Abstract
Background: Excess iodine inhibits thyroid follicular cell proliferation associated with TGFβ pathway activation, although thyroid cancers are frequently refractory to TGFβ signaling. The TGFβ pathway is predicted to be regulated by miR-17-92 cluster microRNAs. MicroRNAs are small noncoding RNAs that inhibit target mRNA translation and have emerged as potent modulators of tumorigenesis. Although the BRAFV600E mutation is the most prevalent alteration in thyroid cancer, the impact of iodine intake on BRAF-mediated oncogenesis remains unclear. Therefore, the aim of this study was to investigate the influence of high iodine on miR-17-92 transcriptional regulation and expression in thyroid cells expressing activated BRAF.
Methods: Rat thyroid follicular cells that conditionally express BRAFV600E under doxycycline stimulation (PC-BRAFV600E-6) were derived from the PCCl3 line. These cells were treated with doxycycline for two days, in the absence or presence of 10 μM sodium iodide. The thyroid cancer cell lines BCPAP and KTC2 were also analyzed. Expression of the miR-17-92 cluster and Notch1 was analyzed by quantitative polymerase chain reaction, and expression of these genes was modulated by anti-miR or anti-Notch1 siRNAs transfection. Protein expression was assessed by Western blot. Luciferase assays were used to quantify Smad4 3′-UTR/miR-19 interaction and Notch signaling activation. TGFβ responsiveness was evaluated by cell cycle analysis of TGFβ-treated cells.
Results: High iodine blocked BRAFV600E-induced upregulation of miR-17-92, including miR-19a/b. miR-17-92 promoter region analysis revealed a putative binding site for Hes1, a transcription factor responsive to Notch signaling. Notch-1 overexpression resulted in miR-19 upregulation in normal thyroid cells, while Notch-1 knockdown blocked BRAF-induced miR-19 expression. Moreover, in anaplastic thyroid cancer cells, Notch-1 knockdown reduced miR-19. Expression of BRAFV600E decreased Smad4 protein in normal thyroid cells. Smad4 was validated as a miR-19 target by luciferase assays, which revealed reduced luminescence associated with miR-19 interaction in Smad4 3′-UTR. Iodine treatment restored Smad4 levels in BRAF-activated cells, resulting in enhanced G1-cell cycle arrest in response to TGFβ. Moreover, this effect was mimicked in papillary thyroid cancer cells treated with anti-miR-19.
Conclusion: High iodine abrogates BRAFV600E-induced activation of miR-19, a newly identified Smad4 regulator, through Notch pathway inhibition and restores responsiveness to TGFβ signaling. Our results indicate that iodine exerts protective effects in thyroid cells, attenuating acute BRAF oncogene-mediated microRNA deregulation.
Introduction
Thyroid follicular cells have the ability to concentrate iodide in order to produce thyroid hormones. Excessive iodine levels in the serum exert an autoregulatory influence on the thyroid gland, blocking cell proliferation and thyroid function (1,2). These effects have been partially attributed to increased TGFβ expression (3,4). Deregulation of TGFβ signaling is observed in thyroid cancer (5,6) and is associated with loss of responsiveness to the antiproliferative effects of TGFβ during the early stages of epithelial cell tumorigenesis (7). Although the influence of iodine in thyroid cancer has not yet been clarified, excess iodine exerts a protective effect during oncogenic RET/PTC3 activation in thyroid follicular cells, blocking ERK phosphorylation and delaying loss of thyroid differentiation markers (8).
In silico analyses have revealed that the TGFβ signaling pathway is targeted by the miR-17-92 cluster (9), which is comprised of seven microRNAs (miRNA): miR-17 (5p and 3p strands), miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92-1. This cluster of genes is frequently deregulated in cancers, including thyroid cancer (10–13). miRNAs are small noncoding RNAs (∼22 nucleotides) that negatively regulate translation of target mRNAs and have emerged as important modulators of the oncogenic process (14).
The BRAF1799T>A mutation, resulting in the BRAFV600E oncoprotein, is highly prevalent in papillary thyroid cancer (PTC), the most common histotype of thyroid cancer. This alteration is also observed in anaplastic thyroid cancers (ATC) derived from PTCs (15,16). Expression of the BRAFV600E oncoprotein deregulates a plethora of signaling pathways to promote thyroid cancer. One such pathway is the Notch pathway, which is activated by the BRAFV600E oncoprotein in thyroid follicular cells and is upregulated in human PTC (17). Interestingly, deregulation of the Notch signaling pathway in cancer is not only associated with tumor initiation, but also tumor progression via epithelial to mesenchymal transition (18).
