Abstract
High light acclimation depends on retrograde control of nuclear gene expression. Retrograde regulation uses multiple signalling pathways and thus exploits signal patterns. To maximally challenge the acclimation system, Arabidopsis thaliana plants were either adapted to 8 (low light (L-light)) or 80 µmol quanta m−2 s−1 (normal light (N-light)) and subsequently exposed to a 100- and 10-fold light intensity increase, respectively, to high light (H-light, 800 µmol quanta m−2 s−1), for up to 6 h. Both L → H- and N → H-light plants efficiently regulated CO2 assimilation to a constant level without apparent damage and inhibition. This experimental set-up was scrutinized for time-dependent regulation and efficiency of adjustment. Transcriptome profiles revealed that N-light and L-light plants differentially accumulated 2119 transcripts. After 6 h in H-light, only 205 remained differently regulated between the L → H- and N → H-light plants, indicating efficient regulation allowing the plants to reach a similar transcriptome state. Time-dependent analysis of transcripts as markers for signalling pathways, and of metabolites and hormones as possibly involved transmitters, suggests that oxylipins such as oxophytodienoic acid and jasmonic acid, metabolites and redox cues predominantly control the acclimation response, whereas abscisic acid, salicylic acid and auxins play an insignificant or minor role.
Keywords: photosynthesis, redox regulation, retrograde signalling, oxylipin, transcript profiling
1. Introduction
Acclimation to changing environmental conditions optimizes plant growth and fitness. Plants encounter biotic and abiotic stress if such changes occur suddenly and with large amplitude. Stress is defined as pronounced deviation from optimum growth condition and is caused by direct effects of the stressor or as indirect consequence of interference of the stressor with metabolism. The acclimation response must be specific in order to react in an appropriate manner and to preserve resources. This specificity is realized by signal integration in signalling cascades that deploy distinct input signals.
High light (H-light) conditions trigger over-reduction of the photosynthetic electron transport (PET) chain with subsequent formation of reactive oxygen species (ROS) [1]. Non-photochemical quenching (NPQ) reduces light-driven electron pressure and ROS generation at photosystems PSII and PSI [2,3]. Singlet oxygen is generated at PSII and superoxide anions at PSI [4,5]. Detoxification of ROS counteracts redox imbalances and maintains plant fitness. In the chloroplast, superoxide dismutase catalyses the synthesis of H2O2 and O2 from two molecules of O2−. H2O2 is detoxified by the ascorbate-dependent pathway using ascorbate peroxidase (Apx) [6,7] or the ascorbate-independent pathway using thiol oxidases such as peroxiredoxins [8,9].
Acclimation to H-light or excess light has been studied in quite some detail owing to its ecological significance, but also because it is experimentally straightforward with a precisely defined starting point. Transcripts of Apx 1 and Apx 2 are upregulated within short time periods [10]. Systemic signalling is indicated by regulation of Apx in shaded distant leaves [11]. ROS amounts determine whether ROS act as signalling molecules or react with sensitive cell components, e.g. proteins, DNA or cell walls [12]. In addition, all chloroplast antioxidative enzymes are encoded in the nucleus [13] and have to be transported to the organelles [14]. The separation of the sites of ROS production and nuclear gene expression reinforces the need for retrograde signalling for information exchange to optimally balance gene activity with chloroplast metabolic state [15–19]. In contrast to earlier concepts that assumed the participation of only few signalling pathways, nowadays a signalling pattern is discussed to indicate retrograde signalling. Among the interesting cues are abscisic acid (ABA) [20], jasmonic acid (JA) [21], H2O2 [22,23] and ↑O2 [24], the PET redox state, including the plastoquinone pool [25,26], and associated components such as STN7 [27]. Precursors that are conditionally synthesized in the chloroplast and afterwards transferred to the cytosol or other compartments such as xanthoxin for ABA synthesis [28] or oxophytodienoic acid (OPDA) for JA synthesis in the peroxisome [29] might be good candidates for signal transmission. Up to now, a ‘plastid factor’ as a distinct signalling molecule with a master function in retrograde signalling has not been identified. Thus, retrograde signalling more likely is recognized as a complex network of fine-tuned mechanisms with crosstalk and different pathways of signal transmission [30].
In a previous study, Arabidopsis (A.) thaliana was grown under normal light (N-light) or acclimatized to low light (L-light), and both transferred to high light (H-light) that corresponded to a 10-fold or 100-fold light shift. Analyses were performed after 6 and 24 h. The transcript abundances of the water–water cycle enzymes were highly similar 6 h after transfer, irrespective of whether plants were subjected to a 10-fold or 100-fold shift, despite extremely different starting points from L- or N-light acclimatized conditions [31]. To understand the dynamic nature of acclimation, we focused our research on the involvement of possible signalling pathways, the time-resolved kinetics of responses and the degree of acclimation at the global transcript scale.
2. Results
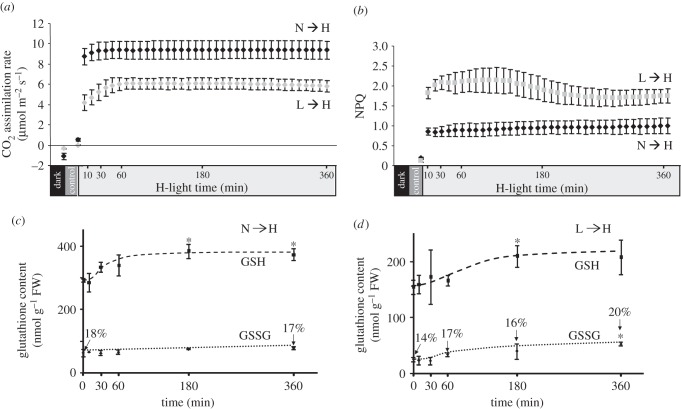
H-light acclimation proceeds in various steps and on different time scales. In the first step, light-dependent photochemical and metabolic events are converted into signalling cues. As basic characterization, we measured CO2 assimilation, quantum yield of PSII, photochemical quenching (qP) and NPQ in a time period between 0 and 6 h after transfer to H-light [31]. The CO2 assimilation rate increased rapidly within 10 min and reached a constant level after 30 min in the N → H-light transfer, and after 60 min in the L → H-light transfer. The rates per leaf area were about 30% lower in the previously L-light acclimatized plants, but were constant until the end of the measuring time at 6 h (figure 1a). Apparently, neither plant suffered severe inhibition. Likewise, the photosynthetic yield declined as expected directly after the H-light shift, but remained mostly constant during the subsequent 6 h (see electronic supplementary material, figure S1). The yield and qP were remarkably lower after the L → H shift, accordingly NPQ was higher for the L-light plants (figure 1b). Constant values were measured for the N → H-light plants, whereas there was a peak between 60 and 120 min in the L → H plants, indicating some relaxation of photoinhibition to maintain constant CO2-fixation rate.
