Abstract
The development and severity of inflammatory bowel diseases (IBD) and other chronic inflammatory conditions can be influenced by host genetic and environmental factors, including signals derived from commensal bacteria1–6. However, the mechanisms that integrate these diverse cues remain undefined. Here we demonstrate that mice with an intestinal epithelial cell-specific deletion of the epigenome-modifying enzyme histone deacetylase 3 (HDAC3ΔIEC mice) exhibited extensive dysregulation of IEC-intrinsic gene expression, including decreased basal expression of genes associated with antimicrobial defense. Critically, conventionally-housed HDAC3ΔIEC mice demonstrated loss of Paneth cells, impaired IEC function and alterations in the composition of intestinal commensal bacteria. In addition, HDAC3ΔIEC mice exhibited significantly increased susceptibility to intestinal damage and inflammation, indicating that epithelial expression of HDAC3 plays a central role in maintaining intestinal homeostasis. Rederivation of HDAC3ΔIEC mice into germ-free conditions revealed that dysregulated IEC gene expression, Paneth cell homeostasis, and intestinal barrier function were largely restored in the absence of commensal bacteria. While the specific mechanisms through which IEC-intrinsic HDAC3 expression regulates these complex phenotypes remain to be elucidated, these data indicate that HDAC3 is a critical factor that integrates commensal bacteria-derived signals to calibrate epithelial cell responses required to establish normal host-commensal relationships and maintain intestinal homeostasis.
Chronic inflammatory diseases, including asthma, allergy, diabetes, and IBD, are multifactorial diseases that develop as a result of complex gene-environment interactions1–6. Genome-wide association studies have identified more than 160 genes or loci that are associated with IBD7. In addition, signals derived from intestinal commensal microbial communities are not only required for normal intestinal function, but also act as environmental cues that influence IBD in genetically susceptible hosts1–3, 8, 9. Intestinal epithelial cells (IECs) function as a crucial cell lineage that integrates microbial signals from the intestinal microenvironment to regulate gene expression and intestinal homeostasis10, 11, however the mechanisms that coordinate these processes remain undefined. HDACs are epigenome-modifying enzymes that alter gene expression and can be regulated by endogenous factors, dietary components, synthetic inhibitors, and bacteria-derived signals and synthetic inhibitors12–17. The class I HDAC, HDAC3 alters transcription through histone deacetylation, and may also mediate the activity of other HDACs, deacetylate non-histone targets, and possess enzyme-independent effects18–21. Tissue-specific deletion of HDAC3 in murine models has suggested critical roles for HDAC3 in complex diseases such as diabetes and heart failure22, 23, however the functional roles of HDAC3 in regulating intestinal homeostasis in the context of health and disease are unknown.
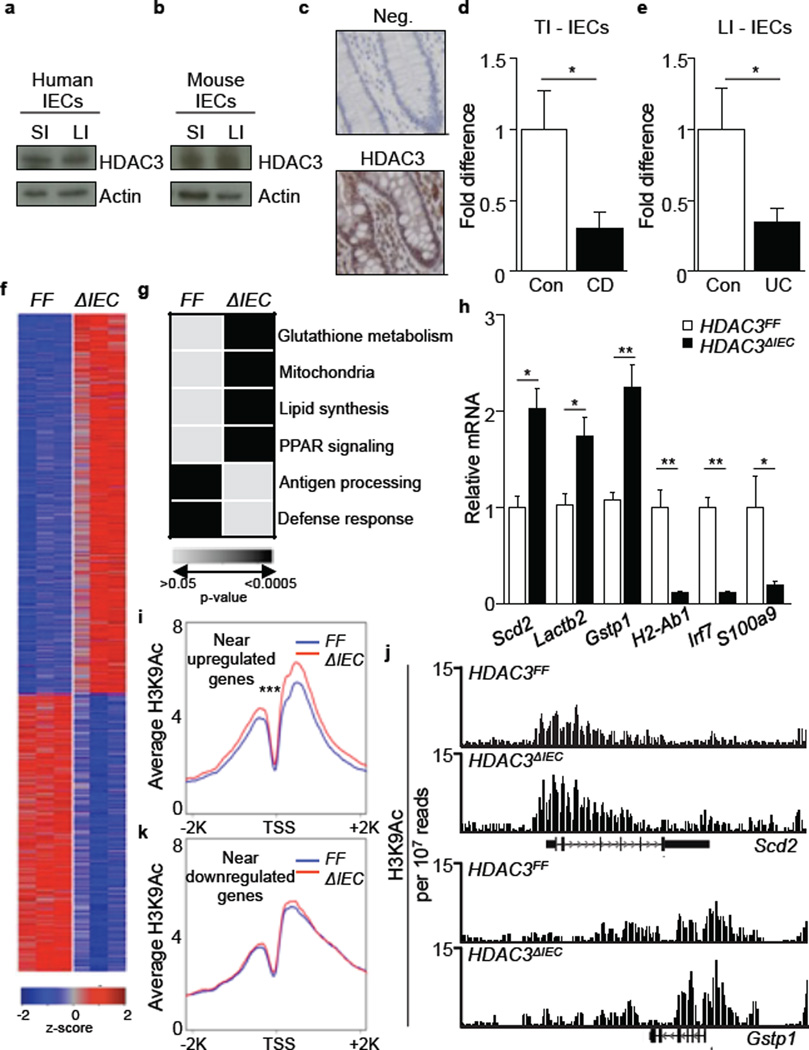
In order to characterize HDAC3 expression in the intestinal epithelium, intestinal samples from healthy humans and mice were evaluated. HDAC3 protein was expressed in IECs from human and mouse small and large intestine, and immunohistochemistry revealed nuclear localization of HDAC3 in healthy human colonic IECs (Fig. 1a-c). IECs were also isolated from IBD patients with either Crohn’s disease (CD), which commonly targets the terminal ileum, or ulcerative colitis (UC), which is restricted to the large intestine. HDAC3 expression was significantly decreased in IECs isolated from the terminal ileum of CD patients (Fig. 1d) and the large intestine of UC patients (Fig. 1e) compared to control patients, suggesting that dysregulated expression of HDAC3 in IECs may be associated with regions of active disease in both forms of IBD.
