Abstract
Infections with West Nile virus (WNV) are typically asymptomatic, but some patients experience severe neurological disease and even death. Over 1500 fatalities have resulted from the more than 37,000 WNV cases in the USA between 1999 and 2012. While it is clear that age is a significant risk factor, markers of immune status associated with susceptibility to severe infections are incompletely defined. We have taken advantage of stable characteristics of individual status to profile immune markers from a stratified cohort of healthy subjects with a history of asymptomatic or severe infection with WNV. We characterized individual variations in antibody and serum cytokine levels and genome-wide transcriptional profiles of peripheral blood cells (PBMCs). While antibody levels were not significantly different between cohorts, we found that subjects with a history of severe infection had significantly lower levels of serum IL-4, and that these changes in IL-4 levels were associated with altered gene expression patterns in PBMCs. In addition, we identified a signature of 105 genes that displayed altered expression levels when comparing subjects with a history of asymptomatic or severe infection. These results suggest that systems-level analysis of immune system status can be used to identify factors relevant for susceptibility to severe infections, and specifically point to an important contribution for IL-4 in resistance to WNV infection.
Introduction
West Nile virus (WNV) is a mosquito-borne enveloped positive-strand RNA virus belonging to the family Flaviviridae, which includes yellow fever and dengue viruses (12). From 1999–2012, 37,088 cases were reported to the Centers for Diseases Control, including 1549 fatalities, and the cumulative incidence of WNV infection may include as many as three million people (27). WNV infections are typically asymptomatic, but patients may experience fever and myalgias to meningoencephalitis and death with severe symptoms being more common in older patients (>55 years old) (4,6,12). Individual variations in immune status and function shape responses to infection and contribute to disease severity and outcome. Markers of immune status associated with susceptibility to severe WNV infection include advanced age, and polymorphisms in several genes, including HLA, and interferon response pathway elements OAS, IRF3, and MX-1 (4). However, variability in individual responses that contribute to susceptibility to WNV infection remain incompletely defined.
Serum cytokines and immune cell gene expression of healthy individuals have intrinsic variation but are remarkably stable characteristics of individual status (40,41,44). Embedded in this normal variation of immune mediators are elements relevant to susceptibility or resistance to infection. In an effort to define markers of susceptibility to WNV, we recruited healthy subjects with a documented history of infection with WNV—asymptomatic or severe—and profiled markers that define individual immune status and may be associated with infection outcome.
Materials and Methods
Human subjects
Blood from study volunteers was obtained with written informed consent under approved protocols following the guidelines of the Human Investigations Committees of The University of Texas Health Science Center, Baylor College of Medicine, and Yale University School of Medicine. The Human Investigations Committee of each institution approved this study. Severity of WNV infection was determined at the time of acute illness according to established CDC guidelines (http://www.cdc.gov/ncidod/dvbid/westnile/clinicians/clindesc.htm) and described in detail previously (25,26). Asymptomatic subjects were identified by Gulf Coast Regional Blood Center (25,26) or by immunoblot (16) and absence of illness history was confirmed by study coordinators. Study participants (n=102) were 41.1% female and 86.3% white and included asymptomatic and severe patients (Table 1). At the time of sample collection, study participants had no acute illness, and took no antibiotics or nonsteroidal anti-inflammatory drugs. Subjects with a history of severe disease (n=59) were enrolled after recovery from illness, with a mean of 6.16±3.1 years between disease onset and sample collection (range 0.66–9.85 years). Samples from asymptomatic subjects (n=43) for whom onset date could be established were collected a minimum of 6 months after enrollment at a mean of 3.55±2.6 years following detection of infection (range 0.51–9.82). Due to limitations of sample size, it was not possible to assess each donor for each assay. Samples were randomly chosen for experiments over >2 years for assays under study at the time of recruitment and sample number is indicated in each figure legend.
Table 1.
Demographic Information for Healthy Subjects with a History of WNV Infection
| Asymptomatic (N=43) | Severe (N=59) | Total (N=102) | P value* | |
|---|---|---|---|---|
| Age (y), mean(SD) (range) | 50 (18.6) | 60 (13.7) | 56 (16.7) | 0.002 |
| (22–88) | (31–86) | (22–88) | ||
| Female gender, N (%) | 20 (46.5) | 22 (37.3) | 42 (41.1) | 0.42 |
| Race, N (%) | ||||
| White | 38 (88.4) | 50 (84.8) | 88 (86.3) | 0.36 |
| Black | 2 (4.7) | 7 (11.9) | 9 (8.8) | |
| Other | 3 (7.0) | 2 (3.4) | 5 (4.9) | |
| Hispanic | 1 (2.3) | 5 (8.5) | 6 (5.8) | 0.40 |
The p values were calculated based on T-test for continuous variables and Fisher exact tests for categorical variables.
Preparation of cells and serum
Blood samples from both recruiting sources were collected in CPT tubes (Becton Dickinson and Co., Franklin Lakes, NJ), processed according to the manufacturer's instructions (including centrifugation within 2 hours of collection), and cells were harvested the following day. Peripheral blood mononuclear cells (PBMCs) were isolated from CPT tubes, washed in phosphate-buffered saline, and suspended in RPMI-1640 containing L-Glutamine supplemented with 10% (v/v) human serum (Lonza, MD), 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen, Carlsbad, CA) (16). Whole blood without anticoagulant was collected for preparation of serum that was stored at −80°C until assay.