In this study, we show that BRAFV600E activates miR-17-92 via upregulation of Notch signaling. We also demonstrate that miR-19 targets Smad4 in thyroid follicular cells. Moreover, high iodine reduces the oncogenic effects of BRAFV600E by blocking Notch signaling activation and miR-19 upregulation, restoring TGFβ inhibitory signaling. Thus, we report a new protective role for iodine in cells undergoing BRAF activation.
Materials and Methods
Cell lines
PC-BRAFV600E-6 cells, derived from PCCl3 rat thyroid follicular cells (19), conditionally express BRAFV600E under doxycycline treatment. These cells were cultivated in F-12 Coon's Modification medium (Sigma, St. Louis, MO) supplemented with 5% fetal bovine serum (FBS), 1 mU/mL bovine thyrotropin (TSH; Sigma), 10 μg/mL insulin (Sigma), 5 μg/mL transferrin (Sigma), 10 nM hydrocortisone (Sigma), 300 μg/mL neomycin (Invitrogen, Carlsbad, CA), and 100 μg/mL hygromycin (Invitrogen). PC-Notch1 cells (17), also derived from PCCl3 cells, constitutively express the Notch intracellular domain and were cultivated in the same medium described above, but in the absence of hygromycin.
Nthy-ori 3-1 cells (Sigma), derived from human normal thyroid follicular cells immortalized with the SV40 virus, were cultivated in RPMI (Invitrogen) supplemented with 10% FBS and 2 mM l-glutamine (Invitrogen). BCPAP cells (BRAFWT/V600E), derived from PTC, were cultivated in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% FBS, and KTC-2 cells (BRAFWT/V600E), derived from ATC, were cultivated in RPMI supplemented with 5% FBS.
This study was performed in accordance with guidelines from the Ethical Committee of the Institute of Biomedical Sciences (no. 134/F93/L2), University of São Paulo, Brazil.
Cell treatments
Sodium iodide and doxycycline
High concentration iodine treatment was performed by diluting a 1 M stock solution of sodium iodide (NaI) in the medium to a final concentration of 10 μM NaI. PC-BRAFV600E-6 cells were treated with 10 μM iodine for five days. BRAFV600E activation was performed by adding 1 μg/mL doxycycline (Calbiochem, San Diego, CA) to the culture medium for 48 hours. PC-BRAFV600E-6 cell groups were described as follows. The Control group includes cells without any treatment; the BRAF group includes cells in which BRAF1799T>A was induced by doxycycline treatment; the Iodine group contains cells treated with 10 μM NaI; and the BRAF/Iodine group contains cells treated with both doxycycline and NaI.
Recombinant TGFβ1
PC-BRAFV600E-6 and BCPAP cells were treated for 24 hours with exogenous recombinant TGFβ (rTGFB; Peprotech, Rocky Hill, NJ) in the culture medium at doses of 1 ng/mL and 10 ng/mL respectively.
BRAFV600E inhibitor PLX4032
Inhibition of BRAFV600E was performed by treatment of KTC2 cells with 1 μM PLX4032 (Selleck Chemicals, Houston, TX) for 24 hours. Control cells were treated with the vehicle (DMSO).
miR-19 mimics and anti-miR
miR-19 modulation was performed using the mirVana™ miRNA mimic (Ambion, Austin, TX), which overexpresses hsa-miR-19a-3p and hsa-miR-19b-3p, or the mirVana™ miRNA inhibitor (Ambion) containing LNA modification, which inhibits hsa-miR-19a-3p and hsa-miR-19b-3p (anti-miRs). All transfections were performed using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's instructions.
Notch1 siRNA (small interfering RNA)
Notch1 knockdown in PC-BRAFV600E-6 and KTC2 cells was performed by transient transfection (48 hours) of 10 nM or 60 nM NOTCH1 siRNA (esiRNA human NOTCH1-EHU150431; Sigma) respectively. Control cells were transfected with an EGFP siRNA (EGFP-EHUGFP; Sigma). All transfections were performed using Lipofectamine™ 2000 according to the manufacturer's instructions. Validation of decreased NOTCH1 expression was performed as previously described (17).
Analysis of microRNA expression
RNA extraction
Small RNA (sRNA) was isolated using the miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. sRNA integrity was analyzed using the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) in combination with the Agilent Small RNA kit.