Figure 1. Decreased expression of HDAC3 in IECs is associated with global alterations in gene expression and histone acetylation.
(a, b) HDAC3 in (a) human and (b) mouse IECs by Western analysis (small (SI) and large (LI) intestine). (c) HDAC3 expression in human colon. No primary antibody (Neg). Bars, 25µm. (d, e) HDAC3 mRNA in IECs from (d) terminal ileum (TI) or (e) LI of control (Con), Crohn’s disease (CD), or ulcerative colitis (UC) patients. *p<0.05, Mann Whitney (TI: 8 Con, 9 CD; LI: 10 Con, 8 UC). (f) Expression heat-map in sorted EpCAM+ IECs from the large intestine of HDAC3FF(FF) versus HDAC3ΔIEC(ΔIEC) mice (fold change > 1.5, row-normalized Z-score). (g) Enriched pathways using DAVID. (h) mRNA in large intestinal IECs. (i) Average profile of H3K9Ac near upregulated genes in HDAC3ΔIEC mice. ***p = 8.64e-44. (j) Representative distribution of H3K9Ac at select genes from (i). (k) Average profile of H3K9Ac near downregulated genes in HDAC3ΔIEC mice. H3K9Ac signals are normalized to reads per 10 million mapped reads. n=3 mice per group. Results are shown as mean ± SEM. *p< 0.05.
To investigate the in vivo functions of IEC-intrinsic HDAC3 expression, IEC-specific HDAC3 deficient (HDAC3ΔIEC) mice were generated. HDAC3ΔIEC mice were born at normal Mendelian frequencies, and deletion of HDAC3 was confirmed in IECs Extended Data Fig. 1a, b). Genome-wide transcriptional profiling on sort-purified live, EpCAM+ IECs from the large intestine revealed that in vivo deletion of HDAC3 resulted in substantial alterations in IEC-intrinsic gene expression (Extended Data Fig. 1c, Fig. 1f). The majority of genes that exhibited dysregulated expression were upregulated compared to HDAC3FF mice, consistent with a role for HDAC3 in transcriptional repression (Fig. 1f). DAVID and gene-set enrichment analyses revealed several HDAC3-dependent pathways in IECs, including those involved in glutathione metabolism, mitochondria, lipid biosynthesis, PPAR signaling, antigen processing and defense response (Fig. 1g, Extended Data Fig. 1d). Altered expression of representative genes in these pathways was confirmed by real-time PCR analysis (Fig. 1h). Collectively, these genome-wide analyses implicate a central role for HDAC3 in coordinating a network of IEC-intrinsic transcriptional pathways that regulate multiple cellular processes.
Analyses of histone acetylation in primary IECs from HDAC3FF and HDAC3ΔIEC mice were conducted utilizing genome-wide chromatin immunoprecipitation-sequencing (ChIP-seq) for H3K9Ac, a histone mark that can be a target for HDAC3 at repressed target genes24. ChIP-seq analyses revealed that H3K9Ac levels were significantly increased near genes that were upregulated in IECs isolated from HDAC3ΔIEC mice (Fig. 1i, Extended Data Fig. 1e). The distribution of H3K9Ac at two representative genes, Scd2 and Gstp1, demonstrated multiple sites of acetylation (Fig. 1j) and increased levels of H3K9Ac in multiple genes from HDAC3-deficient IECs were confirmed by ChIP-qPCR (Extended Data Fig. 1f). Collectively, these data suggest that alterations in histone acetylation occur at upregulated genes in IECs deficient in HDAC3. Significant changes in H3K9Ac were not observed near genes whose expression was decreased in IECs from HDAC3ΔIEC mice (Fig. 1k), indicating that these genes are unlikely to be direct targets of HDAC3 enzymatic activity. Further analysis will be required to determine whether HDAC3-dependent histone acetylation, deacetylation of non-histone proteins, or enzyme-independent processes are required for specific transcriptional changes observed in IECs isolated from HDAC3ΔIEC mice.