Microsphere assay
Microspheres (Luminex Corporation) were coupled to recombinant WNV proteins: envelope (E; L2 Diagnostics); nonstructural protein 1, NS1 (generous gift of Dr. Michael Diamond, Washington University, St. Louis, MO); and NS5 as described (43). WNV-infected subjects provided serum samples at multiple time points from 0.6–10 years following onset of infection (26). De-identified serum samples from these study participants, asymptomatic subjects, and laboratory standard sera were assessed for anti-WNV antibodies by microsphere immunoassay using RPE conjugates to anti-human IgG+IgA+IgM as described (42). Assays were repeated twice.
Assays of serum cytokines
Serum cytokines were quantified using the Luminex Human High Sensitivity Cytokine/Chemokine multianalyte ELISA kit (HSCYTMAG-60SK-09) according to the manufacturer's instructions (EMD Millipore Corporation, Billerica, MA). When samples were available from multiple dates from severely infected subjects, the most recent sample was used to match the sampling interval of asymptomatic subjects. Several safeguards were included to minimize variation in cytokine quantitation, including use of five batched control sera in duplicate in each plate; randomization of sera to plates; conducting serum assays with one lot of reagents; including ‘bridging’ sera from 1 asymptomatic and 1 severe WNV study subject in each plate. Validation of interplate reproducibility showed that values from control sera were reproducible across plates with a coefficient of variation between 0.15 to 0.5 for control sera and 0.15 to 0.68 for bridging patient sera (for cytokines >10 pg/mL). The concentration levels of all samples were mean normalized by a plate-specific normalization factor that was calculated by taking the mean across all cytokines.
Expression microarray
Total RNA from freshly isolated unfrozen PBMCs (2×106 cells) was harvested using the RNeasy mini kit (Qiagen, Alameda, CA) and processed for the Illumina HumanHT-12 v4 BEADCHIP whole human genome expression array for probes derived from the NCBI RefSeq Release 38 (November 7, 2009) and selected from GenBank®, dbEST, and RefSeq. Arrays were processed at Yale's Keck Biotechnology Resource Laboratory. The microarray analysis was carried out using packages in R (35). Raw expression data were normalized using the quantile method provided by the lumi package in R/Bioconductor (8). Differential gene expression between samples from asymptomatic and severe disease subjects was defined by two criteria: (1) an absolute log2 fold-change (FC) ≥0.26, and (2) a statistically-significant change in expression as determined by LIMMA (31) using a Benjamani-Hochberg false discovery rate cutoff of 0.25. Potential batch effects and age-related effects were included as independent variables in the linear model of LIMMA. Age was incorporated as a factor and was defined as young 21–39, middle aged 40–59, and older >60 years. Gene set enrichment analysis (GSEA) was conducted on a set of immune-related KEGG pathways (34). Pre-ranked GSEA was run by ranking the genes according to the t-statistic from LIMMA and signed log2 (FC). False discovery rates were based on 3000 permutations of the gene sets, and a cutoff of 0.05 was used to define significance. Microarray data files have been deposited in a public database (Gene Expression Omnibus (GEO), http://www.ncbi.nlm.nih.gov/projects/geo/) and are accessible through GEO Series accession number GSE43190.
Statistical analysis
The distribution of cytokine levels and antibody titers was compared between groups using the nonparametric Mann-Whitney test. To control for age, a generalized linear model was constructed including age as a factor. To account for measurements below the detection limit, cytokine levels were ranked and transformed into a normal distribution using the pnorm function (Normal package in R). Correlation between cytokines was measured by Pearson correlation coefficients. Associations between gene clusters and serum cytokine levels were identified by performing a weighted correlation network analysis (19). In this case, gene expression data was first processed using ComBat (14) to correct for any batch effects.
Results
Longevity of WNV antibody response in subject cohorts
To investigate whether markers of immune status may be associated with susceptibility to severe WNV infection, we enrolled a cohort of healthy study participants (n=102) with a defined history of WNV infection. Subjects were stratified into asymptomatic and severe disease cohorts as defined by clinical criteria and/or laboratory testing at the time of infection (24,26). Subjects were enrolled long after resolution of the acute response (0.6–10 years after recovery from infection), when these measurements reflect baseline immune status (3). Participants were of both genders (41.1% female) and were predominantly white (86.3%; Table 1).
