Allen MB, Whatley FR. 1954. Photosynthesis by isolated chloroplasts. Nature 174, 394–396. ( 10.1038/174394a0) [DOI] [PubMed] [Google Scholar]
- 2.Hill R, Bendall F. 1960. Function of the two cytochrome components in chloroplasts—a working hypothesis. Nature 186, 136–137. ( 10.1038/186136a0) [DOI] [Google Scholar]
- 3.Nelson N, Ben-Shem A. 2004. The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell. Biol. 5, 971–982. ( 10.1038/nrm1525) [DOI] [PubMed] [Google Scholar]
- 4.Dekker JP, Boekema EJ. 2005. Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706, 12–39. ( 10.1016/j.bbabio.2004.09.009) [DOI] [PubMed] [Google Scholar]
- 5.Fork DC, Herbert SK. 1993. Electron transport and photophosphorylation by photosystem I in vivo in plants and cyanobacteria. Photosynth. Res. 36, 149–168. ( 10.1007/BF00033035) [DOI] [PubMed] [Google Scholar]
- 6.Munekage Y, Hashimoto M, Miyake C, Tomizawa K, Endo T, Tasaka M, Shikanai T. 2004. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429, 579–582. ( 10.1038/nature02598) [DOI] [PubMed] [Google Scholar]
- 7.Hüner NP, Bode R, Dahal K, Hollis L, Rosso D, Krol M, Ivanov AG. 2012. Chloroplast redox imbalance governs phenotypic plasticity: the ‘grand design of photosynthesis’ revisited. Front. Plant Sci. 3, 255 ( 10.3389/fpls.2012.00255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eberhard S, Finazzi G, Wollman FA. 2008. The dynamics of photosynthesis. Annu. Rev. Genet. 42, 463–515. ( 10.1146/annurev.genet.42.110807.091452) [DOI] [PubMed] [Google Scholar]
- 9.Bonardi V, Pesaresi P, Becker T, Schleiff E, Wagner R, Pfannschmidt T, Jahns P, Leister D. 2005. Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437, 1179–1182. ( 10.1038/nature04016) [DOI] [PubMed] [Google Scholar]
- 10.Fristedt R, Willig A, Granath P, Crèvecoeur M, Rochaix JD, Vener AV. 2009. Phosphorylation of photosystem II controls functional macroscopic folding of photosynthetic membranes in Arabidopsis. Plant Cell 21, 3950–3964. ( 10.1105/tpc.109.069435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foyer CH, Noctor G. 2013. Redox signaling in plants. Antioxid. Redox Signal. 18, 2081–2090. ( 10.1089/ars.2013.5278) [DOI] [PubMed] [Google Scholar]
- 12.Kasahara M, Kagawa T, Oikawa K, Suetsugu N, Miyao M, Wada M. 2002. Chloroplast avoidance movement reduces photodamage in plants. Nature 420, 829–832. ( 10.1038/nature01213) [DOI] [PubMed] [Google Scholar]
- 13.Wada M, Kagawa T, Sato Y. 2003. Chloroplast movement. Annu. Rev. Plant Biol. 54, 455–468. ( 10.1146/annurev.arplant.54.031902.135023) [DOI] [PubMed] [Google Scholar]
- 14.Zacks DN, Derguini F, Nakanishi K, Spudich JL. 1993. Comparative study of phototactic and photophobic receptor chromophore properties in Chlamydomonas reinhardtii. Biophys. J. 65, 508–518. ( 10.1016/S0006-3495(93)81067-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kröger P, Hegemann P. 1994. Photophobic responses and phototaxis in Chlamydomonas are triggered by a single rhodopsin photoreceptor. FEBS Lett. 341, 5–9. ( 10.1016/0014-5793(94)80229-7) [DOI] [PubMed] [Google Scholar]
- 16.Bennoun P. 1982. Evidence for a respiratory chain in the chloroplast. Proc. Natl Acad. Sci. USA 79, 4352–4356. ( 10.1073/pnas.79.14.4352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jans F, et al. 2008. A type II NAD(P)H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc. Natl Acad. Sci. USA 105, 20 546–20 551. ( 10.1073/pnas.0806896105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald AE, Ivanov AG, Bode R, Maxwell DP, Rodermel SR, Hüner NP. 2011. Flexibility in photosynthetic electron transport: the physiological role of plastoquinol terminal oxidase (PTOX). Biochim. Biophys. Acta 1807, 954–967. ( 10.1016/j.bbabio.2010.10.024) [DOI] [PubMed] [Google Scholar]