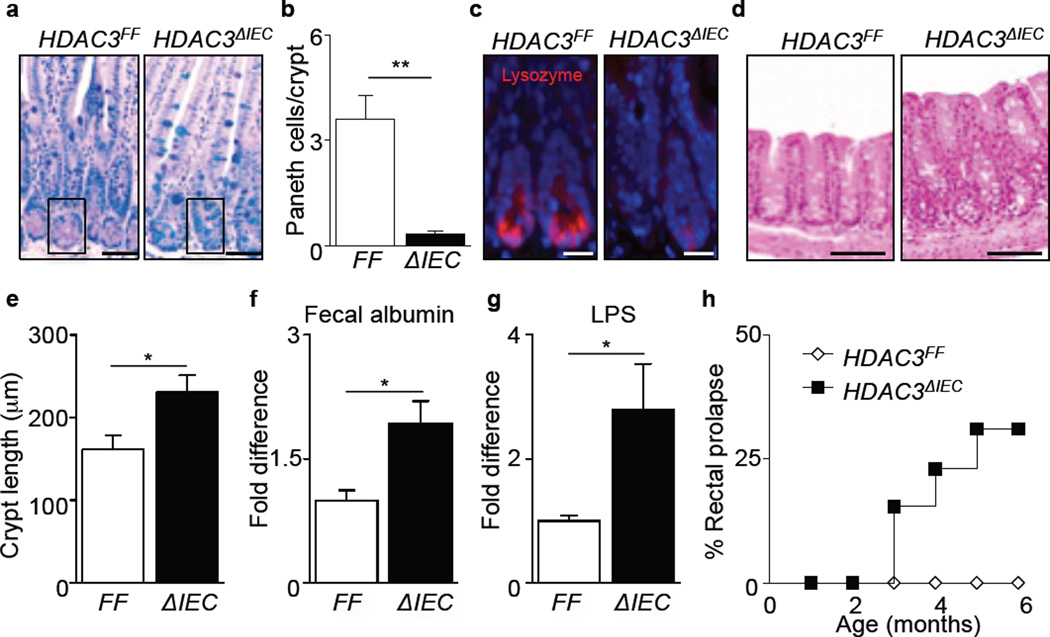
While genome-wide analyses demonstrated significant alterations in gene expression in HDAC3ΔIEC mice, histologic analyses of HDAC3FF versus HDAC3ΔIEC mice revealed normal intestinal architecture in HDAC3ΔIEC mice (Fig. 2a). However, fundamental alterations in Paneth cells were observed in HDAC3ΔIEC mice, as indicated by significantly decreased numbers of these cells and reduced lysozyme expression (Fig. 2a-c). Paneth cells were observed in postnatal 18 day old HDAC3ΔIEC mice (Extended Data Fig. 2a), suggesting that the absence of Paneth cells in adult HDAC3ΔIEC mice did not reflect a primary intrinsic developmental defect. Instead, active caspase-3 staining demonstrated elevated cell death in crypts of adult HDAC3ΔIEC mice (Extended Data Fig. 2b), and electron microscopy revealed the presence of degenerating organelle membranes and loss of granules in Paneth cells of adult HDAC3ΔIEC mice (Extended Data Fig. 2c). Although inhibition of Paneth cell differentiation in adult mice may also occur, these data suggest that impaired Paneth cell survival contributes to altered Paneth cell homeostasis adult HDAC3ΔIEC mice. Further, evaluation of the large intestine revealed that HDAC3ΔIEC mice exhibited crypt elongation (Fig. 2d, e) and more extensive Ki-67 staining within IECs (Extended Data Fig. 2d), indicating that there is increased IEC proliferation in HDAC3ΔIEC mice. Collectively, these findings identify that HDAC3 regulates IEC homeostasis throughout the intestine.
Figure 2. IEC-intrinsic HDAC3 expression regulates intestinal homeostasis.
(a) PAS/Alcian Blue stained ileal sections. Box surrounds Paneth cells. Bars, 50 µm. (b) Paneth cells per crypt. (c) Immunofluorescent staining of lysozyme (pink) and nuclei (blue). Bars, 10µm. (d) Colonic sections. Bars, 100µm. (e) Crypt length in colon. n = 3 mice per group. (f) Albumin from fecal samples of HDAC3FF (n=8) and HDAC3ΔIEC (n=6) mice. (g) LPS levels in mesenteric lymph node (mLN). n=8 mice per group. (h) Development of rectal prolapse in HDAC3ΔIEC mice (n=13). Data are representative of 2–3 independent experiments. Results are shown as mean ± SEM. *p< 0.05, **p<0.01.
We sought to test whether the alterations in IEC homeostasis observed in HDAC3ΔIEC mice were associated with alterations in intestinal barrier function. Naïve HDAC3ΔIEC mice exhibited increased fecal albumin (Fig. 2f), increased plasma levels of FITC following oral administration of FITC-dextran (Extended Data Fig. 3a), and increased LPS in mesenteric lymph nodes (Fig. 2g) compared to HDAC3FF mice, indicating that HDAC3ΔIEC mice exhibit impaired intestinal barrier function and bacterial translocation. Consistent with the lack of Paneth cells, HDAC3ΔIEC mice also exhibited impaired crypt bactericidal activity (Extended Data Fig. 3b) and increased susceptibility to oral Listeria monocytogenes infection (Extended Data Fig. 3c, d). Further, as HDAC3ΔIEC mice matured, they demonstrated an increased prevalence of rectal prolapse (Fig. 2h, Extended Data Fig. 3e), and colons from these mice exhibited increased inflammation (Extended Data Fig. 3f, g) and elevated disease score (Extended Data Fig. 3h). Collectively, these findings demonstrate that loss of HDAC3 expression in IECs results in impaired barrier function and development of spontaneous intestinal inflammation.
To assess the significance of IEC-intrinsic HDAC3 expression in the context of intestinal damage and inflammation, mice were treated with the dextran sodium sulfate (DSS) for 5 days. While HDAC3FF mice were minimally affected, HDAC3ΔIEC mice exhibited profound weight loss (Fig. 3a), increased disease severity (Fig. 3b), colonic shortening (Fig. 3c, d), increased infiltration of neutrophils and macrophages in the intestine (Fig. 3e), and extensive intestinal ulceration, loss of crypt architecture, edema and inflammation (Fig. 3f), indicating that IEC-intrinsic HDAC3 expression is critical for limiting DSS-induced intestinal damage and inflammation. In addition to IECs, macrophages provide an important link between the microbiota and intestinal homeostasis25, 26. Remarkably, unlike HDAC3ΔIEC mice, exposure of HDAC3ΔLysM mice24 to DSS did not result in significant weight loss, disease, or intestinal inflammation (Extended Data Fig. 4a-d), indicating that IEC-intrinsic, but not LysM-expressing myeloid cell-intrinsic, expression of HDAC3 is critical in maintaining intestinal homeostasis and limiting DSS-induced inflammation.
Figure 3. IEC-intrinsic HDAC3 expression regulates susceptibility to DSS-induced intestinal damage and inflammation.