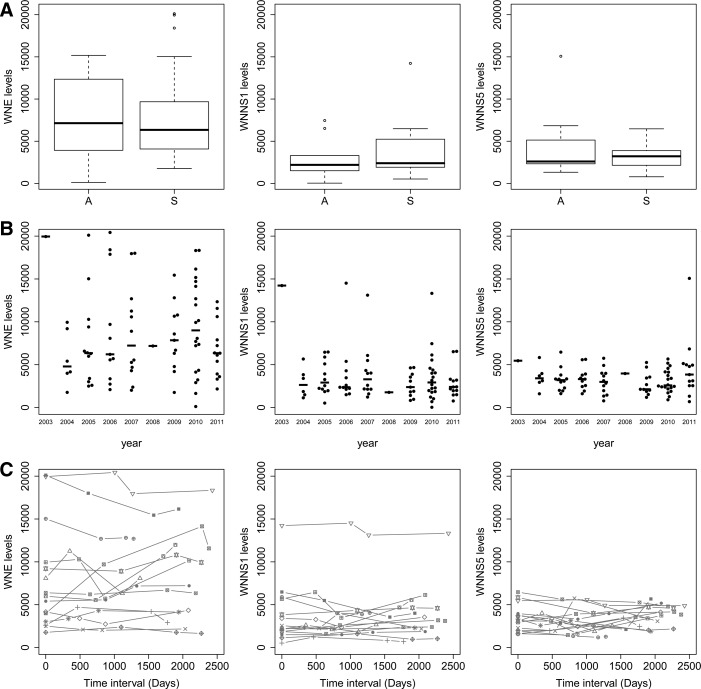
To investigate markers of the immune status associated with B cell responses and humoral immunity, we quantified antibody levels to WNV antigens. We employed a fluorescent microsphere assay that reliably discriminates between an inactivated flavivirus vaccine and natural WNV infection, and between infections with WNV or dengue virus (43). We quantified anti-WNV antibodies from a subset of symptomatic WNV subjects (n=17 severe) who had serum collected on four dates reflecting an average of 7.3 years of follow-up surveillance (range 4.8–8.6 years) and from asymptomatic subjects (single date, n=10) drawn during the same sampling period (Fig. 1). We detected high antibody levels to WNV envelope (E) protein (range 121–20,432), and somewhat lower levels for nonstructural protein 1 (NS1, range 34–14,517) and WNV NS5 (range 691–15,075). Moreover, the levels of antibody titers against WNV E and NS1 proteins were highly correlated with one another (R=0.73), suggesting that these proteins may have similar exposure in the host.
FIG. 1.
Antibody levels in WNV subject sera. Sera from healthy subjects with a history of WNV infection (n=17 severe, n=10 asymptomatic) were assessed by microsphere immunoassay for reactivity to WNV antigens E, NS1, NS5. Data represent fluorescent units for (A). Antibody levels against WNV antigens by cohort of severity of infection for asymptomatic (A) and severely infected (S) subjects; (B) Antibody levels assessed by year of sample collection; (C) Antibody levels assessed by duration of follow-up.
When we compared cohorts of participants by disease severity, we noted no significant differences between study groups whether they experienced asymptomatic or severe infection (Fig. 1A), showing that both severe and asymptomatic subjects induce a humoral response. Our data suggest that the level of the antiviral titers is not a driving feature of disease severity, which is similar to findings of antibody levels in human subjects infected with the related flavivirus hepatitis C virus, for which antibody responses do not predict clearance of infecting virus (2,29). Notably, the kinetics of antibody development during infection may play a role in response to infection and this was not assessed in our cohort. Among study participants with severe disease, females showed higher titers against WNV E than males (data not shown, p<0.05) consistent with general findings of higher antibody responses in females (15).
Differences in severity of clinical disease may reflect differences in infecting strains of virus in different years. However, we found no significant differences in the antibody levels related to year of sample collection (Fig. 1B), consistent with the observation that strains of WNV circulating in the continental USA have been largely unchanging since 2002 (5). Moreover, antibody levels were remarkably stable over time (Fig. 1C). Thus, the range of antibody levels appears to reflect inherent immune responses in the subjects whenever it occurs, rather than any difference in viral strains, duration, or other environmental variations.
Low basal levels of IL-4 in serum are associated with severity
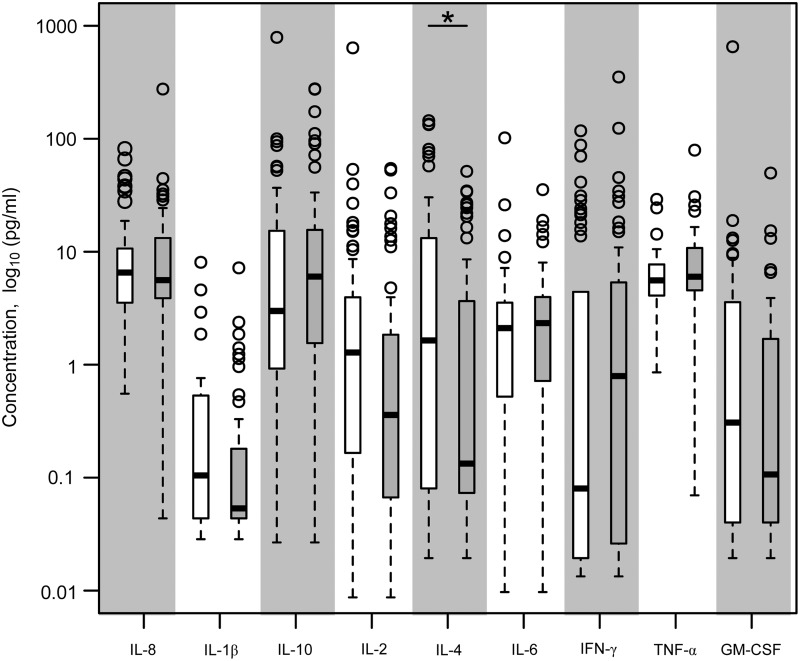