Figure 1.
Time kinetics of photosynthetic parameters and the glutathione pool following the shift to H-light. (a) CO2 assimilation rate and (b) non-photochemical quenching (NPQ) were determined in L-light and N-light plants after transfer to H-light. Prior to the measurement, plants had been dark-acclimatized for 30 min. (c,d) Reduced glutathione (GSH) and oxidized glutathione (GSSG) were quantified in (c) (N → H)- and (d) (L → H) plants, respectively. Data are means ± s.d. of n = 3 independent experiments. In the case of glutathione, asterisks indicate significant differences to the controls, Student's t-test, p < 0.05.
Reduced and oxidized ascorbate (ASC/DHA) and glutathione (GSH/GSSG) are metabolites with functions as antioxidants, cosubstrates and regulators [6,7]. Their levels provide a basic characterization of cell redox state. Fresh-weight related GSH levels of N-light plants exceeded that of L-light plants by a factor of 3.5. Following the N → H shift and after a short delay, GSH increased by about 20%, which at t = 180 and 360 min was significantly different from t = 0 min. GSSG levels also increased slightly with time, thus the GSH/GSSG ratio remained unaffected (figure 1c). In a converse manner, GSSG increased more than GSH after L → H shift, and thus the GSH/GSSG ratio increased from 14 to 20%. Total ascorbate levels were more than twofold higher in N-light plants compared with L-light plants. After N → H shift, only a slight increase until t = 30 min could be detected. The proportion of DHA remained constant at 31% (see electronic supplementary material, figure S1c), similar to L → H shifted plants. Both ASC and DHA levels increased in L → H plants towards the end of the experiment (see electronic supplementary material, figure S1d).
Transcriptome profiling was intended to define the acclimation state after 6 h at a global scale. Thus, leaves from four conditions were harvested at 15.00, 6 h after the onset of H-light, namely L-, L → H, N- and N → H-light plants. Overall, approximately 12 times more genes were upregulated in their expression after the 100-fold light shift compared with the 10-fold light shift (see electronic supplementary material, figure S2a), and the number of downregulated genes was even 20-fold higher in the 100-fold shift (see electronic supplementary material, figure S2b). Categorization in functional groups with MapMan software [32] (see electronic supplementary material, figure S2e,f) revealed that most upregulated transcripts belong to the GO group of nucleic acid and protein synthesis. Surprisingly, stress-dependent genes were upregulated in N → H plants more than in L → H plants. The first analysis focused on 67 genes involved in ROS detoxification [33] and compared the transcript levels before the light-shift (N/L), during the light-shift (N → H/N, L → H/L; both compared with the untreated controls) and after the light shift (N → H/L → H; electronic supplementary material, figure S3). The transcript levels of water–water cycle enzymes were higher in N-light compared with L-light plants and both showed an upregulation after 6 h of H-light treatment. In comparison, NADPH oxidases showed mostly no changes in the transcript levels. A downregulation of the transcript amount was observed for cat3 and most glutathione peroxidases during both shifts. Overall, only small differences in transcript levels of the ROS network were detected 6 h after the light shift of L-light and N-light plants.
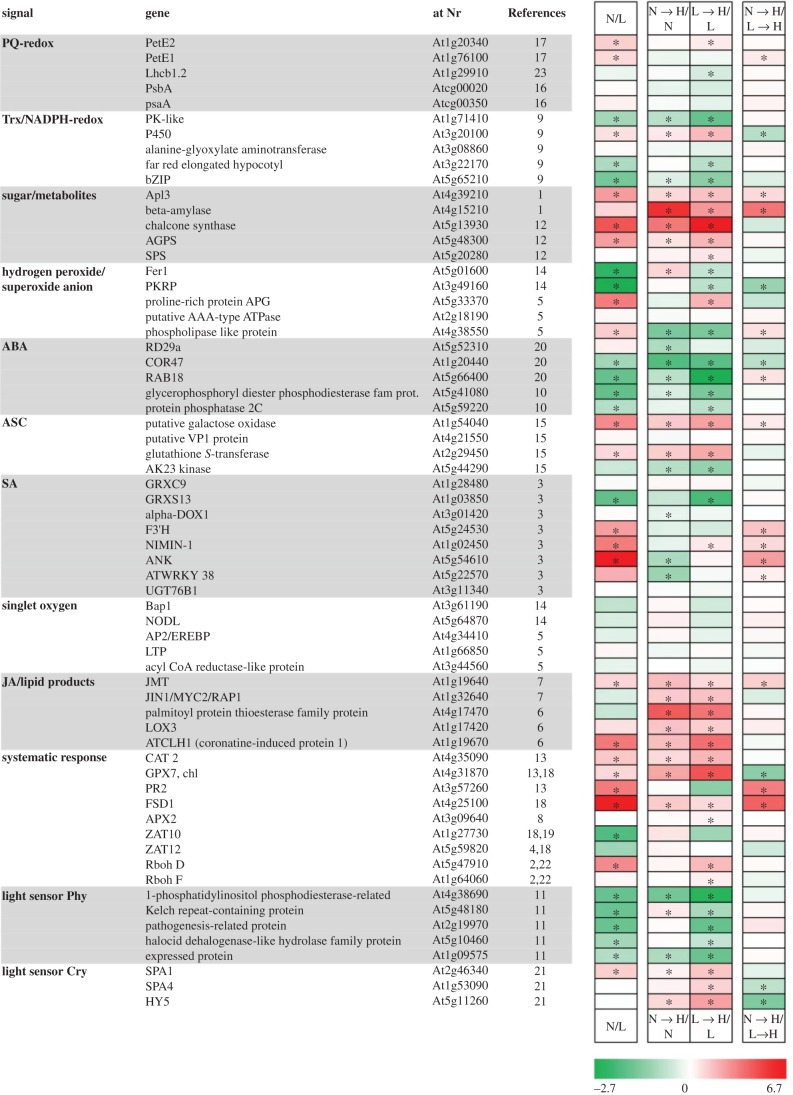
Reference genes were selected from literature dealing with retrograde signalling and possible messenger molecules. The transcript abundances of these genes were assumed to give hints to involved retrograde signalling pathways after 6 h at H-light (figure 2). Only minor differential transcript accumulation was detected for selected marker genes of plastoquinone- and singlet oxygen-dependent signalling such as PetE and Bap1, respectively. Strongest regulation occurred for markers of sugar- and metabolite-signalling such as β-amylase and chalcone synthase, and also for oxylipin-dependent signalling as indicated by lipoxygenase LOX3. Some upregulation was seen for salicylic acid (SA) and ascorbate markers. Unexpectedly, considerable downregulation was observed for marker genes of ABA-, systemic and phytochrome-dependent signalling. At the end of the 6 h period of H-light acclimation, only 19 of the 64 transcripts were still differentially accumulated between former L-light and N-light plants; 12 were higher in N → H compared with L → H plants.