(a) Changes in body weight following 2.5% DSS. (b) Total disease score and (c, d) colon length (% naive) on day 5 of DSS. (e) Frequencies of neutrophils (CD11b+ Ly6G+) and macrophages (CD11b+ Ly6G−) in the colonic lamina propria. (f) Intestinal sections (Cecum: top; Colon: bottom; inset: 40x of crypt abscess). (g) Changes in body weight for tamoxifen-induced mice. (h) Total disease score on day 5 of DSS. Data are representative of four independent experiments containing 4 mice per group. (i) Comparison of stool bacterial communities at multiple timepoints (red circles: 6 weeks, blue squares: 8 weeks, orange triangle: 10 weeks). (j) Phylum comparison of compiled samples from (i). (k, l) Average UniFrac distance between HDAC3FF and HDAC3ΔIEC mice (FF-ΔIEC) or within genotypes (FF-FF or ΔIEC-ΔIEC) based on 16S rRNA gene sequences from (k) small or (l) large intestinal luminal samples. n=3 mice per group. (m, n) Changes in body weight of mice (m) cross-fostered(CF) or (n) co-housed(CH) with HDAC3FF or HDAC3ΔIEC mice prior to DSS treatment. (o) Changes in body weight following DSS and (p) ileal sections from colonized germ-free (GF) mice. Data are representative of 2–3 independent experiments containing 3 mice per group. Bars, 50µm. Results are shown as mean ± SEM *p<0.05, **p<0.01.
To test how IEC-intrinsic HDAC3 expression functions in adult mice and to eliminate potential developmental effects of constitutive deletion, we generated an inducible tamoxifen-dependent IEC-specific HDAC3 knockout mouse model (HDAC3ΔIEC-IND) (Extended Data Fig. 5a) in which depletion of HDAC3 in IECs could be detected after 5 days of tamoxifen treatment (Extended Data Fig. 5b, c). Repeated administration of tamoxifen resulted in decreased Paneth cells and increased caspase-3 staining in the ileal crypts of HDAC3ΔIEC-IND mice (Extended Data Fig. 5d), suggesting that Paneth cells in adult HDAC3ΔIEC-IND mice exhibit altered homeostasis, similar to mice with constitutive deletion of HDAC3 (Extended Data Fig. 2b, c). Further, increased fecal albumin (Extended Data Fig. 5e) and FITC-dextran permeability (Extended Data Fig. 5f) were observed following deletion of HDAC3, demonstrating that IEC-intrinsic expression of HDAC3 dynamically regulates intestinal barrier function in the adult intestine. Tamoxifen-treated HDAC3ΔIEC-IND mice subjected to DSS exhibited profound weight loss (Fig. 3g), increased disease severity (Fig. 3h), exacerbated colonic shortening (Extended Data Fig. 6a, b), increased inflammatory cell infiltrates (Extended Data Fig. 6c), and histologic lesions in both the large and small intestine (Extended Data Fig. 6d). Collectively, these results identify a critical role for HDAC3 in actively regulating IEC function and tissue homeostasis in adult mice.
Dysregulation of intestinal homeostasis and susceptibility to intestinal inflammation are often associated with alterations in commensal bacterial populations27–30. Therefore, pyrosequencing was employed to interrogate temporal and spatial differences in microbial diversity within intestinal bacterial communities. Bacterial communities differed significantly between fecal and intestinal samples of HDAC3FF and HDAC3ΔIEC littermate mice, whereas mice within the same genotype had a more similar bacterial composition (Fig. 3i-l, Extended Data Fig. 7a-c). Most notably, HDAC3ΔIEC mice consistently exhibited increased levels of Proteobacteria (Fig. 3j, Extended Data Fig. 7b,c). Further, analyses of HDAC3ΔIEC-IND mice revealed that alterations in the composition of intestinal bacterial communities occur secondary to deletion of IEC-intrinsic HDAC3 in adult mice (Extended Data Fig. 7d, e). Recent studies have demonstrated that the colitogenic activity of specific microbial communities can result in exacerbated susceptibility to colitis27–29. However, unlike HDAC3ΔIEC mice, wildtype mice that were cross-fostered (Fig. 3m) or co-housed (Fig. 3n) with HDAC3ΔIEC mice did not demonstrate increased susceptibility to DSS (Fig. 3m, n). Furthermore, germ-free wildtype mice colonized with either the HDAC3FF or HDAC3ΔIEC microbiota did not exhibit differences in susceptibility to DSS-induced inflammation (Fig. 3o) or Paneth cell homeostasis (Fig. 3p).