As humoral responses did not correlate with infection outcome, we quantified serum cytokines to assess variations in circulating cytokines across the cohort that might identify factors relevant to susceptibility to infection. We investigated cytokine levels from sera of WNV-exposed subjects (n=88) drawn 0.6–10 years after recovery from WNV infection, after baseline levels are restored (3), as a reflection of variation in individual host milieu that might be relevant to antiviral immune pathways. For this purpose, all subjects being compared share a history of exposure to WNV, which supports more relevant and pertinent comparison groups than using unexposed healthy controls, whose susceptibility to WNV is undefined. Serum cytokines were quantified using a high sensitivity multiplex ELISA format and in-depth analysis of bridging control sera showed high reproducibility across different plates in the assays (see Methods). When study participants were compared for baseline levels of cytokines, significant differences were noted between age groups with older donors showing elevations in TNFα (p<0.05), consistent with the pro-inflammatory milieu in normal aging described as ‘inflammaging’ (9). Comparison of baseline cytokine levels between asymptomatic and symptomatic subjects revealed that several cytokines, IL-1ß, IL-2, and IL-4, were significantly higher in asymptomatic subjects (p<0.05). However, as fewer asymptomatic patients were older (4), we corrected for age, and found that after correcting for age, only higher levels of IL-4 in asymptomatic subjects remained significant (Fig. 2, p<0.05). This cytokine is involved in proinflammatory signaling pathways and in differentiation and survival of naive helper T cells (10), suggesting that asymptomatic subjects, who mount an effective anti-WNV response, may be more efficient in initiating adaptive immune pathways. While serum IL-4 levels varied across the cohort, all subjects with levels of IL-4 greater than 54 pg/mL were from the asymptomatic cohort, while 50% of the subjects in the severe cohort had levels of IL-4 less than 4 pg/mL.
FIG. 2.
Serum cytokines from WNV subject cohorts. Sera were collected from a cohort of healthy subjects with a defined history of WNV infection (n=41 asymptomatic, n=47 severe) for multiplex cytokine determination. The coefficient of variation across different plates was between 0.15 to 0.5 for control sera and 0.15 to 0.68 for bridging patient sera. Data shown are cytokine concentration (log10) showing median, 25th and 75th percentiles, and outliers (o) for asymptomatic (open bars) and severely infected subjects (filled bars); *p<0.05 after correcting for age effects.
Gene expression patterns correlate with cytokine levels
Many host genes could contribute to and/or be affected by the differential levels of IL-4 noted in serum from our cohort. To investigate how altered levels of IL-4 may influence susceptibility to WNV, we investigated association of serum cytokine levels with transcriptional profiles of peripheral blood mononuclear cell (PBMC) populations. We examined baseline global gene expression from a subset of these subjects (severe disease, n=21; asymptomatic, n=18) by microarray analysis of 31,427 genes using Illumina Human HT-12 v4 beadchip. In light of the significant elevation of IL-4 in the serum of asymptomatic subjects, we were particularly interested in examining IL-4 in our cohorts. However, basophils, a major circulating source of IL-4, would be excluded from the CPT cell preparations employed to collect subject samples (32). Indeed, the gene expression intensities of IL-4 itself were low in all subjects, and below detection in microarray samples from 12/39 subjects (detection p value>0.05 for probe ILMN_1669174). However, we found that TNF and IL-6 gene expression values were significantly correlated with their serum cytokine levels across subjects (p<0.05). This suggests that one or more PBMC subsets are a main source for these cytokines. Additionally, levels of some serum cytokines were significantly correlated with their receptors, namely IL-1ß with IL1R1 expression and TNF with TNF receptors (TNFRSF12A and TNFRSF9), suggesting a possible correlation between receptor mediated signaling and serum cytokine levels.