Figure 2.
Transcript amounts of genes selected as markers for signalling pathways. Sixty-four reference genes known to respond to specific signalling cues were selected from the literature in order to determine their transcript amounts after 6 h of H-light treatment. Comparison before the light shift (N/L), after the light shift (N → H/N, L → H/L) and between the H-light treatments (N → H/L → H) was based on the data from the ATH1 whole genome microarray. log2-fold; green, downregulation; red, upregulation; asterisks indicate significant differences, Student's t-test, p < 0.005, n = 3. PQ, plastoquinone; Trx, thioredoxin. References for selected marker genes: 1: Baier et al. [34]; 2: Bechthold et al. [35]; 3: Blanco et al. [36]; 4: Davletova et al. [37]; 5: Gadjev et al. [38]; 6: Goda et al. [39]; 7: cited in Goda et al. [39]; 8: Karpinski et al. [11]; 9: Lintala et al.[40]; 10: Matsui et al. [41]; 11: Mazella et al. [42]; 12: Muller et al. [43]; 13: Mullineaux et al. [44]; 14: op den Camp et al. [24]; 15: Pastori et al. [45]; 16: Pfannschmidt et al. [46]; 17: Pfannschmidt et al. [47]; 18: Pogson et al. [17]; 19: Rossel et al. [25]; 20: Sanchez et al. [48]; 21: Sellaro et al. [49], 22: Torres et al. [50]; 23: Yang et al. [51]. (Online version in colour.)
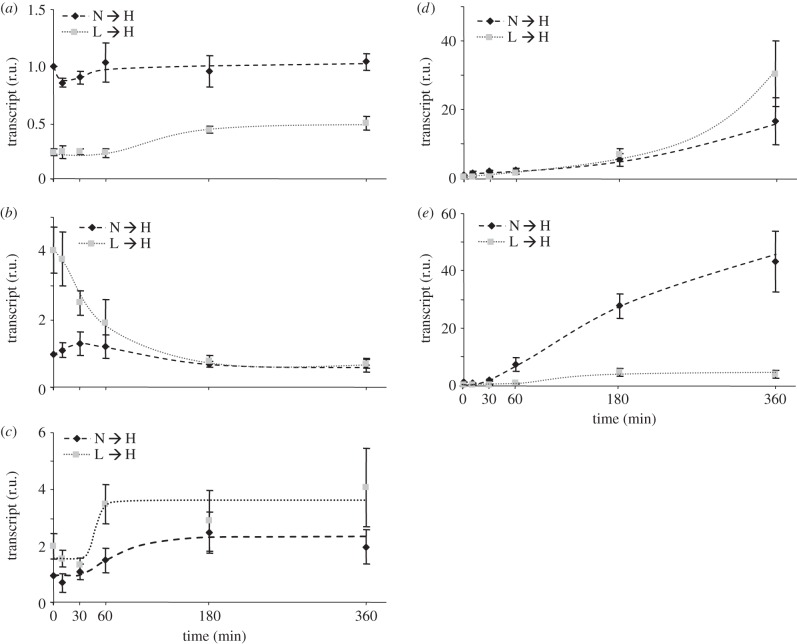
Five selected markers were analysed in detail to gain insights into the kinetics of regulation. Monodehydroascorbate reductase (MDHAR, At1g63940) as marker for the water–water cycle (figure 3a) doubled within 3 h after the L → H transfer, whereas little transcriptional change occurred after the N → H shift. The transcript level of the ABA-dependent reference gene COR47 (At1g20440) (figure 3b) was downregulated after the L → H transfer and reached the level of the N → H-shifted plants. The transcript of the H2O2-dependent marker PKRP (At3g49160) was twofold upregulated upon both light shifts (figure 3c). Interestingly, the response was delayed in both plants by about 30 min, and the rate of increase was slower in the L → H plants. The palmitoyl protein thioesterase family protein (PPTE, At4g17470) used as reference transcript for oxylipin signalling accumulated with ongoing H-light exposure and reached a greater than 20-fold (N → H) or greater than 30-fold (L → H) higher level in comparison with N- or L-light plants, respectively (figure 3d). The transcript level of the sugar marker β-amylase (At4g15210) increased more than 40-fold after the 10-fold shift but only approximately fivefold upon the 100-fold shift (figure 3e).
Figure 3.
Time-resolved abundances of marker transcripts. Reference transcripts were quantified following the N → H- or L → H shift by qPCR (a) monodehydroascorbate reductase (MDHAR), (b) ABA-dependent cold regulated 47 (COR47), (c) ROS-dependent pyruvate kinase related protein (PKRP), (d) oxylipin-indicating palmitoyl protein thioesterase family protein (PPTE) and (e) the sugar marker β-amylase. Data are means ± s.d. of n = 3 independent experiments, each with replicates.