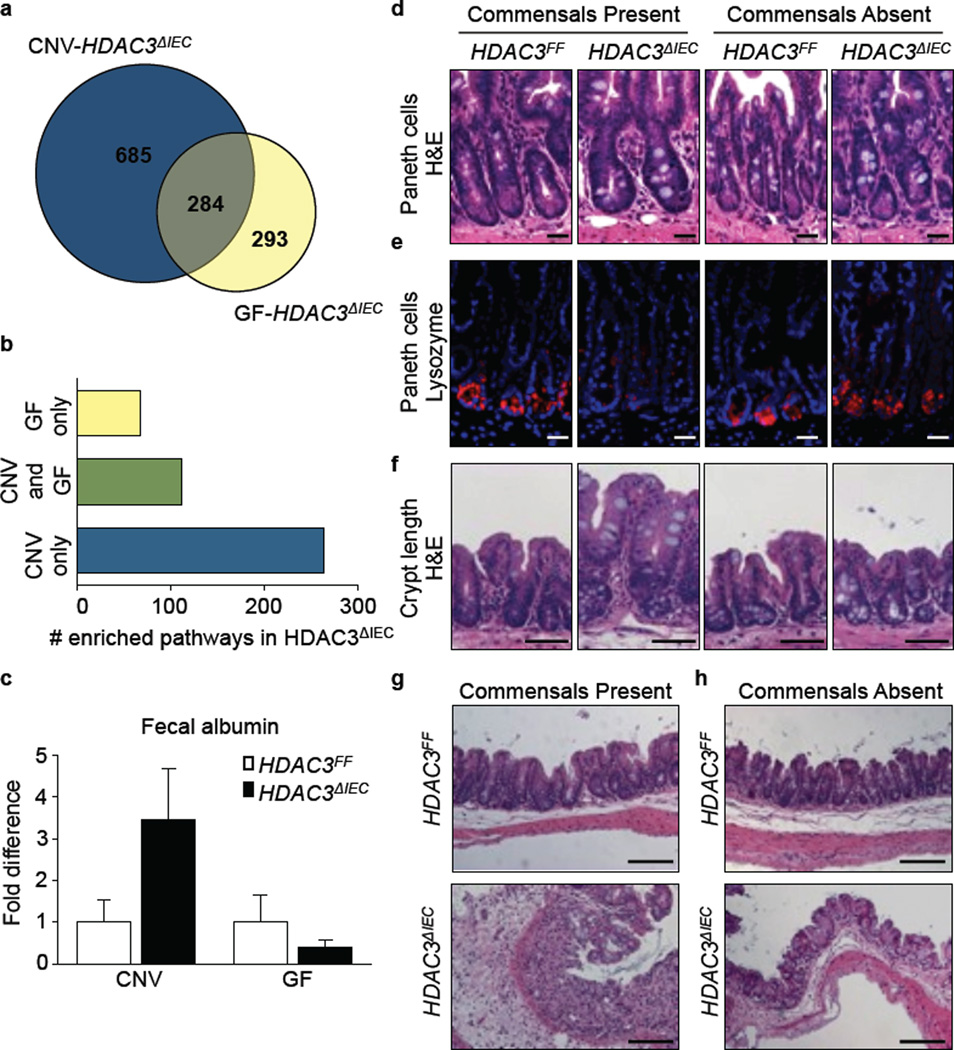
These results indicate that the intestinal dysbiosis alone is insufficient to cause the dysregulation in IEC homeostasis or susceptibility to intestinal inflammation that occurs in HDAC3ΔIEC mice. Therefore, we hypothesized that HDAC3 expression may be required to integrate signals derived from commensal bacteria to regulate intestinal homeostasis. To test this, HDAC3ΔIEC mice were rederived into germ-free (GF) conditions. Genome-wide transcriptional profiling comparing differentially expressed genes of conventionally-housed (CNV)-HDAC3ΔIEC mice versus GF-HDAC3ΔIEC mice revealed that a significant proportion of HDAC3-dependent gene expression relies on the presence of live commensal bacteria (Fig. 4a). Further, alterations in the majority of HDAC3-dependent transcriptional pathways, including those involved in anti-microbial defense, occurred only in HDAC3ΔIEC mice housed under CNV conditions (Fig. 4b, Extended Data Fig. 8), indicating that a dominant role for HDAC3 in regulation of IEC-intrinsic transcriptional pathways requires commensal bacteria-derived signals. Consistent with this, while fecal albumin levels (Fig. 4c), Paneth cell homeostasis (Fig. 4d, e), and crypt length (Fig. 4f) were dysregulated in CNV-HDAC3ΔIEC mice compared to CNV-HDAC3FF mice, these differences were significantly abrogated between GF-HDAC3FF versus GF-HDAC3ΔIEC mice (Fig. 4c-f). In addition, while CNV-HDAC3ΔIEC mice were more susceptible to DSS-induced intestinal inflammation compared to CNV-HDAC3FF mice (Fig. 4g), minimal differences in intestinal inflammation were observed between GF-HDAC3FF versus GF-HDAC3ΔIEC mice (Fig. 4h).
Figure 4. HDAC3-dependent regulation of intestinal homeostasis depends on integration of commensal bacteria-derived signals.
(a) Venn diagram showing overlap of differentially expressed genes in EpCAM+ IECs isolated from conventionally housed (CNV)-HDAC3ΔIEC mice versus Germ-free (GF)-HDAC3ΔIEC mice. Numbers of genes were determined by comparison of HDAC3ΔIEC mice to respective CNV- or GF-HDAC3FF controls. n=3 mice per group. (b) Number of significantly enriched pathways in HDAC3ΔIEC mice using DAVID pathway analysis (p<0.05). (c) Albumin measured by ELISA from fecal samples. n=3 mice per group. (d) H&E and (e) lysozyme stained small intestine. Bars, 20µm. (f) H&E stained large intestine. Bars, 50µm. (g, h) H&E stained large intestine from (g) CNV- or (h) GF-HDAC3FF or HDAC3ΔIEC mice after 4 days of 2.5% DSS administration. Bars, 100µm. Data are representative of two independent experiments. Results are shown as mean ± SEM.
Collectively, these data indicate that expression of HDAC3 in IECs is critical in orchestrating networks of gene expression that regulate IEC function and tissue homeostasis in the presence of commensal bacteria. Remarkably, under GF conditions, HDAC3 expression was dispensable for regulation of epithelial barrier integrity and intestinal homeostasis, indicating that HDAC3 integrates commensal bacteria-derived signals to maintain normal host-commensal relationships (Extended Data Fig. 9). Therefore, in addition to established pathways of immune recognition of commensal bacteria via germ-line encoded pattern recognition receptors, IEC-intrinsic expression of HDAC3 may have an evolutionarily conserved role in regulating host-commensal relationships. In this context, IEC-intrinsic HDAC3 may influence susceptibility to multiple systemic chronic inflammatory diseases that are influenced by both host genetic and microbe-derived factors.