- 19.Mehler AH. 1957. Studies on reactions of illuminated chloroplasts. I. Mechanism of the reduction of oxygen and other Hill reagents. Arch. Biochem. Biophys. 33, 65–77. ( 10.1016/0003-9861(51)90082-3) [DOI] [PubMed] [Google Scholar]
- 20.Horton P, Ruban AV, Walters RG. 1996. Regulation of light harvesting in green plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 655–684. ( 10.1146/annurev.arplant.47.1.655) [DOI] [PubMed] [Google Scholar]
- 21.Niyogi KK. 1999. Photoprotection revisited: genetic and molecular approaches. Annu. Rev. Plant. Physiol. Plant Mol. Biol. 50, 333–359. ( 10.1146/annurev.arplant.50.1.333) [DOI] [PubMed] [Google Scholar]
- 22.Horton P, Hague A. 1988. Studies on the induction of chlorophyll fluorescence in isolated barley protoplasts: IV. Resolution of non-photochemical quenching. Biochim. Biophys. Acta 932, 107–115. ( 10.1016/0005-2728(88)90144-2) [DOI] [Google Scholar]
- 23.Pfündel EE, Dilley RA. 1993. The pH dependence of violaxanthin deepoxidation in isolated pea chloroplasts. Plant Physiol. 101, 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilmore AM. 1997. Mechanistic aspects of xanthophyll cycle-dependent photoprotection in higher plant chloroplasts and leaves. Physiol. Plant. 99, 197–209. ( 10.1111/j.1399-3054.1997.tb03449.x) [DOI] [Google Scholar]
- 25.Müller P, Li XP, Niyogi KK. 2001. Non-photochemical quenching. A response to excess light energy. Plant Physiol. 125, 1558–1566. ( 10.1104/pp.125.4.1558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hope AB. 1993. The chloroplast cytochrome b6f complex: a critical focus on function. Biochim. Biophys. Acta 1143, 1–22. ( 10.1016/0005-2728(93)90210-7) [DOI] [PubMed] [Google Scholar]
- 27.Rochaix JD. 2011. Regulation of photosynthetic electron transport. Biochim. Biophys. Acta 1807, 375–383. ( 10.1016/j.bbabio.2010.11.010) [DOI] [PubMed] [Google Scholar]
- 28.Rochaix JD, Lemeille S, Shapiguzov A, Samol I, Fucile G, Willig A, Goldschmidt-Clermont M. 2012. Protein kinases and phosphatases involved in the acclimation of the photosynthetic apparatus to a changing light environment. Phil. Trans. R. Soc. B 367, 3466–3474. ( 10.1098/rstb.2012.0064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aro EM, Kettunen R, Tyystjärvi E. 1992. ATP and light regulate D1 protein modification and degradation. Role of D1* in photoinhibition. FEBS Lett. 297, 29–33. ( 10.1016/0014-5793(92)80320-G) [DOI] [PubMed] [Google Scholar]
- 30.Bailey S, et al. 2008. Alternative photosynthetic electron flow to oxygen in marine Synechococcus. Biochim. Biophys. Acta 1777, 269–276. ( 10.1016/j.bbabio.2008.01.002) [DOI] [PubMed] [Google Scholar]
- 31.Cardol P, et al. 2008. An original adaptation of photosynthesis in the marine green alga Ostreococcus. Proc. Natl Acad. Sci. USA 105, 7881–7886. ( 10.1073/pnas.0802762105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li XP, Björkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK. 2000. A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403, 391–395. ( 10.1038/35000131) [DOI] [PubMed] [Google Scholar]
- 33.Bonente G, Passarini F, Cazzaniga S, Mancone C, Buia MC, Tripodi M, Bassi R, Caffarri S. 2008. The occurrence of the psbS gene product in Chlamydomonas reinhardtii and in other photosynthetic organisms and its correlation with energy quenching. Photochem. Photobiol. 84, 1359–1370. ( 10.1111/j.1751-1097.2008.00456.x) [DOI] [PubMed] [Google Scholar]
- 34.Koziol AG, Borza T, Ishida K, Keeling P, Lee RW, Durnford DG. 2007. Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms. Plant Physiol. 143, 1802–1816. ( 10.1104/pp.106.092536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delosme R, Olive J, Wollman FA. 1996. Changes in light energy distribution upon state transitions: an in vivo photoacoustic study of the wildtype and photosynthesis mutants from Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1273, 150–158. ( 10.1016/0005-2728(95)00143-3) [DOI] [Google Scholar]