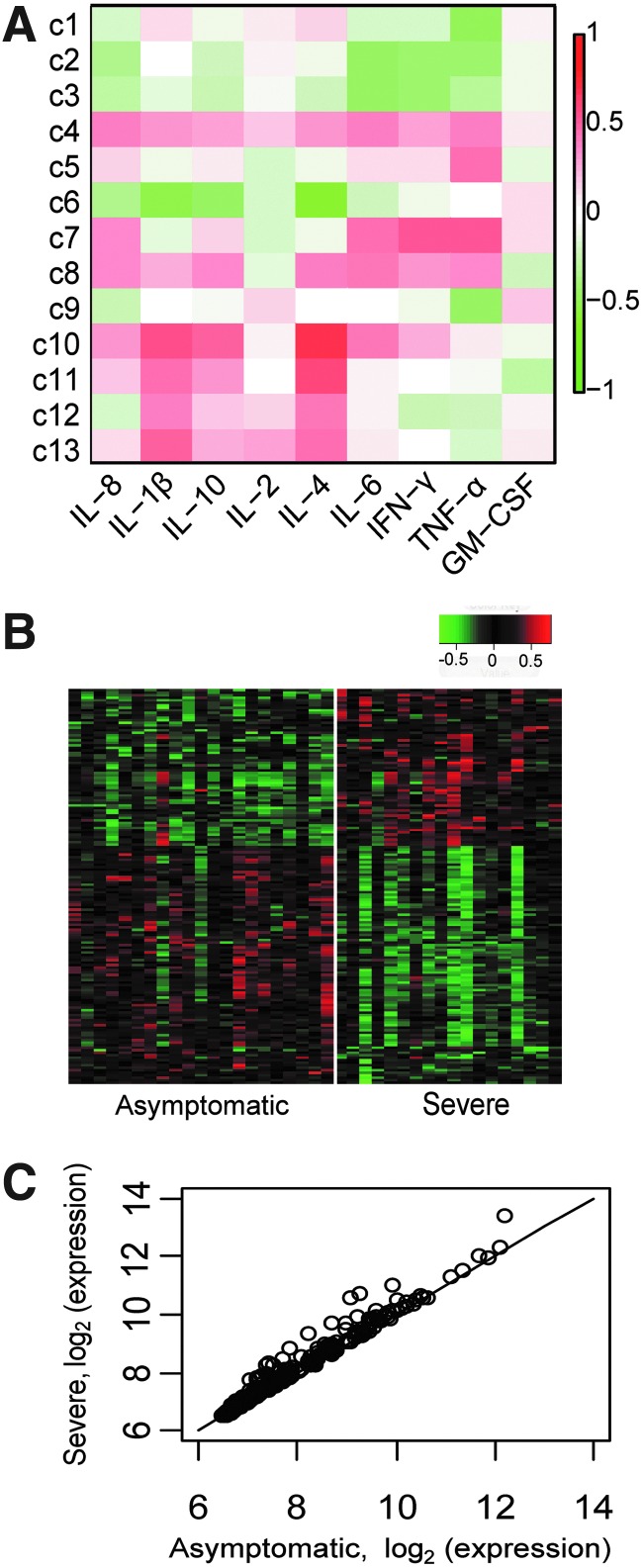
To investigate the impact of differential IL-4 levels on the immune system, we employed a weighted correlation network approach (19) to identify clusters of genes that were associated with serum cytokine levels. Significant associations were found for two clusters (Fig. 3A, c6 and c10). The larger cluster c10 includes 397 genes and is highly correlated with IL-4 (R=0.74, FDR <10−4) (Supplementary Fig. F1 and Supplementary Table S1; supplementary material is available online at www.liebertpub.com).
FIG. 3.
Clusters of co-expressed genes are associated with serum cytokine levels. (A) The weighted correlation network approach was applied to identify clusters of genes (rows) whose expression was correlated with serum cytokine levels (columns). Coloring indicates the degree of positive (red) or negative (green) correlation, with white indicating no relationship. (B) Row-normalized log2 expression levels for genes differentially expressed between subjects with a history of asymptomatic and severe disease. Coloring indicates the Z-score ranging from lower (green) to higher (red) relative expression levels. (C) Average expression of genes that are part of cluster c10 from (A).
One strongly correlated gene is the WNT signaling protein WNT5A, which has been shown to be critical for T lymphocyte differentiation and survival (11). The cluster also includes chemokines (CCL19, CCL20, CCL23), which play a role in flavivirus replication in neurons (22,36); SERPINB7, which was shown to play a role in viral resistance in our whole genome RNAi study in HeLa cells (17); and metalloproteinases (MMPs 1, 10, 14), which be may be relevant to entry of WNV into the brain, shown for MMP9 (39).
Differential gene expression between severe and asymptomatic subjects
To test whether altered transcriptional profiles were directly related to infection susceptibility, differential expression analysis was carried out to compare gene expression between subjects with a history of asymptomatic or severe WNV infection. After correcting for age, we identified 105 genes that were differentially expressed between asymptomatic and severe WNV subject cohorts (log2 (fold-change) ≥0.26 and p<0.002) (Fig. 3B; Supplementary Table S2). Severe disease subjects had higher expression levels of several cytokine and chemokine genes (IL-8, TNF), antiviral signaling genes including TMEM158 (or STING), an endoplasmic reticulum adaptor protein that plays a pivotal role in antiviral immune signaling (13), and CD69, a C-type lectin that is involved in T cell activation, proliferation, and signaling (23). In addition to higher expression of pro-inflammatory genes, subjects with history of severe infection also showed higher levels of anti-inflammatory genes such as PI3, a protease inhibitor that acts to reduce inflammation (37), and TNFAIP3, which inhibits NF-kappa B activation (33). Gene set enrichment analysis (GSEA) (34) identified several immune-related pathways that were differentially expressed between asymptomatic and severe subjects (FDR<0.05). Pathways associated with inflammation such as cytokines, cytokine receptors, and viral-recognition signaling were significantly enriched among subjects with a history of severe disease (Table 2). Enrichment of these pathways may reflect an inflammatory state of the host that would contribute to viral susceptibility.
Table 2.