METHODS
Mice
Previously described HDAC3FF mice24 were bred to C57BL/6 mice expressing Cre-recombinase31 or tamoxifen-dependent Cre recombinase32 under the control of the villin promoter (Jackson Laboratory) to generate HDAC3ΔIEC and HDAC3ΔIEC-IND mice, respectively. Genotypes were determined by PCR. Deletion of HDAC3 in HDAC3ΔIEC-IND mice was induced by intraperitoneal injection of 1 mg of tamoxifen (Sigma) once per day for 5 days post–weaning to adult mice that ranged from 8–10 weeks old. For Paneth cell examination in HDAC3ΔIEC-IND mice, tamoxifen was administered for three 5 day intervals during a 30 day period. HDAC3ΔLysM mice have been described previously24 and express Cre recombinase under the control of the Lysozyme M promoter. Germ-free (GF) HDAC3ΔIEC mice were rederived at the University of Pennsylvania Gnotobiotic Mouse Facility. For co-housing experiments, 4 week old WT and HDAC3FF or HDAC3ΔIEC mice were co-housed for 4 weeks. For cross-fostering experiments, newborn HDAC3FF mice were transferred to a litter with a HDAC3ΔIEC dam and HDAC3ΔIEC pups at birth. Colonization of 8 week old WT GF mice was performed with cecal contents harvested from either HDAC3FF or HDAC3ΔIEC mice. All mice were used at 7–12 weeks old, and age- and gender- matched mice were used for all experiments. Animals were housed up to 5 per cage in a ventilated isolator cage system in a 12 hr light/dark cycle, with free access to water and chow. Animals requiring medical attention were provided with appropriate veterinary care by a licensed veterinarian and were excluded from the experiments described. No other exclusion criteria existed. All experiments were done according to the guidelines of the University of Pennsylvania Institutional Animal Care and Use Committee. Germ-free mice were maintained in plastic isolator units, were fed autoclaved feed and water and were routinely monitored to ensure the absence of microbial contamination.
Human intestinal samples, IEC harvest, RNA, ChIP, real-time PCR, Western blotting
De-identified human intestinal tissue from the terminal ileum and rectum were obtained from the University of Pennsylvania IBD Immunology Initiative (IRB 814428). All IBD participants had a confirmed diagnosis of CD or UC by a gastroenterologist and had provided informed consent. Patients younger than 18 years of age were excluded. Controls samples were obtained from consenting men and women older than 18 years of age with no history of a diagnosis of inflammatory bowel disease, microscopic colitis, or ischemic colitis. Human IECs were purified from biopsy samples by incubating tissue in 1 mM EDTA/1 mM DTT and 5% FCS at 37°C for 20–30 minutes and vortexing every 5 minutes. Human IECs were 99.3% negative for CD45+ cells. IECs were isolated from murine samples by shaking intestinal tissue in 1 mM EDTA/1 mM DTT and 5% FCS at 37°C for 10 minutes, resulting in 80–90% EpCAM+ IEC purity. RNA was isolated from cells using the RNeasy Kit (Qiagen) then subjected to reverse transcription with Superscript reverse transcriptase (Invitrogen). ChIP was performed as described previously20, 24 with few modifications. Briefly, cells were fixed in 1% PFA for 10 min and quenched with glycine. Total cell extracts were sonicated using a Bioruptor (Diagenode) and appropriate sonication was confirmed using an Agilent bioanalyzer. Extracts were immunoprecipitated with rabbit anti-H3K9ac (Millipore; 06-942). Real-time PCR was performed using SYBR green chemistry (Applied Biosystems), commercially available primer sets (Qiagen) or custom made primer pairs (Invitrogen). Reactions were run on a real-time PCR system (ABI7500; Applied Biosystems) and data were analyzed with a threshold set in the linear range of amplification and processed based on a standard curve of serial ten-fold dilutions for each primer set. Samples were normalized to an unaffected endogenous control gene and plotted as mean fold difference (+/− SEM) relative to control mice. For Western blot analysis, IECs were lysed in a modified RIPA buffer and lysates were subjected to immunoblot analysis. Blots were probed with rabbit anti-HDAC3 (Santa Cruz) and mouse anti-Actin (Cell Signaling).
Flow cytometry
To isolate lamina propria immune cells, IEC and intraepithelial lymphocyte layers were first stripped by shaking sections of large intestine in 5 mM EDTA/1 mM DTT. Remaining tissue was then digested with collagenase (0.5 mg/ml) to obtain single cell suspensions. For flow cytometry, cells were stained with a combination of the following fluorescence-conjugated mAbs: phycoerythrin (PE)–Texas red conjugated anti-CD11b (Invitrogen), PE-Cy7 conjugated anti-F4/80 (eBioscience), Alexa Fluor 700 conjugated anti-Ly6G (Biolegend), allophycocyanin (APC) conjugated anti-CD3 (eBioscience), PE-Cy5 conjugated anti-CD19 (eBioscience), APC-Cy7 conjugated anti-CD11c (eBioscience), eFluor650NC conjugated anti-CD45 (eBioscience), PerCP-Cy5.5 conjugated anti-CD4 (eBioscience), Dead cells were excluded from analysis through the use of a LIVE/DEAD Fixable Aqua Dead Cell Stain kit (Invitrogen). Samples were acquired on an LSR II (BD Biosciences) and were analyzed with FlowJo software (v9.2; TreeStar).