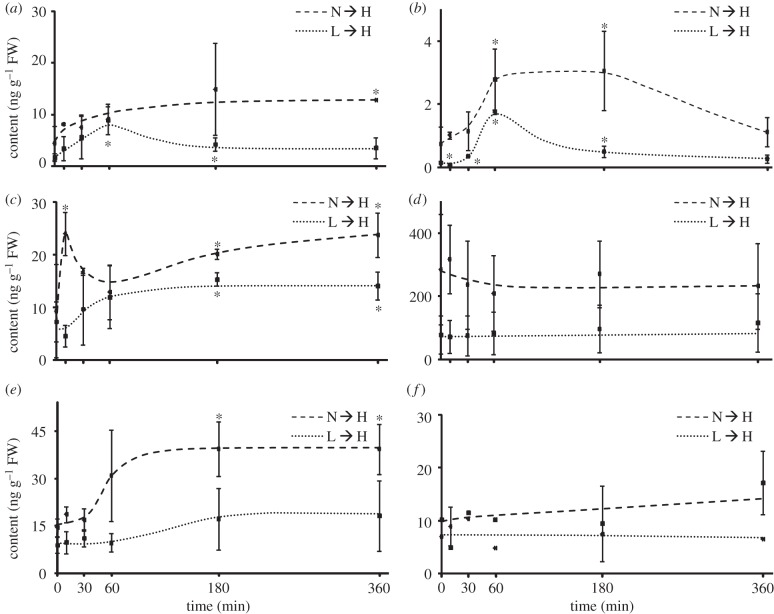
Phytohormones have been suggested to be involved in light acclimation [15,30,45]. Therefore, phytohormone levels were determined in order to identify candidate signals and to mechanistically link or validate our observations. OPDA, JA and its conjugate jasmonyl-isoleucine (JA-Ile) act as signalling molecules to stress [52,53]. Their concentration changed upon transfer to H-light with distinct kinetics and amplitude (figure 4a–c). Upon L → H transfer, OPDA, JA and JA-Ile increased until 60 min. Subsequently, JA and JA-Ile decreased again, whereas OPDA remained at a twofold higher level. In the N → H light experiment, OPDA increased fast, followed by JA and JA-Ile. In comparison, the JA antagonist SA [54] showed no significant regulation; if at all, there was a trend to decrease which will be important for the discussion (figure 4d). ABA plays a crucial role in control of development and stress acclimation [55,56]. The ABA level was unchanged during the first 30 min following the transfer. A significant increase by more than twofold was measured in N → H plants (figure 4e). Indole-3-acetic acid (IAA) levels were unaffected by H-light despite its connection to ROS [57,58] (figure 4f). The minor contribution of ROS signalling was confirmed by comparing the transcriptional regulation in our experiment with published ROS-related arrays using the ROSMETER [59] (see electronic supplementary material, figure S4). The heat map (please note that the colour scale covers only −0.4 to 0.4) indicates that the transcriptional differences between N/L, N → H/N and L → H/L were alike and that they were slightly coregulated with transcripts linked to H2O2 and singlet oxygen, but these differences were levelled in the N → H/L → H comparison as described above. It is also noteworthy that a negative correlation was observed with signatures derived from mitochondrial ROS.
Figure 4.
Changes in phytohormone contents in plants transferred to H-light as quantified by LC–MS/MS. (a) Jasmonic acid (JA) and (b) jasmonyl-Ile, (c) 12-oxophytodienoic acid (OPDA), (d) salicylic acid (SA), (e) abscisic acid (ABA) and (f) indole-3-acetic acid (IAA) were quantified in a time kinetics after transfer to H-light. Data are means ± s.d. of n = 3 independent experiments (IAA n = 2), asterisks indicate significant differences to the controls, Student's t-test, p < 0.1.
3. Discussion
The experimental set-up of shifting plants to the same H-light condition from different preconditioning states was recently designed to maximally challenge the acclimation machinery of leaves [31]. L-light plants growing in less than 10 µmol quanta m−2 s−1 for 10 days are completely shade-acclimatized with only 25% electron transport capacity of N-light plants. Focus of the previous study was directed to regulation of the enzymes of the water–water cycle at the levels of transcript, protein, enzyme activity and metabolite in N → H and L → H plants at 0, 6 and 24 h. The observed remarkably high and efficient plasticity prompted us to direct our attention to a time-resolved analysis. Here, it is shown that the plants reached and maintained constant CO2 fixation rates in H-light within 60 min. This result in combination with the kinetics of NPQ and qP confirmed that activation of quenching mechanisms [60] was perfectly tuned to maintain efficient gross photosynthesis while avoiding damage development.
(a). Conditional acclimation after 6 h of high light treatment
Transcriptome analysis after 6 h H-light revealed differential transcript accumulation of approximately 7.5% in N → H plants and 20% in L → H plants, respectively. The high number of differentially accumulated transcripts by far exceeded that of paraquat-treated flu mutants [24] or H2O2-treated A. thaliana wild-type [37] and indicates a profound reprogramming of L-light plants. The set of differentially (twofold or more) regulated transcripts between N- and L-light plants comprised 2219 before the H-light transfer and had decreased to 205 genes between N → H and L → H plants at t = 6 h. Apparently, the global transcriptome adjustment of L-light plants upon H-light treatment occurred extremely efficiently to a similar state as in N-light plants. In contrast to previously analysed redox- and ROS-related transcriptomes [33], here also a large set of transcripts among the antioxidant network was upregulated. This indicates a role of ROS in response regulation or the preparatory activation of a programme for improved ROS scavenging in dependence on light. Also, in the ROS network, the majority of the significant differences were minimized after 6 h, leading to the hypothesis that ROS-dependent signalling does not play a major role 6 h or more after L → H transfer [31]. However, ROS signalling might contribute to H-light response regulation at earlier time points as suggested previously [10,11]. This is in line with the upregulation of the ROS marker PKRP [24] between 30 and 60 min. The upregulation occurred faster in N → H than in L → H plants (figure 3). The difference in response speed tentatively correlated with lower expression of RbohD and RbohF in L-light plants (figure 2). Likewise, the levels of markers for cell signalling (figure 2) were highly similar between N → H and L → H plants, completing the picture of efficient transcriptional acclimation of A. thaliana to H-light.
(b). Involvement of oxylipins and redox regulators
Up-to-date concepts on retrograde control of nuclear gene expression assume the activation of different signals that combine to a specific pattern which in turn optimizes the acclimation response [15,17,30]. This study presents the kinetic dissection of (i) marker transcript regulation, and (ii) the accumulation of metabolite signals and phytohormones. Except for JA and OPDA, ASC and GSH, which increased readily in N → H plants, all other markers showed a delay of at least 30–60 min. OPDA is synthesized from α-linolenic acid in the chloroplast and exerts signalling function independent of JA and JA-Ile regulation [61,62]. Following binding to cyclophilin 20-3, the Cyp20-3/OPDA complex activates cysteine synthesis, increases thiol levels and reduction state and induces defence gene expression [53]. Accumulation of GSH may be explained by this regulatory mechanism and could be part of a general acclimation response. In line with this result, A. thaliana deficient of Cyp20-3 revealed a H-light sensitive phenotype [63]. Furthermore, OPDA-triggered signalling involves activation of TGA transcription factors [64]. The upregulation of PPTE, a reference gene for oxylipin signalling [39], might be the consequence of higher OPDA levels after the H-light transfer.