Mature miRNA levels
Briefly, 10 ng of sRNA was reverse transcribed using the TaqMan reverse transcription kit (Applied Biosystems, Foster City, CA) in the presence of stem-loop primers. Quantitative PCR was then performed, using the TaqMan® MicroRNA Assays for miR-17-5p (assay 393), miR-18a-5p (assay 2422), miR-19a-3p (assay 395), miR-19b-3p (assay 396), miR-20a-5p (assay 580), miR-92a-3p (assay 430), miR-146b-5p (assay 1097), snoRNA (small nucleolar RNA) (assay 1718), or RNU6B (U6B nuclear RNA; assay 1093; Applied Biosystems); TaqMan® Universal PCR Master Mix; and No AmpErase® UNG (Life Technologies) in a ABI® 7300 Sequence Detection System (PE Applied Biosystems). Gene expression was normalized by comparison to snoRNA or RNU6B levels and calculated using the QGene program (20).
Western blot analyses
Total protein was extracted from cell lines using RIPA buffer (20 mM Tris, pH 7.5; 150 mM NaCl; 1% Nonidet P-40; 0.5% sodium deoxycholate; 1 mM EDTA; and 0.1% SDS) containing 10% protease inhibitor cocktail (Sigma). Protein concentration was determined using the Bradford assay (Bio-Rad Laboratories, Hercules, CA), and 40 μg of each sample was resolved on a 10% acrylamide gel by SDS-PAGE and blotted to a Hybond-ECL nitrocellulose membrane (Amersham Biosciences, Little Chalfont, United Kingdom). Nonspecific binding sites were blocked using 5% nonfat dry milk in Tris-buffered saline containing 0.1% Tween-20. The following primary antibodies were used: rabbit anti-SMAD4 (sc-7866), rabbit anti-NOTCH1 (sc-6014), mouse anti-α-tubulin (sc-5286), and mouse anti-β-actin (sc-47778; Santa Cruz, Santa Cruz, CA). The rabbit anti-Nis antibody was kindly donated by Dr. Sissy Jhang. The antigen–antibody complexes were detected with corresponding horseradish peroxidase-conjugated secondary antibodies. Chemiluminescence emission was visualized with luminol and p-cumaric acid (Sigma) in the presence of H2O2 using an ImageQuant LAS4000 imaging system (GE Healthcare, Little Chalfont, United Kingdom).
Luciferase assay
miR-19 target validation
A segment of the rat Smad4 3′-untranslated region (UTR) containing the predicted binding site for miR-19a/miR-19b (position 1297-1303) based on TargetScan database (www.targetscan.org) was cloned into the pmiR-Glo plasmid (pmiR-Glo-Smad4-wt plasmid; Promega, Madison, WI) according to the manufacturer's instructions. A second version of the Smad4 3′-UTR containing a mutated binding site was also cloned into the same vector (pmiR-Glo-Smad4-mut; Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/thy). N-thy ori-3.1cells were transiently transfected with pmiR-Glo-Smad4-wt or pmiR-Glo-Smad4-mut in the presence of 10 nM miR-19a/ miR-19b mimics and/or 10 nM anti-miR-19a/miR-19b antisense oligonucleotides. In order to evaluate miR-19 site specificity, we also overexpressed let-7a via the pH1-RNA-puro-let7a plasmid. Forty-eight hours after transfection, the cell lysate was collected, and firefly luciferase activity was measured in a SpectraMax L luminometer (Molecular Devices, Sunnyvale, CA) using the Dual Luciferase® Reporter Assay System (Promega).
Notch signaling activation
Activation of Notch signaling results in cleavage of the Notch receptor and release of the Notch intracellular domain (NICD), which translocates into the nucleus and interacts with specific factors, such as CBF1, to bind and regulate target gene expression. In order to evaluate Notch signaling activation, we used the reporter plasmid 4xwtCBF1Luc, which contains four CBF1 binding sites, and 4xmutCBF1Luc, which contains mutated binding sites and served as a control (21). PC-BRAFV600E-6 cells were transfected with 4xwtCBF1Luc or 4xmutCBF1Luc and pRL plasmid, and lysate was collected 48 hours after transfection. Luminescence was measured as described above.
TGFβ responsiveness
Cells were detached by trypsinization and fixed with 70% ethanol. After hydration in PBS, cells were treated with RNase (100 μg/mL). The cell cycle was evaluated by analyzing DNA content of cells stained with propidium iodide (50 μg/mL) by flow cytometry (10,000 events, red emission) using Guava EasyCyte Mini (Guava Technologies, Hayward, CA).