- 36.Rintamaki E, Martinsuo P, Pursiheimo S, Aro EM. 2000. Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proc. Natl Acad. Sci. USA 97, 11 644–11 649. ( 10.1073/pnas.180054297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinsuo P, Pursiheimo S, Aro EM, Rintamaki E. 2003. Dithiol oxidant and disulfide reductant dynamically regulate the phosphorylation of lightharvesting complex II proteins in thylakoid membranes. Plant Physiol. 133, 37–46. ( 10.1104/pp.103.027268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemeille S, Willig A, Depège-Fargeix N, Delessert C, Bassi R, Rochaix JD. 2009. Analysis of the chloroplast protein kinase Stt7 during state transitions. PLoS Biol. 7, e45 ( 10.1371/journal.pbio.1000045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allen JF, Bennett J, Steinback KE, Arntzen CJ. 1981. Chloroplast protein phosphorylation couples plastoquinone redox state to distribution of excitation energy between photosystems. Nature 291, 25–29. ( 10.1038/291025a0) [DOI] [Google Scholar]
- 40.Vener AV, Van Kan PJ, Gal A, Andersson B, Ohad I. 1995. Activation/deactivation cycle of redox-controlled thylakoid protein phosphorylation. Role of plastoquinol bound to the reduced cytochrome bf complex. J. Biol. Chem. 270, 25 225–25 232. ( 10.1074/jbc.270.42.25225) [DOI] [PubMed] [Google Scholar]
- 41.Zito F, Finazzi G, Delosme R, Nitschke W, Picot D, Wollman FA. 1999. The Qo site of cytochrome b6f complexes controls the activation of the LHCII kinase. EMBO J. 18, 2961–2969. ( 10.1093/emboj/18.11.2961) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapiguzov A, Ingelsson B, Samol I, Andres C, Kessler F, Rochaix JD, Vener AV, Goldschmidt-Clermont M. 2010. The PPH1 phosphatase is specifically involved in LHCII dephosphorylation and state transitions in Arabidopsis. Proc. Natl Acad. Sci. USA 107, 4782–4787. ( 10.1073/pnas.0913810107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pribil M, Pesaresi P, Hertle A, Barbato R, Leister D. 2010. Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biol. 8, e1000288 ( 10.1371/journal.pbio.1000288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Avenson TJ, Cruz JA, Kanazawa A, Kramer DM. 2000. Regulating the proton budget of higher plant photosynthesis. Proc. Natl Acad. Sci. USA 102, 9709–9713. ( 10.1073/pnas.0503952102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foyer CH, Neukermans J, Queval G, Noctor G, Harbinson J. 2012. Photosynthetic control of electron transport and the regulation of gene expression. J. Exp. Bot. 63, 1637–1661. ( 10.1093/jxb/ers013) [DOI] [PubMed] [Google Scholar]
- 46.DalCorso G, Pesaresi P, Masiero S, Aseeva E, Schünemann D, Finazzi G, Joliot P, Barbato R, Leister D. 2008. A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132, 273–285. ( 10.1016/j.cell.2007.12.028) [DOI] [PubMed] [Google Scholar]
- 47.Takahashi S, Milward SE, Fan DY, Chow WS, Badger MR. 2009. How does cyclic electron flow alleviate photoinhibition in Arabidopsis? Plant Physiol. 149, 1560–1567. ( 10.1104/pp.108.134122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allorent G, et al. 2013. A dual strategy to cope with high light in Chlamydomonas reinhardtii. Plant Cell 25, 545–557. ( 10.1105/tpc.112.108274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finazzi G, Furia A, Barbagallo RP, Forti G. 1999. State transitions, cyclic and linear electron transport and photophosphorylation in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1413, 117–129. ( 10.1016/S0005-2728(99)00089-4) [DOI] [PubMed] [Google Scholar]
- 50.Finazzi G, Rappaport F, Furia A, Fleischmann M, Rochaix JD, Zito F, Forti G. 2002. Involvement of state transitions in the switch between linear and cyclic electron flow in Chlamydomonas reinhardtii. EMBO Rep. 3, 280–285. ( 10.1093/embo-reports/kvf047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwai M, Takizawa K, Tokutsu R, Okamuro A, Takahashi Y, Minagawa J. 2010. Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature 464, 1210–1213. ( 10.1038/nature08885) [DOI] [PubMed] [Google Scholar]