The JA-antagonist SA decreased during N → H and remained constant during L → H, which argues against SA as transmitter in H-light acclimation. SA suppresses JA signalling [65] by inhibition of the E3 ubiquitin-ligase SCF (COI1) which is required for JA signalling [66]. Suppression of SA may enable oxylipin signalling in the H-light response [54,67]. RBOHD/F activity, ROS accumulation and SA control systemic H-light acclimation and the cell death programme [68]. The lack of upregulation in the L → H transfer experiments indicates that retrograde signalling can use different signalling patterns.
In N → H plants, ABA increased to about twice the control level after 3 h. The trend to increase was also seen in L → H plants, but was insignificant and occurred with a slight delay. Surprisingly, an anti-parallel response could be detected between ABA-regulated COR47 [48] and ABA amount. The discrepancy could be explained by alkaline trapping of deprotonated ABA in the stroma in H-light as modelled by Slovik & Hartung [69] and discussed before [31]. As a consequence of ABA anion accumulation in the alkaline stroma, the effective ABA concentration in the nucleoplasm and cytosol could be low despite the overall increase in ABA accumulation of the leaves (figure 4e).
Acclimation to H-light involves diverse mechanisms at different time scales such as rapid photoinhibition and antioxidant expression, and long-term acclimation, including adjustment of chlorophyll synthesis [3,10,70,71]. The complex acclimation process, here underscored by reorganization of 20% of the transcriptome in L → H plants, suggests that no single and exclusive pathway mediates the response. ROS [10], SA [68], ABA [72], redox signals [46] and others have been suggested to mediate the acclimation. Our study provides a detailed inventory after 6 h following the N → H and L → H transfer and a framework of the timing of signalling events. The increase in oxylipin levels was among the fast responses followed by an increase in GSH levels in the N → H plants. The glutathione increase was delayed in L-light acclimatized plants, possibly owing to resource limitation and starvation. Likely, L-light plants build up cell constituents such as carbohydrates and proteins before investing in defence. This is also indicated by the delayed upregulation in L → H plants of β-amylase, a reliable sugar marker [34]. Interestingly, also the ROS marker PKRP revealed the faster and more intense response in the N → H plants. SA, ABA and IAA appear to play no major role in our experimental system. We conclude from this study that oxylipins, metabolites and redox cues, but possibly less ROS, should be investigated in the future to understand rapid processes in H-light acclimation.
4. Methods
(a). Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia (wild-type) was grown as described in [31] (10 h light with 80 µmol quanta m−2 s−1, and 21°C/14 h dark and 18°C, 50% relative humidity). After three weeks, plants were either transferred to 8 (low light, L-light) or kept at 80 µmol quanta m−2 s−1 (normal light, N-light) for 10 days. Plants (4.5 week old) were transferred to high light (800 µmol quanta m−2 s−1, H-light) for time periods as indicated, or remained in L- or N-light. Rosettes were harvested at 15.00 (±30 min), frozen in liquid nitrogen and stored at −80°C.
(b). Assimilation rate and photosynthetic parameters
Assimilation rate, PSII quantum yield, qP and NPQ were determined with the gas exchange fluorescence system GFS-3000 according to the manufacturer's instructions [31] (Walz, Germany) with plants acclimatized to darkness for 30 min. Following 10 min at 8 and 80 µmol quanta m−2 s−1 for L- and N-light controls, respectively, the light intensity was increased to 800 µmol quanta m−2 s−1 for H-light treatment.
(c). Whole genome ATH1 microarray
Total RNA was isolated using RNeasy mini kit (Qiagen). KFB (Regensburg, Germany) was commissioned for hybridization of the Affymetrix ATH1 genome array. Each condition was hybridized three times from independent experiments. Microarray data were imported into ROBIN (MPI Golm, Potsdam, Germany) and normalized according to the RMA algorithm [73,74]. Differentially regulated subsets were defined as those with twofold or greater difference and an adjusted p-value cut-off of less than or equal to 0.005 [75,76]. The gene list was imported into the MapMan ImageAnnotator 1.6.3 [32] to analyse pathways. The transcriptome data were analysed for ROS signatures by the ROSMETER according to Rosenwasser et al. [59] as implemented in the bioinformatics tool that is available at http://app.agri.gov.il/noa/ROS_desc.php.
(d). Transcript profiling
RNA isolation and cDNA synthesis were performed as by Wormuth et al. [77], and quantitative real-time PCR (qPCR) as by Oelze et al. [31] with the single difference of using MESA blue MasterMix plus SYBR assay (Eurogentec, Belgium). Actin2 (At3g18780) was used as reference gene.
(e). Ascorbate and glutathione content
Reduced (ASC) and oxidized ascorbic acid (DHA) were determined as by Horling et al. [78]. For the determination of reduced and oxidized glutathione (GSH/GSSG), an enzyme cycling assay based on sequential oxidation of GSH by 2,2′-dinitro-5′5-dithiodibenzoic acid (DTNB) and reduction by NADPH in the presence of glutathione reductase was performed as by Oelze et al. [31].
(f). Analysis of oxylipins, salicylic acid, indole acetic acid and abscisic acid in Arabidopsis thaliana
The analysis of hormones was performed according to Stingl et al. [79]. Plant material was extracted with 950 µl ethyl acetate/formic acid (99:1, v/v). Dihydrojasmonic acid (50 ng), JA-nor-Val (50 ng), [18O2]OPDA (50 ng), [D6]ABA (50 ng), [D4]SA (50 ng) and [D2]IAA (50 ng) were added as internal standards. Samples were homogenized with a ball mill for 3 min at 20 Hz. Pellets following centrifugation were re-extracted, and the pooled supernatants were dried in a vacuum concentrator. After evaporation of the solvent, samples were dissolved in 40 µl acetonitrile/water (50:50, v/v) and analysed by liquid chromatography (LC)–tandem mass spectrometry (MS/MS).
A. thaliana ID numbers: Actin (At3g18780), β-amylase (At4g15210), COR47 (At1g20440), MDHAR (At1g63940), PKRP (At3g49160), PPTE (At4g17470).