Bioinformatics
miR-19a and miR-19b target prediction analysis was performed using the TargetScan database (22). The rat and human miR-17-92 promoter regions (5 kb upstream) were analyzed for potential transcription factor binding sites using Transcription Element Search System (TESS) software available online (23).
Statistical analyses
Results are presented as the mean±standard deviation (SD). Significant differences were assessed by analysis of variance followed by the Tukey test to compare more than two groups or the t-test to compare two groups. p-Values of <0.05 were considered significant.
Results
High iodine blocks BRAFV600E-induced miR-17-92 expression
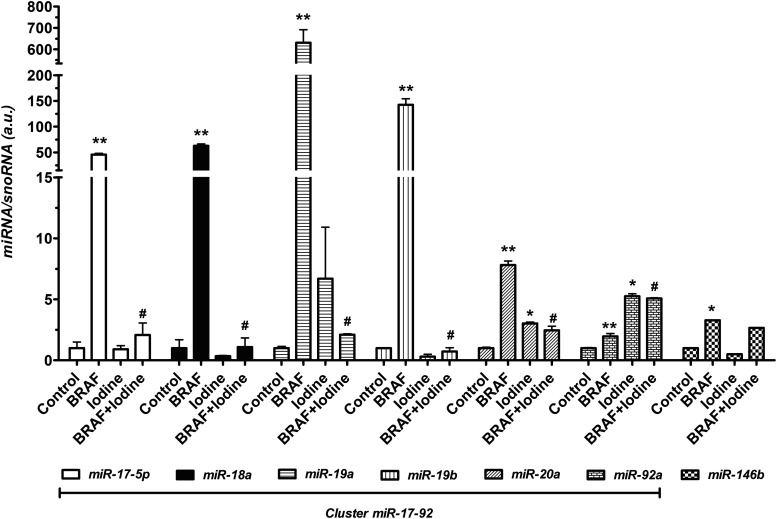
In this study, we assessed the effects of iodine treatment on miRNA expression during acute BRAFV600E oncogene activation in thyroid follicular cells (Fig. 1). We observed that induction of BRAFV600E expression in PC-BRAFV600E-6 cells by doxycycline strongly increased the levels of each of the components of the miR-17-92 cluster after 48 hours of treatment (Fig. 1). miRNAs belonging to the miR-17-92 cluster were differentially induced by expression of the BRAF oncogene, as miR-17-5p, miR-18a, miR-19a, miR-19b, miR20a, and miR-92a exhibited approximately 45, 62, 630, 142, 7, and 2-fold increase, respectively, relative to the control group. In contrast, treatment of PC-BRAFV600E-6 cells with high iodine prior to BRAF induction (BRAF/Iodine group) blocked the increase of miR-17-5p, miR-18a, and miR-19b, partially blocked that of miR-19a and miR-20a, but had no effect on miR-92a expression. Interestingly, treatment with high iodine alone (Iodine group) induced expression of miR-20a and miR-92a (Fig. 1). We also assessed levels of miR-146b, a hallmark miRNA for PTC, and observed that expression of BRAF increased miR-146b expression, but iodine treatment did not reverse this induction (Fig. 1).
FIG. 1.
Effect of BRAFV600E modulation on miR-17-92 expression. Conditional induction of BRAFV600E for 48 hours activates cluster miR-17-92 and miR-146b expression in PC-BRAFV600E-6 cells. Moreover, iodine treatment prior to BRAF activation reduces the expression of miR-17-92 components except for miR-92a, but does not modulate miR-146b. *p<0.01, **p<0.002 vs. Control; #p<0.002 vs. BRAF. Data are expressed as mean±standard error (SE). Representative results of two independent experiments.
High iodine blocks BRAF-induced activation of Notch signaling
In order to determine whether the miR-17-92 gene underwent transcriptional regulation in response to BRAF expression, we investigated its putative promoter region for predicted transcription factor binding sites. In silico analysis of 5 kb of sequence upstream of the rat miR-17-92 gene using the TESS tool (23) revealed several potential transcription factor binding sites in this region, including five putative binding sites for Hes1, a downstream target of Notch signaling. In the sequence 5 kb upstream the human miR-17-92 gene, we found two putative HES1 binding sites. Moreover, recently, it was shown that Notch1 is activated in human PTC and that BRAFV600E induction stimulates Notch signaling in thyroid follicular cells (17).