Pathway Analysis
| APathway name | # genes | FDR |
|---|---|---|
| KEGG_APOPTOSIS | 88 | 4.7E-04 |
| KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | 265 | 2.1E-03 |
| KEGG_T_CELL_RECEPTOR_SIGNALING_PATHWAY | 108 | 2.5E-03 |
| KEGG_CHEMOKINE_SIGNALING_PATHWAY | 188 | 3.0E-03 |
| KEGG_HEMATOPOIETIC_CELL_LINEAGE | 87 | 6.7E-03 |
| KEGG_NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY | 137 | 5.8E-03 |
| KEGG_NOD_LIKE_RECEPTOR_SIGNALING_PATHWAY | 59 | 1.6E-02 |
| KEGG_CELL_ADHESION_MOLECULES_CAMS | 134 | 2.1E-02 |
| KEGG_UBIQUITIN_MEDIATED_PROTEOLYSIS | 137 | 3.2E-02 |
| KEGG_JAK_STAT_SIGNALING_PATHWAY | 155 | 3.0E-02 |
| KEGG_RIG_I_LIKE_RECEPTOR_SIGNALING_PATHWAY | 69 | 3.4E-02 |
| KEGG_CYTOSOLIC_DNA_SENSING_PATHWAY | 55 | 4.5E-02 |
| KEGG_CELL_CYCLE | 126 | 4.6E-02 |
| BPathway name | # Genes | FDR |
|---|---|---|
| KEGG_LYSOSOME | 121 | 3.3E-04 |
| KEGG_OXIDATIVE_PHOSPHORYLATION | 120 | 3.3E-04 |
| KEGG_CITRATE_CYCLE_TCA_CYCLE | 32 | 3.0E-03 |
GSEA was used to identify KEGG pathways that were significantly enriched in subject cohorts (FDR<0.05). (A) Subjects with a history of severe WNV infection compared to those with asymptomatic infection; and (B) subjects with a history of asymptomatic WNV infection compared to those with severe infection.
Asymptomatic subjects, who mount an effective antiviral response, showed elevations in a distinct set of genes, generally not associated with antiviral activity, although elevated levels were detected for TRIM25, which has been shown to be involved in ubiquitinylation of RIG-I and promoting anti-WNV pathways (38); autophagy gene ATG4C, one of a family of proteins that facilitate sensing of intracellular viral elements by pattern-recognition receptors (7,21); and other less recognized genes, such as MS4A6A and CHST13, which were detected by our recent RNA-Seq-based high-throughput gene expression analysis of macrophages from healthy donors infected in vitro with WNV (30). GSEA identification of genes differentially expressed by asymptomatic subjects revealed enrichment of the lysosome pathway and two metabolic pathways (Table 2).
Since IL-4 levels in serum were significantly different in our subject cohorts, and we found a positive correlation between serum IL-4 level and expression of genes from cluster c10, we assessed whether genes in the cluster were differentially expressed between subject cohorts. While many genes in the cluster were expressed equally by subjects in both cohorts, a subset of immune pathway genes were higher in subjects with severe disease (Fig. 3C). Comparison of the expression patterns of the two subject groups suggests differences in factors relevant to viral infection that may play a role in susceptibility.
Discussion
In an effort to identify markers of immune status that are associated with susceptibility to severe infection, we performed a systems level analysis of characteristic stable individual immune parameters in a cohort of healthy subjects with a defined history of WNV infection. Control of WNV infection by the immune system is multifactorial and relies on elements including viral recognition receptors (Toll-like receptors, TLRs; Rig-I like receptors, RLRs), control of permeability of the blood brain barrier, and both innate and adaptive immune determinants (4). No significant difference in antibody levels was observed between subjects with a history of asymptomatic or severe WNV infection, suggesting that susceptibility was not a consequence of an inability to mount a humoral response. This has been noted previously from comparisons of asymptomatic and mild WNV patients (20) and in patients infected with the related hepatitis C virus, for which antibody levels do not correlate with viral clearance (2,29). In our subjects, anti-E antibody serum levels were relatively constant for up to 8 years after infection, consistent with previous reports of persistence of anti-WNV antibodies for >1 year (3,28). Moreover, humoral responses to other viral antigens have also been shown to be long-lasting, in contrast to antibody responses to nonreplicating protein antigens, which tend to wane over time (1).
We investigated serum cytokines from our stratified cohort and found that while many cytokines were not different, asymptomatic subjects had elevated baseline levels of the proinflammatory cytokine IL-4, which is important in enhancing immune responses. This difference remained significant after controlling for age. Notably, severe WNV infections are defined by meningitis and/or encephalitis and IL-4 has recently been reported to serve an anti-inflammatory effect in the brain, perhaps mediated by skewing of meningeal macrophages to an alternatively activated phenotype (10). It is possible that the higher levels of IL-4 in asymptomatic subjects contribute to their lower incidence of encephalitis, although our studies of convalescent sera do not address acute infection or whether lower levels of IL-4 in severe subjects may result from WNV infection in the central nervous system or effects of persistent antigen (24). Moreover, we identified a cluster of genes whose expression in PBMCs is correlated with the serum levels of IL-4, and immune response against flavivirus, of which some were differentially expressed between our subject groups. Thus, our results suggest an important role for IL-4 in contribution to a more favorable outcome to WNV infection. Notably, a comparison of patients infected with the related dengue flavivirus also shows elevations of IL-4 and IL-1 in patients with the milder form of dengue fever compared to more severe infection during acute infection (although later time points differ) (18), and such individual variations in the host immune milieu may reflect patterns with more successful resistance to flavivirus infections.