Acknowledgements
M.O.V. designed the research, analysed the microarray data and wrote the paper. M.L.O. designed the research, analysed photosynthetic parameters and prepared the transcript profiling. K.A., M.M. and K.K. performed measurements, analysed and interpreted data. N.S. and M.J.M. performed the LC–MS/MS analyses and discussed the data. H.F. analysed and interpreted data. K.J.D. designed, supervised and discussed the study, and wrote the paper.
Funding statement
The authors acknowledge funding by the Deutsche Forschungsgemeinschaft (FOR 804). K.A. acknowledges support by Mu'tah University (Jordan). Professional technical assistance by Petra Witte-Brüggemann and Heike Bogunovic is highly appreciated.
References
- 1.Walters RG, Ruban AV, Horton P. 1996. Identification of proton-active residues in a higher plant light-harvesting complex. Proc. Natl Acad. Sci. USA 93, 14 204–14 209. ( 10.1073/pnas.93.24.14204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Demmig-Adams B, Adams WW. 1996. The role of xanthophylls cycle carotenoids in the protection of photosynthesis. Trends Plant Sci. 1, 21–26. ( 10.1016/S1360-1385(96)80019-7) [DOI] [Google Scholar]
- 3.Niyogii KK. 1999. Photoprotection revisited: genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 333–359. ( 10.1146/annurev.arplant.50.1.333) [DOI] [PubMed] [Google Scholar]
- 4.Mehler AH. 1951. Studies on reactions of illuminated chloroplasts. I. Mechanism of the reduction of oxygen and other Hill reagents. Arch. Biochem. Biophys. 33, 65–77. ( 10.1016/0003-9861(51)90082-3) [DOI] [PubMed] [Google Scholar]
- 5.Owens TG. 1994. Processing of excitation energy by antenna pigments. In Photosynthesis and environment (ed. Baker NR.), pp. 1–23. Dordrecht, The Netherlands: Kluwer. [Google Scholar]
- 6.Asada K. 1999. The water–water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess light. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 601–639. ( 10.1146/annurev.arplant.50.1.601) [DOI] [PubMed] [Google Scholar]
- 7.Asada K. 2000. The water–water cycle as alternative photon and electron sinks. Phil. Trans. R. Soc. Lond. B 355, 1419–1431. ( 10.1098/rstb.2000.0703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dietz KJ, Jacob S, Oelze ML, Laxa M, Tognetti V, de Miranda SM, Baier M, Finkemeier I. 2006. The function of peroxiredoxins in plant organelle redox metabolism. J. Exp. Bot. 57, 1697–1709. ( 10.1093/jxb/erj160) [DOI] [PubMed] [Google Scholar]
- 9.Muthuramalingam M, Seidel T, Laxa M, Nunes de Miranda SM, Gärtner F, Ströher E, Kandlbinder A, Dietz KJ. 2009. Multiple redox and non-redox interactions define 2-Cys peroxiredoxin as a regulatory hub in the chloroplast. Mol. Plant 2, 1273–1288. ( 10.1093/mp/ssp089) [DOI] [PubMed] [Google Scholar]
- 10.Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM. 1997. Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9, 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karpinski S, Reynolds H, Karpinska B, Wingsle G, Creissen G, Mullineaux P. 1999. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284, 654–657. ( 10.1126/science.284.5414.654) [DOI] [PubMed] [Google Scholar]
- 12.Møller IM, Jensen PE, Hansson A. 2007. Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 58, 459–481. ( 10.1146/annurev.arplant.58.032806.103946) [DOI] [PubMed] [Google Scholar]
- 13.Sakamoto W, Miyagishima SY, Jarvis P. 2008. Chloroplast biogenesis: control of plastid development, protein import, division and inheritance. The Arabidopsis book 6, e00110 ( 10.1199/tab.0110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foyer CH, Noctor G. 2003. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol. Plant. 119, 355–364. ( 10.1034/j.1399-3054.2003.00223.x) [DOI] [Google Scholar]
- 15.Baier M, Dietz KJ. 2005. Chloroplasts as source and target of cellular redox regulation: a discussion on chloroplast redox signals in the context of plant physiology. J. Exp. Bot. 56, 1449–1462. ( 10.1093/jxb/eri161) [DOI] [PubMed] [Google Scholar]
- 16.Nott A, Jung HS, Koussevvitzky S, Chory J. 2006. Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57, 739–759. ( 10.1146/annurev.arplant.57.032905.105310) [DOI] [PubMed] [Google Scholar]
- 17.Pogson BJ, Nick SW, Förster B, Small I. 2008. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 13, 602–609. [DOI] [PubMed] [Google Scholar]
- 18.Chi W, Sun X, Zhang L. 2013. Intracellular signaling from plastid to nucleus. Annu. Rev. Plant. Biol. 64, 559–582. ( 10.1146/annurev-arplant-050312-120147) [DOI] [PubMed] [Google Scholar]
- 19.Szechynska-Hebda M, Karpinski S. 2013. Light intensity-dependent retrograde signalling in higher plants. J. Plant Physiol. 170, 1501–1516. ( 10.1016/j.jplph) [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Jia W, Yang J, Ismael AM. 2006. Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res. 97, 111–119. ( 10.1016/j.fcr.2005.08.018) [DOI] [Google Scholar]
- 21.Reinbothe S, Reinbothe C, Parthier B. 1993. Methyl jasmonate-regulated translation of nuclear-encoded chloroplast proteins in barley (Hordeum vulgare L. cv Salome). J. Biol. Chem. 268, 10 606–10 611. [PubMed] [Google Scholar]
- 22.Desikan R, Mackerness S, Hancock J, Neill S. 2001. Regulation of Arabidopsis transcriptome by oxidative stress. Plant Physiol. 127, 159–172. ( 10.1104/pp.127.1.159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maruta T, Noshi M, Tanouchi A, Tamoi M, Yabuta Y, Yoshimura K, Ishikawa T, Shigeoka S. 2012. H2O2-triggered retrograde signaling from chloroplasts to nucleus plays specific role in response to stress. J Biol. Chem. 287, 11 717–11 729. ( 10.1074/jbc.M111.292847) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Op den Camp RGL, et al. 2003. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15, 2320–2332. ( 10.1105/tpc.014662) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossel JB, et al. 2007. Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell 19, 4091–4110. ( 10.1105/tpc.106.045898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lepetit B, Sturm S, Rogato A, Gruber A, Sachse M, Falciatore A, Kroth PG, Lavaud J. 2013. High light acclimation in the secondary plastids containing diatom Phaeodactylum tricornutum is triggered by the redox state of the plastoquinone pool. Plant Physiol. 161, 853–865. ( 10.1104/pp.112.207811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pesaresi P, et al. 2009. Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 21, 2402–2423. ( 10.1105/tpc.108.064964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong L, Zhu JK. 2003. Regulation of abscisic acid biosynthesis. Plant Physiol. 133, 29–36. ( 10.1104/pp.103.025395) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wasternack C, Kombrink E. 2010. Jasmonates: structural requirements for lipid-derived signals active in plant stress responses and development. Am. Chem. Soc. J. 5, 63–77. [DOI] [PubMed] [Google Scholar]