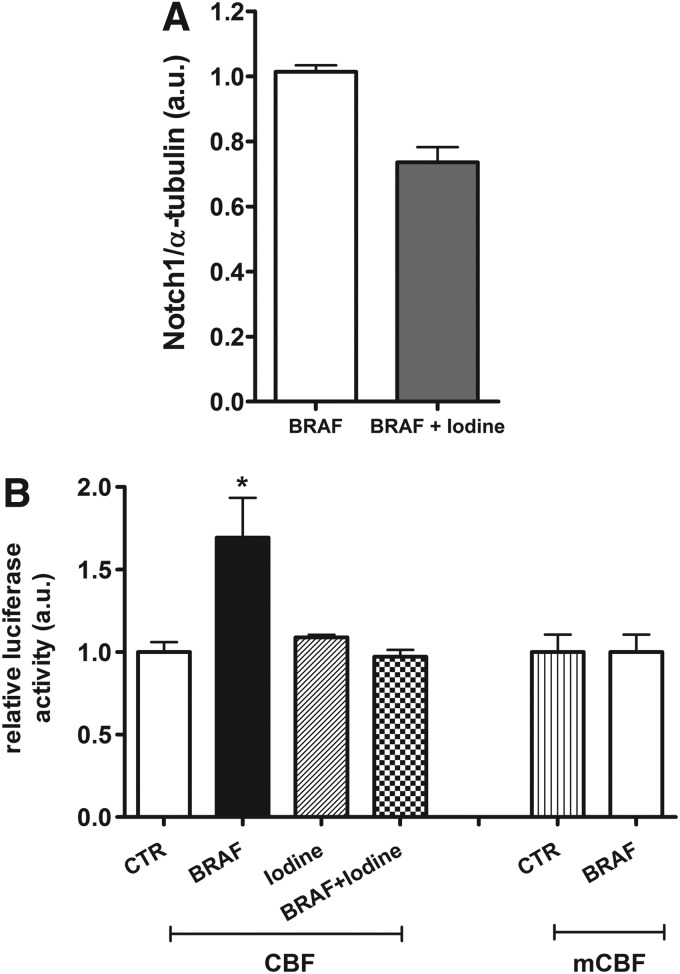
Therefore, we next analyzed expression of Notch1, the transmembrane receptor for Notch signaling, and observed that iodine treatment reduced Notch1 protein levels during BRAF induction (Fig. 2A). In order to analyze Notch signaling activity during high iodine treatment of BRAF-induced cells (BRAF/Iodine group), we transfected cells with a reporter plasmid containing four CBF binding sites (4xCBF1Luc). We observed that iodine treatment blocked BRAF-mediated activation of Notch signaling (Fig. 2B).
FIG. 2.
Iodine influence on Notch signaling. (A) High iodine treatment decreased Notch1 protein in BRAF-activated thyroid cells analyzed by Western blot. (B) BRAF oncogene activation increased Notch signaling through Notch-CBF1 binding to CBF plasmid and enhanced luciferase activity. Iodine treatment blocked BRAF-induced Notch signaling activation in PC-BRAFV600E-6 cells. Mutated CBF binding site plasmid (mCBF) was used as binding specificity control. Luminescence (arbitrary units, a.u.) was normalized by cotransfection of pRL-CMV plasmid that encodes Renilla luciferase. *p<0.05 vs. Control. Data are expressed as mean±SE (n=3). Representative results of two independent experiments.
Notch signaling influences miR-19 expression
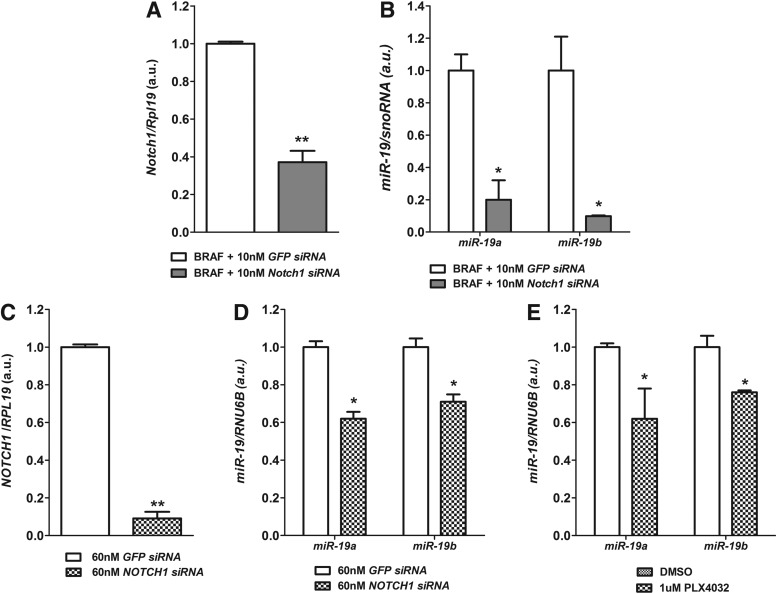
By transfecting cells with targeted small interfering RNAs (siRNAs), we observed that the knockdown of Notch1 in BRAF-activated thyroid cells (BRAF group; Fig. 3A) resulted in decreased expression of miR-19a and miR-19b (Fig. 3B). Moreover, knockdown of NOTCH1 in the anaplastic thyroid cell line KTC2 (Fig. 3C) decreased miR-19 levels (Fig. 3D). Next, we analyzed the expression of miR-19 in KTC2 cells treated with the BRAFV600E specific inhibitor PLX4032. Inhibition of BRAFV600E with 1 μM PLX4032 resulted in partial inhibition of miR-19 cluster expression after 24 hours of treatment (Fig. 3E).