- 52.Minagawa J. 2011. State transitions—the molecular remodeling of photosynthetic supercomplexes that controls energy flow in the chloroplast. Biochim. Biophys. Acta 1807, 897–905. ( 10.1016/j.bbabio.2010.11.005) [DOI] [PubMed] [Google Scholar]
- 53.Takahashi H, Clowez S, Wollman FA, Vallon O, Rappaport F. 2013. Cyclic electron flow is redox-controlled but independent of state transition. Nat. Commun. 4, 1954 ( 10.1038/ncomms2954) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Locke JC, Kozma-Bognár L, Gould PD, Fehér B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ. 2006. Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. 2, 59 ( 10.1038/msb4100102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laisk A, Eichelmann H, Oja V. 2006. C3 photosynthesis in silico. Photosynth. Res. 90, 45–66. ( 10.1007/s11120-006-9109-1) [DOI] [PubMed] [Google Scholar]
- 56.Samaga R, Klamt S. 2013. Modeling approaches for qualitative and semi-quantitative analysis of cellular signaling networks. Cell Commun. Signal. 11, 43 ( 10.1186/1478-811X-11-43) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ebenhöh O, Houwaart T, Lokstein H, Schlede S, Tirok K. 2011. A minimal mathematical model of nonphotochemical quenching of chlorophyll fluorescence. Biosystems 103, 196–204. ( 10.1016/j.biosystems.2010.10.011) [DOI] [PubMed] [Google Scholar]
- 58.Zaks J, Amarnath K, Kramer DM, Niyogi KK, Fleming GR. 2012. A kinetic model of rapidly reversible nonphotochemical quenching. Proc. Natl Acad. Sci. USA 109, 15 757–15 762. ( 10.1073/pnas.1211017109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, Hippler M, Niyogi KK. 2009. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462, 518–521. ( 10.1038/nature08587) [DOI] [PubMed] [Google Scholar]
- 60.Bulte L, Gans P, Rebeille F, Wollman FA. 1990. ATP control on state transitions in vivo in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1020, 72–80. ( 10.1016/0005-2728(90)90095-L) [DOI] [Google Scholar]
- 61.Bonaventura C, Myers J. 1969. Fluorescence and oxygen evolution from Chlorella pyrenoidosa. Biochim. Biophys. Acta 189, 366–383. ( 10.1016/0005-2728(69)90168-6) [DOI] [PubMed] [Google Scholar]
- 62.Rintamaki E, Salonen M, Suoranta UM, Carlberg I, Andersson B, Aro EM. 1997. Phosphorylation of light-harvesting complex II and photosystem II core proteins shows different irradiance-dependent regulation in vivo. Application of phosphothreonine antibodies to analysis of thylakoid phosphoproteins. J. Biol. Chem. 272, 30 476–30 482. ( 10.1074/jbc.272.48.30476) [DOI] [PubMed] [Google Scholar]
- 63.Lemeille S, Rochaix JD. 2010. State transitions at the crossroad of thylakoid signalling pathways. Photosynth. Res. 106, 33–46. ( 10.1007/s11120-010-9538-8) [DOI] [PubMed] [Google Scholar]
- 64.Vink M, Zer H, Alumot N, Gaathon A, Niyogi K, Herrmann RG, Andersson B, Ohad I. 2004. Light-modulated exposure of the light-harvesting complex II (LHCII) to protein kinase(s) and state transition in Chlamydomonas reinhardtii xanthophyll mutants. Biochemistry 43, 7824–7833. ( 10.1021/bi030267l) [DOI] [PubMed] [Google Scholar]
- 65.Owens TG. 1986. Light-harvesting function in the diatom Phaeodactylum tricornutum. II. Distribution of excitation energy between the photosystems. Plant Physiol. 80, 739–746. ( 10.1104/pp.80.3.739) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lavaud J, Rousseau B, van Gorkom HJ, Etienne AL. 2002. Influence of the diadinoxanthin pool size on photoprotection in the marine planktonic diatom Phaeodactylum tricornutum. Plant Physiol. 129, 1398–1406. ( 10.1104/pp.002014) [DOI] [PMC free article] [PubMed] [Google Scholar]