Further investigation of gene expression by PBMCs of subjects from these cohorts revealed 105 genes that were differentially expressed between subjects with a history of asymptomatic and severe infections. Analysis of these genes showed that susceptibility to severe disease is associated with higher levels of genes in viral-recognition and antiviral pathways. Some of these differences may be expected from mechanistic studies in vitro, which have identified an important role for chemokines in flaviviral response signatures and WNV pathogenesis (22), and higher levels of these immune-pathways at baseline might suggest disregulation of the pathways. Further, genomic studies of WNV patient cohorts have identified IFN response elements and the chemokine CCR5 to be involved in susceptibility (4). Additional investigation of these genes may provide insights into mechanistic roles in anti-WNV immune responses.
The majority of people exposed to WNV mount successful immune responses and may be unaware of being infected. Through investigation of stable characteristics of individual immune milieu in a stratified cohort of subjects, we have identified serum cytokine and gene expression patterns that are associated with susceptibility to severe WNV infection. These results suggest that a systems-level analysis can be used to identify factors relevant for susceptibility to severe infections, and specifically point to an important contribution for IL-4 in resistance to WNV infection.
Supplementary Material
Acknowledgments
Funding: This work was supported in part by the National Institutes of Health (HHS N272201100019C, U19AI089992, R01AI091816), the Gillson Longenbaugh Foundation, and by Cooperative Agreement Numbers U50CI223671 and U50CK000263 from the Centers for Disease Control and Prevention. The funders have no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Certain data used in this study were obtained from the Department of Public Health of the State of CT, which approved this study.
The authors are grateful to Dr. Michael Diamond for the kind gift of recombinant WNV NS1, to Lesley Devine and Sui Tsang for valuable assistance, and the Yale Human Immunophenotyping Consortium (HIPC) team for insightful discussions.
Author Disclosure Statement
The authors assume full responsibility for analyses and interpretation of the data and declare they do not have a commercial or other association that might pose a conflict of interest for this study. The manuscript contents are solely the responsibility of the authors and do not necessarily represent the official views of the Centers for Disease Control and Prevention.
References
- 1.Amanna IJ, Carlson NE, and Slifka MK. Duration of humoral immunity to common viral and vaccine antigens. N Eng J Med 2007;357:1903–1915 [DOI] [PubMed] [Google Scholar]
- 2.Armstrong GL, Wasley A, Simard EP, McQuillan GM, Kuhnert WL, and Alter MJ. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann Int Med 2006;144:705–714 [DOI] [PubMed] [Google Scholar]
- 3.Busch MP, Kleinman SH, Tobler LH, et al. Virus and antibody dynamics in acute West Nile virus infection. J Infect Dis 2008;198:984–993 [DOI] [PubMed] [Google Scholar]
- 4.Colpitts TM, Conway MJ, Montgomery RR, and Fikrig E. West Nile virus: Biology, transmission and human infection. Clin Microbiol Rev 2012;25:635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis CT, Ebel GD, Lanciotti RS, et al. Phylogenetic analysis of North American West Nile virus isolates, 2001–2004: Evidence for the emergence of a dominant genotype. Virology 2005;342:252–265 [DOI] [PubMed] [Google Scholar]
- 6.Debiasi RL, and Tyler KL. West Nile virus meningoencephalitis. Nat Clin Prac 2006;2:264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreux M, and Chisari FV. Viruses and the autophagy machinery. Cell Cycle 2010;9:1295–1307 [DOI] [PubMed] [Google Scholar]
- 8.Du P, Kibbe WA, and Lin SM. Lumi: A pipeline for processing Illumina microarray. Bioinformatics 2008;24:1547–1548 [DOI] [PubMed] [Google Scholar]
- 9.Franceschi C, Capri M, Monti D, et al. Inflammaging and anti-inflammaging: A systemic perspective on aging and longevity emerged from studies in humans. Mech Ageing Dev 2007;128:92–105 [DOI] [PubMed] [Google Scholar]
- 10.Gadani SP, Cronk JC, Norris GT, and Kipnis J. IL-4 in the brain: A cytokine to remember. J Immunol 2012;189:4213–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gattinoni L, Ji Y, Restifo NP. Wnt/beta-catenin signaling in T-cell immunity and cancer immunotherapy. Clin Cancer Res 2010;6:4695–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gubler DJ. The continuing spread of West Nile virus in the western hemisphere. Clin Infect Dis 2007;45:1039–1046 [DOI] [PubMed] [Google Scholar]
- 13.Ishikawa H, and Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008;455:674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson WE, Li C, and Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–127 [DOI] [PubMed] [Google Scholar]