FIG. 3.
Effect of Notch1 inhibition on miR-19 expression. (A, B) Notch1 siRNA treatment during 48-hour BRAF activation in PC-BRAFV600E-6 cells reduced Notch1 mRNA and inhibited miR-19a and miR-19b. (C, D) Knockdown of NOTCH1 in KTC2 cells resulted in decreased levels of miR-19a and miR-19b after 48 hours transfection. *p<0.05, **p<0.01 vs. siRNA EGFP. (E) BRAFV600E inhibition with specific inhibitor PLX4032 (1 μM for 24 hours) resulted in reduction of miR-19 levels in KTC2 cells. Data are expressed as mean±SE (n=3). *p<0.05 vs. DMSO.
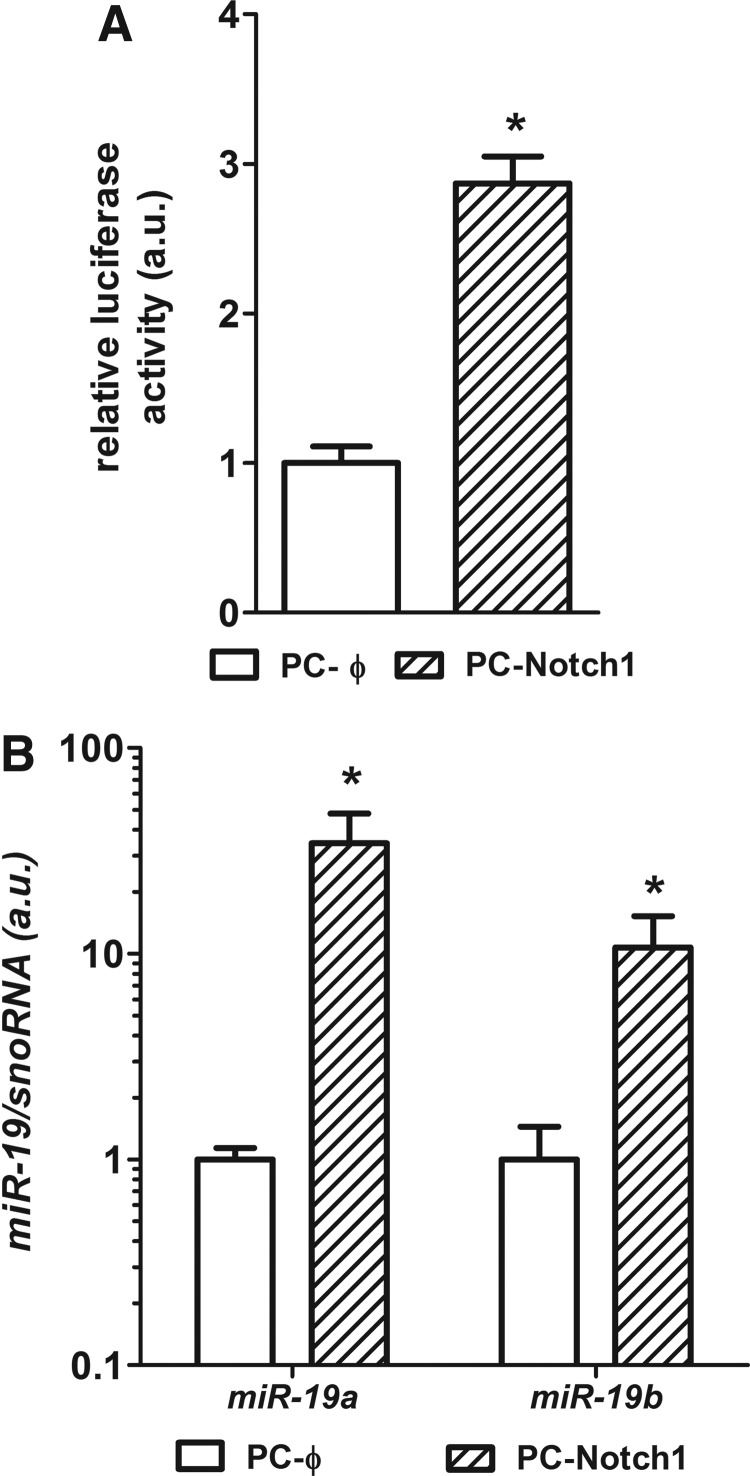
In order to understand further the role of the Notch pathway in regulation of transcription of miR-17-92, we used the PC-Notch1 cell line (17), which overexpresses the Notch1 intracellular domain (NICD). PC-Notch1 cells exhibited activation of Notch signaling, observed as enhanced luminescence in a Notch1-CBF luciferase reporter assay (Fig. 4A). Analysis of miR-19 expression in PC-Notch1 cells revealed that increased Notch1 signaling strongly induces miR-19a and miR-19b expression (Fig. 4B).