Microarray analysis
EpCAM+ IECs were sorted as DAPI−, Lin− (CD45, CD4, CD8, CD11b, CD19), EpCAM+ from large intestinal IEC preparations using a BD Aria II with a 100µm nozzle. EpCAM staining was performed using APC conjugated anti-EpCAM (eBioscience; G8.8). Three biological replicates were collected for each genotype and condition, each containing 1.0 – 1.7× 105 cells sorted to a purity of 99%. Total RNA was prepared using TRIzol (Invitrogen) and analyzed by the Microarray Core at the University of Pennsylvania. cDNA was amplified using NuGen WT Ovation Pico kit and hybridized to an Affymetrix GeneChip (Mouse Gene 1.0ST). Affymetrix Power Tools software was used for processing and quantile normalization of fluorescence hybridization signals. Transcripts were log2-normalized and average values were obtained for analysis of expression. Detailed microarray data have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO series accession number GSE50190. Genes with greater than 1.5 fold change were analyzed within DAVID (Database for Annotation, Visualization and Integrated Discovery). The R package ComBat was implemented to eliminate non-biological experimental artifacts33 and the Broad Institute GSEA software was used for gene-set enrichment analysis as described34.
ChIP-seq
ChIP DNA libraries from three biological replicates for each genotype were prepared for sequencing according to the multiplex amplification protocol from Illumina. Sequencing was performed by the The Functional Genomics Core of the Penn Diabetes Research Center. Sequence reads of 50 base pairs (bp) were obtained using the Solexa Analysis Pipeline. ChIP-seq reads for H3K9ac were mapped to the mouse genome (mm9) using bowtie by allowing up to two mismatches35. Reads were normalized to reads per kilobase per 10 million mapped reads and clonal reads were removed. Average H3K9Ac signals for each gene at the TSS +/−2kb region were calculated from 3 independent biologic replicates and p values were determined using a paired t-test. ChIP-seq data have been deposited in GEO and are accessible through GEO series accession number GSE50453.
Histology, immunohistochemistry, immunofluoresence
Sections of intestine were fixed in 4% paraformaldehyde, paraffin embedded, sectioned, and stained with hematoxylin and eosin (H&E), periodic acid-Schiff (PAS)/Alcian blue, anti-HDAC3 (Santa Cruz), anti-Ki67 (AbCam), or anti-caspase 3 (R&D systems) for immunohistochemistry, or anti-Lysozyme (Santa Cruz) and DAPI for immunofluoresence. Pathology was scored based on edema (1–4) and inflammation (1–4). For EM, intestinal tissues were fixed with 2.5% glutaraldehyde, 2.0% paraformaldehyde in 0.1M sodium cacodylate buffer, pH 7.4. After buffer washes, the samples were post-fixed in 2.0% osmium tetroxide for 1 h at room temperature, and rinsed in dH2O prior to en bloc staining with 2% uranyl acetate. After dehydration through a graded ethanol series, the tissue was infiltrated and embedded in EMbed-812 (Electron Microscopy Sciences). Thin sections were stained with uranyl acetate and lead citrate and examined with a JEOL 1010 electron microscope fitted with a Hamamatsu digital camera and AMT Advantage image capture software.
In vivo intestinal barrier function assays
Mice were fasted overnight and FITC-dextran (0.6mg/g; Sigma) diluted in PBS was gavaged the following day. Fluorescence intensity of plasma samples was measured (excitation 485 nm/emission, 535 nm) 4 h after gavage. For fecal albumin assays, fecal pellets were weighed and homogenized in diluent (PBS, 1%BSA, 0.05% Tween20). Albumin levels in fecal homogenates were quantified by ELISA according to the manufacturer’s protocol (Bethyl Laboratories). LPS levels in mesenteric lymph node (mLN) homogenates were assayed via the Limulus amebocyte lysate (LAL) test according to the manufacturer’s protocol (Lonza). Albumin and LPS levels were normalized to fecal or tissue weight and presented as fold difference relative to HDAC3FF mice.
Murine colitis model
DSS (MP Biomedicals) was added to drinking water at 2.5% weight/volume for 5 days. Disease was scored as follows: (a) weight loss (no change = 0; < 5% =1; 6 – 10% = 2; 11 – 20% = 3; > 20% = 4), (b) feces (normal = 0; pasty, semiformed= 2; liquid, sticky, or unable to defecate after 5 min = 4), (c) blood (no blood = 0; visible blood in rectum = 1; visible blood on fur = 2), and (d) general appearance (normal = 0; piloerection = 1; lethargy and piloerection = 2; motionless = 4).
16S rRNA gene pyrosequencing
Stool and intestinal contents total DNA was extracted using the QIAamp DNA Stool Mini Kit (Qiagen). DNA samples were amplified using bar coded V1-V2 region primers targeting bacterial 16S rRNA gene and sequenced using 454/Roche Titanium technology. Sequence analysis was carried out using the QIIME pipeline36 with default settings and data was submitted to the Sequence Read Archive (SRA) and are accessible through accession number SRP029234. Taxonomic assignments were carried out using RDP. Community structure comparisons were carried out using UniFrac37, 38 and principal coordinate analysis. For distance analysis in UniFrac, p values were determined using label permutation as implemented in the QIIME package. Pyrosequencing parameters including number of reads, alpha diversity, rarefaction curves, additional PCA plots, and clustering dendrograms are displayed in Extended Data Figure 10.
Oral L. monocytogenes infection and bactericidal assays
Mice were infected orally with Streptomycin-resistant L. monocytogenes with 3×108 colony forming units (cfu) and weighed daily. 72 hours after infection, mLNs were homogenized in PBS, and serial dilutions of the homogenates were plated on LB plates containing 100µg/ml streptomycin, incubated at 37 °C and cfu were counted. For bactericidal assays, small intestinal crypts were isolated, stimulated with 10µm carbamyl choline (CCh) and assayed against Salmonella typhimurium as described previously39.