- 30.Leister D. 2012. Retrograde signaling in plants: from simple to complex scenarios. Front. Plant Sci. 3, 135 ( 10.3389/fpls.2012.00135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oelze ML, Vogel MO, Alsharafa K, Kahmann U, Viehhauser A, Maurino VG, Dietz KJ. 2012. Efficient acclimation of the chloroplast antioxidant defence of Arabidopsis thaliana leaves in response to a 10- or 100-fold light increment and the possible involvement of retrograde signals. J. Exp. Bot. 63, 1297–1313. ( 10.1093/jxb/err356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thimm O, et al. 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–39. ( 10.1111/j.1365-313X.2004.02016.x) [DOI] [PubMed] [Google Scholar]
- 33.Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. 2004. Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. [DOI] [PubMed] [Google Scholar]
- 34.Baier M, Hemmann G, Holman R, Corke F, Card R, Smith C, Rook F, Bevan MW. 2004. Characterization of mutants in Arabidopsis showing increased sugar-specific gene expression, growth, and developmental responses. Plant Physiol. 134, 81–91. ( 10.1104/pp.103.031674) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bechtold U, Richard O, Zamboni A, Gapper C, Geisler M, Pogson B, Karpinski S, Mullineaux PM. 2008. Impact of chloroplastic- and extracellular-sourced ROS on high light-responsive gene expression in Arabidopsis. J. Exp. Bot. 59, 121–133. ( 10.1093/jxb/erm289) [DOI] [PubMed] [Google Scholar]
- 36.Blanco F, Salinas P, Cecchini NM, Jordana X, Van Hummelen P, Alvarez ME, Holuigue L. 2009. Early genomic responses to salicylic acid in Arabidopsis. Plant Mol. Biol. 70, 79–102. ( 10.1007/s11103-009-9458-1) [DOI] [PubMed] [Google Scholar]
- 37.Davletova S, Rizhsky L, Liang HJ, Zhong SQ, Oliver DJ, Shulaev V, Schlauch K, Mittler R. 2005. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17, 268–281. ( 10.1105/tpc.104.026971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gadjev I, et al. 2005. Transcriptomic footprints disclose specificity of reactive oxygen species signalling in Arabidopsis. Plant Physiol. 141, 436–445. ( 10.1104/pp.106.078717) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goda H, et al. 2008. The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 55, 526–542. ( 10.1111/j.1365-313X.2008.03510.x) [DOI] [PubMed] [Google Scholar]
- 40.Lintala M, Allahverdiyeva Y, Kangasjarvi S, Lehtimaki N, Keranen M, Rintamaki E, Aro EM. 2009. Comparative analysis of leaf-type ferredoxin-NADP(+) oxidoreductase isoforms in Arabidopsis thaliana. Plant J. 57, 1103–1115. ( 10.1111/j.1365-313X.2008.03753.x) [DOI] [PubMed] [Google Scholar]
- 41.Matsui A, et al. 2008. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 49, 1135–1149. ( 10.1093/pcp/pcn101) [DOI] [PubMed] [Google Scholar]
- 42.Mazella MA, et al. 2005. Phytochrome control of the Arabidopsis transcriptome anticipates seedling exposure to light. Plant Cell 17, 2507–2516. ( 10.1105/tpc.105.034322) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller R, Nilsson L, Nielsen LK, Nielsen TH. 2005. Interaction between phosphate starvation signalling and hexokinase-independent sugar sensing in Arabidopsis leaves. Physiol. Plant. 124, 81–90. ( 10.1111/j.1399-3054.2005.00496.x) [DOI] [Google Scholar]
- 44.Mullineaux PM, Ball L, Escobar C, Karpinska B, Creissen G, Karpinski S. 2000. Are diverse signalling pathways integrated in the regulation of Arabidopsis antioxidant defence gene expression in response to excess excitation energy? Phil. Trans. R. Soc. Lond. B 355, 1531–1540. ( 10.1098/rstb.2000.0713) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pastori GM, Kiddle G, Antoniw J, Bernard S, Veljovc-Jovanovic S, Verrier PJ, Noctor G, Foyer CH. 2003. Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signalling. Plant Cell 15, 939–951. ( 10.1105/tpc.010538) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfannschmidt T, Nilsson A, Allen JF. 1999. Photosynthetic control of chloroplast gene expression. Nature 397, 625–628. ( 10.1038/17624) [DOI] [Google Scholar]
- 47.Pfannschmidt T, Allen JF, Oelmüller R. 2001. Principles of redox control in photosynthesis gene expression. Physiol. Plant. 112, 1–9. ( 10.1034/j.1399-3054.2001.1120101.x) [DOI] [Google Scholar]
- 48.Sanchez JP, Duque P, Chua NH. 2004. ABA activates ADPR cyclase and cADPR induces a subset of ABA-responsive genes in Arabidopsis. Plant J. 38, 381–395. ( 10.1111/j.1365-313X.2004.02055.x) [DOI] [PubMed] [Google Scholar]
- 49.Sellaro R, Hoecker U, Yanovski M, Chory J, Casal JJ. 2009. Synergism of red and blue light in the control of Arabidopsis gene expression and development. Curr. Biol. 19, 1216–1220. ( 10.1016/j.cub.2009.05.062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torres MA, Dangl JL, Jones JD. 2002. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for the accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99, 517–522. ( 10.1073/pnas.012452499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang DH, Andersson B, Aro EM, Ohad I. 2001. The redox state of plastoqinone pool controls the level of light-harvesting chlorophyll a/b protein of photosystem II in response to elevated light intensities. Plant Physiol. 118, 827–834. ( 10.1104/pp.118.3.827) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katsir L, Chung HS, Koo AJK, Howe GA. 2008. Jasmonate signalling: a conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 11, 428–435. ( 10.1016/j.pbi.2008.05.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park SW, et al. 2013. Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc. Natl Acad. Sci. USA 110, 9559–9564. ( 10.1073/pnas.1218872110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niki T, Mitsuhara I, Seo S, Ohtsubo N, Ohashi Y. 1998. Antagonistic effect of salicylic acid and jasmonic acid on the expression of pathogenesis-related (PR) protein genes in wounded mature tobacco leaves. Plant Cell Physiol. 39, 500–507. ( 10.1093/oxfordjournals.pcp.a029397) [DOI] [Google Scholar]