FIG. 4.
Influence of Notch signaling activation on miR-19 expression. (A) Overexpression of Notch1 in PCCl3 cells (PC-Notch1) induced Notch signaling activation observed by increased CBF binding and luminescence in luciferase reporter assay. Luminescence was normalized by cotransfection of pRL-CMV plasmid that encodes Renilla luciferase. (B) Moreover, PC-Notch1 cells exhibited a robust increase in miR-19a and miR-19b. Data are expressed as mean±SE for gene expression and mean±standard deviation (SD) for luminescence (n=3). Representative results of two independent experiments. *p<0.05, **p<0.01 vs. PC-Φ.
BRAF inhibits Smad4 via miR-19 in thyroid cells and impairs the TGFβ signaling response
As miR-19a and miR-19b are the key components required for miR-17-92 cluster oncogenicity (24), we searched for predicted targets of these miRNAs that could cooperate with the BRAF oncogene to promote malignancy. Bioinformatic searches using the TargetScan tool revealed Smad4, a component of the TGFβ signaling pathway, as a potential target of miR-19. The rat and human Smad4 mRNA 3′-UTRs contain one predicted binding site for miR-19, which is conserved among vertebrates.
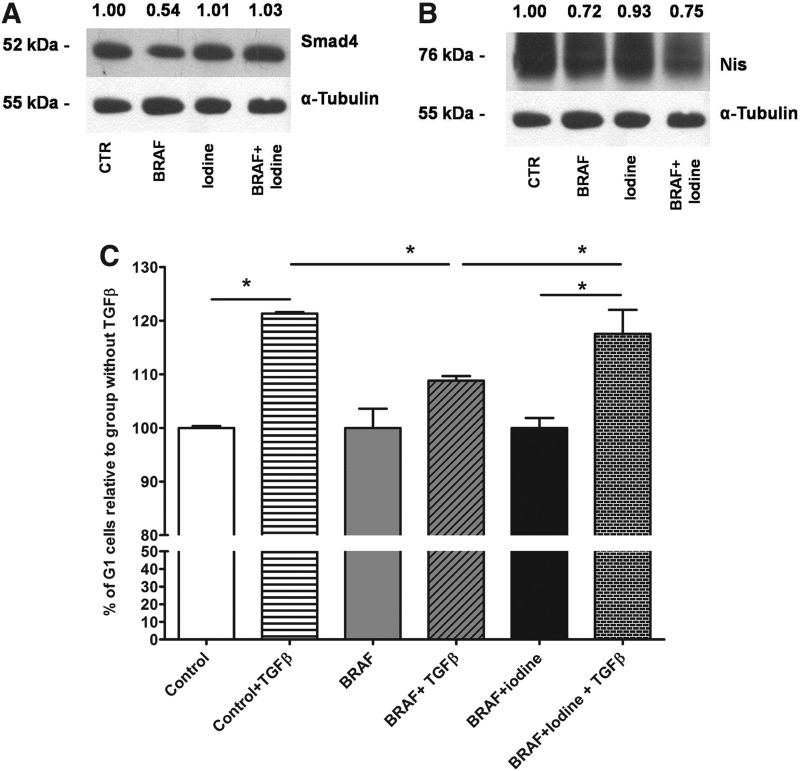
Analysis of protein expression by Western blot revealed that BRAF induction reduced Smad4 levels (Fig. 5A). Furthermore, high iodine treatment blocked BRAF-induced repression of Smad4 (Fig. 5A). We also assessed protein levels of the sodium-iodide symporter Nis and observed that high iodine did not block BRAF-induced Nis repression (Fig. 5B).