Statistics
Results are shown as mean ± SEM. To determine group sizes necessary for adequate statistical power, power analysis was performed using preliminary data sets. Mice of the indicated genotypes were assigned at random to groups. Mouse studies were not performed in a blinded fashion. All inclusion/exclusion criteria were pre-established. Statistical significance was determined with the t-test or Mann-Whitney test. All data meet the assumptions of the statistical tests used. Within each group there is an estimate of variation, and the variance between groups is similar. For each statistical analysis, appropriate tests were selected based on whether the data was normally distributed and whether multiple comparisons were made. Results were considered significant at *p ≤ 0.05; **p ≤ 0.01. Statistical analyses were performed using Prism version 5.0a (GraphPad Software, Inc).
Supplementary Material
Acknowledgements
We thank members of the Artis laboratory for discussions and critical reading of the manuscript. This research is supported by the National Institutes of Health (AI061570, AI095608, AI087990, AI074878, AI095466, AI106697, AI102942 and AI097333 to D.A.; DK043806 to M.A.L.; T32-RR007063, K08-DK093784 to T.A.; DP5OD012116 to G.F.S.; F31-GM082187 to S.A.S.), the Crohns and Colitis Foundation of America (T.A. and D.A.), the Burroughs Wellcome Fund Investigator in Pathogenesis of Infectious Disease Award (D.A.), and the Irvington Institute Postdoctoral Fellowship of the Cancer Research Institute (L.C.O.). We also thank S. Lukovac and R. Aoki (University of Pennsylvania) for technical assistance, the University of Pennsylvania Matthew J. Ryan Veterinary Hospital Pathology Lab, Center for AIDS Research, the Penn Microarray Facility, the NIH/NIDDK Center for Molecular Studies in Digestive and Liver Diseases (P30-DK050306) and its core facilities (Molecular Pathology and Imaging; Molecular Biology; Cell Culture; Transgenic and Chimeric Mouse), the Functional Genomics Core of the Penn Diabetes Research Center (DK19525), the Pathology Core at the Stokes Institute, the Electron Microscopy Resource Laboratory, the Penn IBD Immunology Initiative (I3), and the Mucosal Immunology Studies Team (MIST) of the NIAID for sharing expertise and resources. The authors would also like to thank the Abramson Cancer Center Flow Cytometry and Cell Sorting Resource Laboratory for technical advice and support. The ACC Flow Cytometry and Cell Sorting Shared Resource is partially supported by NCI Comprehensive Cancer Center Support Grant (#2-P30 CA016520). Some human tissue samples were provided by the Cooperative Human Tissue Network (funded by the National Cancer Institute).
Footnotes
Full Methods and any associated references are available in the online version of the paper.
Extended Data Information is available in the online version of the paper.
Author Contributions T.A., L.C.O., S.A.S., D.K., M.W., A.S., P.R.G., K.L.J., and G.F.S. designed and performed the research, T.A. and D.K. rederived the germ-free mice, C.G.K.Z. performed analyses of microarray data, I.C. and K.W. performed analyses of ChIP-seq data, S.G., R.S. and F.D.B. performed and analyzed 454 pyrosequencing, T.B.H. and M.B. provided human tissues, S.E.M., I.E.B., G.F.S., F.D.B., and M.A.L. provided mouse strains, advice or technical expertise, and T.A. and D.A. analyzed the data and wrote the manuscript.
The authors declare no competing financial interests.
REFERENCES
- 1.Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117:514–521. doi: 10.1172/JCI30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renz H, et al. Gene-environment interactions in chronic inflammatory disease. Nat Immunol. 2011;12:273–277. doi: 10.1038/ni0411-273. [DOI] [PubMed] [Google Scholar]
- 5.Slomko H, Heo HJ, Einstein FH. Minireview: Epigenetics of obesity and diabetes in humans. Endocrinology. 2012;153:1025–1030. doi: 10.1210/en.2011-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mukherjee AB, Zhang Z. Allergic asthma: influence of genetic and environmental factors. J Biol Chem. 2011;286:32883–32889. doi: 10.1074/jbc.R110.197046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denson LA, et al. Challenges in IBD research: update on progress and prioritization of the CCFA's research agenda. Inflamm Bowel Dis. 2013;19:677–682. doi: 10.1097/MIB.0b013e31828134b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanov II, Honda K. Intestinal commensal microbes as immune modulators. Cell Host Microbe. 2012;12:496–508. doi: 10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cadwell K, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8:411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 12.Donohoe DR, Bultman SJ. Metaboloepigenetics: interrelationships between energy metabolism and epigenetic control of gene expression. J Cell Physiol. 2012;227:3169–3177. doi: 10.1002/jcp.24054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perissi V, Rosenfeld MG. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol. 2005;6:542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- 14.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim GW, Gocevski G, Wu CJ, Yang XJ. Dietary, metabolic, and potentially environmental modulation of the lysine acetylation machinery. Int J Cell Biol. 2010;2010:632739. doi: 10.1155/2010/632739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dashwood RH, Ho E. Dietary histone deacetylase inhibitors: from cells to mice to man. Semin Cancer Biol. 2007;17:363–369. doi: 10.1016/j.semcancer.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, et al. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc Natl Acad Sci U S A. 2012;109:E2865–E2874. doi: 10.1073/pnas.1121131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 19.You SH, et al. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat Struct Mol Biol. 2013;20:182–187. doi: 10.1038/nsmb.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alenghat T, et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischle W, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 22.Montgomery RL, et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullican SE, et al. Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev. 2011;25:2480–2488. doi: 10.1101/gad.175950.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci U S A. 2005;102:99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diehl GE, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raetz M, et al. Parasite-induced TH1 cells and intestinal dysbiosis cooperate in IFN-gamma-dependent elimination of Paneth cells. Nat Immunol. 2013;14:136–142. doi: 10.1038/ni.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madison BB, et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 32.el Marjou F, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 33.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 34.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ayabe T, et al. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.