- 55.Himmelbach A, Iten M, Grill E. 1998. Signalling of abscisic acid to regulate plant growth. Phil. Trans. R. Soc. Lond. B 353, 1439–1444. ( 10.1098/rstb.1998.0299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roychoudhury A, Paul S, Basu S. 2013. Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep. 32, 985–1006. ( 10.1007/s00299-013-1414-5) [DOI] [PubMed] [Google Scholar]
- 57.Normanly J. 2010. Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harbor Perspect. Biol. 2, a001594 ( 10.1101/cshperspect.a001594) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tognetti V, Mühlenbock P, Van Breusegem F. 2011. Stress homeostasis – the redox and auxin perspective. Plant Cell Environ. 35, 321–333. ( 10.1111/j.1365-3040.2011.02324.x) [DOI] [PubMed] [Google Scholar]
- 59.Rosenwasser S, Fluhr R, Joshi RJ, Leviatan N, Sela N, Hetzroni A, Friedman H. 2013. ROSMETER: a bioinformatic tool for the identification of reactive oxygen species type and origin in plants. Plant Physiol. 163, 1071–1083. ( 10.1104/pp.113.218206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young AJ, Frank HA. 1996. Energy transfer reactions involving carotenoids: quenching of chlorophyll fluorescence. J. Photochem. Photobiol. B 36, 3–15. ( 10.1016/S1011-1344(96)07397-6) [DOI] [PubMed] [Google Scholar]
- 61.Stintzi A, Weber H, Reymond P, Browse J, Farmer EE. 2001. Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc. Natl Acad. Sci. USA 98, 12 837–12 842. ( 10.1073/pnas.211311098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feussner I, Wasternack C. 2002. The lipoxygenase pathway. Annu. Rev. Plant Biol. 53, 275–297. ( 10.1146/annurev.arplant.53.100301.135248) [DOI] [PubMed] [Google Scholar]
- 63.Dominguez-Solis JR, He Z, Lima A, Ting J, Buchanan BB, Luan S. 2008. A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts. Proc. Natl Acad. Sci. USA 105, 16 386–16 391. ( 10.1073/pnas.0808204105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mueller S, Hilbert B, Dueckershoff K, Roitsch T, Krischke M, Mueller MJ, Berger S. 2008. General detoxification and general stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20, 768–785. ( 10.1105/tpc.107.054809) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van der Does D, et al. 2013. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25, 744–761. ( 10.1105/tpc.112.108548) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D. 2002. The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14, 1919–1935. ( 10.1105/tpc.003368) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koornneef A, Leon Reyes HA, Ritsema T, Verhage A, den Otter FC, van Loon LC, Pieterse CMJ. 2008. Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 147, 1358–1368. ( 10.1104/pp.108.121392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mühlenbock P, Szechynska-Hebda M, Plaszczyca M, Baudo M, Mullineaux PM, Parker JE, Karpinska B, Karpinski S. 2008. Chloroplast signaling and lesion simulating disease1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20, 2339–2356. ( 10.1105/tpc.108.059618) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Slovik S, Hartung W. 1992. Compartmental distribution and redistribution of abscisic acid in intact leaves. 3. Analysis of the stress signal chain. Planta 187, 37–47. ( 10.1007/BF00201621) [DOI] [PubMed] [Google Scholar]
- 70.Bonardi V, Pesaresi P, Becker T, Schleiff E, Wagner R, Pfannschmidt T, Jahns P, Leister D. 2005. Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437, 1179–1182. ( 10.1038/nature04016) [DOI] [PubMed] [Google Scholar]
- 71.Kopecná J, Komenda J, Bucinská L, Sobotka R. 2013. Long term acclimation of the cyanobacterium Synechocystis sp. PCC 6803 to high light is accompanied by an enhanced production of chlorophyll that is preferentially channelled to trimeric photosystem I. Plant Physiol. 160, 2239–2250. ( 10.1104/pp.112.207274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Galvez-Valdivieso G, et al. 2009. The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 21, 2143–2162. ( 10.1105/tpc.108.061507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. 2003. Summaries of affymetrix GeneChip probe level data. Nucl. Acids Res. 31, e15 ( 10.1093/nar/gng015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ji X, Dong B, Shiram B, Talbot MJ, Edlington JE, Hughes T, White RG, Gubler F, Dolferus R. 2011. Control of abscisic acid catabolism and abscisic acid homeostasis is important for reproductive stage stress tolerance in cereals. Plant Physiol. 156, 647–662. ( 10.1104/pp.111.176164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. [Google Scholar]
- 76.Benjamini Y, Hochberg Y. 2000. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J. Educ. Behav. Stat. 25, 60–83. [Google Scholar]
- 77.Wormuth D, Baier M, Kandlbinder A, Scheibe R, Hartung W, Dietz KJ. 2006. Regulation of gene expression by photosynthetic signals triggered through modified CO2 availability. BMC Plant Biol. 6, 15–39. ( 10.1186/1471-2229-6-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Horling F, Lamkemeyer P, König J, Finkemeier I, Baier M, Kandlbinder A, Dietz KJ. 2003. Divergent light-, ascorbate-, and oxidative stress-dependent regulation of expression of the peroxiredoxin gene family in Arabidopsis thaliana. Plant Physiol. 131, 317–325. ( 10.1104/pp.010017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stingl N, Krischke M, Fekete A, Mueller MJ. 2013. Analysis of defense signals in Arabidopsis thaliana leaves by ultra-performance liquid chromatography/tandem mass spectrometry: jasmonates, salicylic acid, abscisic acid. Methods Mol. Biol. 1009, 103–113. ( 10.1007/978-1-62703-401-2_11) [DOI] [PubMed] [Google Scholar]