- 15.Klein SL, Jedlicka A, and Pekosz A. The Xs and Y of immune responses to viral vaccines. Lancet Infect Dis 2010;10:338–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong K-F, Delroux K, Wang X, et al. Dysregulation of TLR3 impairs the innate immune response to West Nile virus in the elderly. J Virol 2008;82:7613–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishnan MN, Ng A, Sukumaran B, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature 2008;455:242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar Y, Liang C, Bo Z, Rajapakse JC, Ooi EE, and Tannenbaum SR. Serum proteome and cytokine analysis in a longitudinal cohort of adults with primary dengue infection reveals predictive markers of DHF. PLoS Negl Trop Dis 2012;6:e1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langfelder P, and Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanteri MC, O'Brien KM, Purtha WE, et al. Tregs control the development of symptomatic West Nile virus infection in humans and mice. J Clin Invest 2009;119:3266–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HK, Lund JM, Ramanathan B, Mizushima N, and Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 2007;315:1398–1401 [DOI] [PubMed] [Google Scholar]
- 22.Lim JK, and Murphy PM. Chemokine control of West Nile virus infection. Exper Cell Res 2011;317:569–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin P, and Sanchez-Madrid F. CD69: An unexpected regulator of TH17 cell-driven inflammatory responses. Sci Signal 2011;4:pe14. [DOI] [PubMed] [Google Scholar]
- 24.Murray K, Walker C, Herrington E, Lewis JA, McCormick J, Beasley DW, Tesh RB, and Fisher-Hoch S. Persistent infection with West Nile virus years after initial infection. J Infect Dis 2010;201:2–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray KO, Baraniuk S, Resnick M, et al. Clinical investigation of hospitalized human cases of West Nile virus infection in Houston, Texas, 2002–2004. Vector Borne Zoonotic Dis 2008;8:167–174 [DOI] [PubMed] [Google Scholar]
- 26.Nolan MS, Schuermann J, and Murray KO. West Nile virus infection among Humas, Texas, USA, 2002–2011. Emerg Infect Dis 2013;19:137–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersen LR, Carson PJ, Biggerstaff BJ, Custer B, Borchardt SM, and Busch MP. Estimated cumulative incidence of West Nile virus infection in US adults, 1999–2010. Epidemiol Infect 2013;141:591–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prince HE, Lape-Nixon M, Yeh C, Tobler LH, and Busch MP. Persistence of antibodies to West Nile virus nonstructural protein 5. J Clin Virol 2008;43:102–106 [DOI] [PubMed] [Google Scholar]
- 29.Qian F, Bolen CR, Wang X, et al. Impaired TLR3-mediated interferon responses from macrophages of patients chronically infected with hepatitis C virus. Clin Vaccine Immunol 2013;20:146–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian F, Chung L, Zheng W, et al. Identification of genes critical for resistance to infection by West Nile virus using RNA-Seq analysis. Viruses 2013;5:1664–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyth GK.Limma: Linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, et al. (eds) Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer, New York, 2005; 397–420 [Google Scholar]
- 32.Sokol CL, and Medzhitov R. Role of basophils in the initiation of Th2 responses. Curr Op Immunol 2010;22:73–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song HY, Rothe M, and Goeddel DV. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-kappaB activation. Proc. Nat Acad Sci USA 1996;93:6721–6725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Nat Acad Sci USA 2005;102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2010 [Google Scholar]
- 36.Tobler LH, Cameron MJ, Lanteri MC, et al. Interferon and interferon-induced chemokine expression is associated with control of acute viremia in West Nile virus-infected blood donors. J Infect Dis 2008;198:979–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verrier T, Solhonne B, Sallenave JM, and Garcia-Verdugo I. The WAP protein Trappin-2/Elafin: A handyman in the regulation of inflammatory and immune responses. Int J Biochem Cell Biol 2012;44:1377–1380 [DOI] [PubMed] [Google Scholar]
- 38.Wang P, Arjona A, Zhang Y, Dai J, LeBlanc P, Doiron K, Sultana H, Saleh M, and Fikrig E. Caspase-12 controls West Nile virus infection via the viral RNA receptor RIG-I. Nat Immunol 2010;11:912–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang P, Dai J, Bai F, Kong KF, Wong SJ.Montgomery RR, Madri JA, Fikrig E. 2008Matrix metalloproteinase 9 facilitates West Nile Virus entry into the brain. J. Virol. 2008;82:8978–8985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, and Brown PO. Individuality and variation in gene expression patterns in human blood. Proc Nat Acad Sci USA 2003;100:1896–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong HL, Pfeiffer RM, Fears TR, Vermeulen R, Ji S, and Rabkin CS. Reproducibility and correlations of multiplex cytokine levels in asymptomatic persons. Cancer Epidemiol Biomarkers Prevent 2008;17:3450–3456 [DOI] [PubMed] [Google Scholar]
- 42.Wong SJ, Boyle RH, Demarest VL, et al. Immunoassay targeting nonstructural protein 5 to differentiate West Nile virus infection from dengue and St. Louis encephalitis virus infections and from flavivirus vaccination. J Clin Micro 2003;41:4217–4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong SJ, Demarest VL, Boyle RH, et al. Detection of human anti-flavivirus antibodies with a West Nile virus recombinant antigen microsphere immunoassay. J Clin Micro 2004;42:65–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yaqoob P, Newsholme EA, and Calder PC. Comparison of cytokine production in cultures of whole human blood and purified mononuclear cells. Cytokine 1999;11:600–605 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.