FIG. 5.
Iodine and BRAF effects on TGFβ signaling pathway. (A) Induction of BRAF decreased Smad4 expression, while high iodine treatment blocked this effect in PC-BRAFV600E-6 cells. (B) BRAF oncogene reduced Nis protein expression, an effect not blocked by iodine treatment. High iodine alone slightly decreased Nis levels. (C) BRAF activation impaired responsiveness to rTGFβ shown as reduced number of cells in G1 phase, while iodine restored cell cycle arrest at G1 as analyzed by flow cytometry. Data are expressed as mean±SD. Representative results of two independent experiments performed in triplicate. *p<0.05.
To characterize the impact of reduced Smad4 protein levels in BRAF-activated thyroid cells, we evaluated the response of these cells to treatment with recombinant TGFβ by flow cytometry. We found that BRAF induction (BRAF group) impaired TGFβ-mediated G1-phase arrest, and that treatment with high iodine restored G1 arrest in these cells (Fig. 5C).
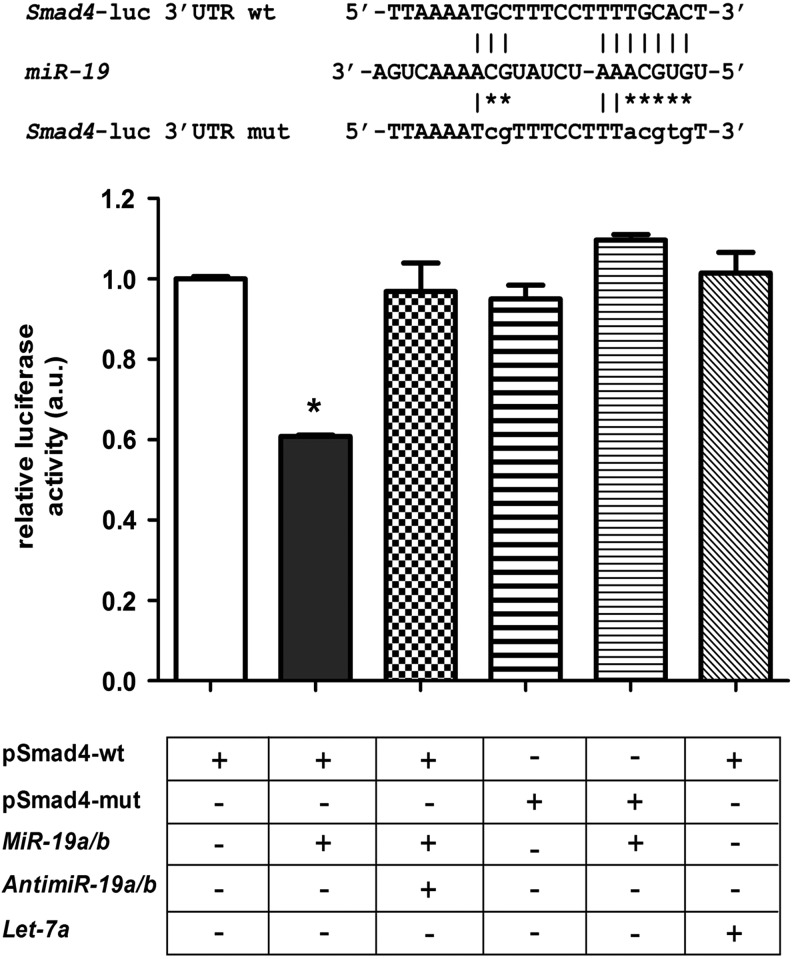
In order to investigate the interaction of miR-19 with the Smad4 mRNA 3′-UTR, we generated a reporter plasmid containing the predicted binding site for miR-19 downstream of the luciferase gene (pmiRGlo-Smad4-wt; Fig. 6). Transient transfection of the pmiRGlo-Smad4-wt construct together with miR-19a/miR-19b mimics in Nthy-ori 3-1 cells resulted in a significant ∼40% reduction in luciferase activity, which could be abolished by the concomitant transfection of anti-miR-19a/anti-miR-19b (Fig. 6). Moreover, mutation of the predicted miR-19 binding site (pmiRGlo-Smad4-mut) abolished the inhibitory effect of miR-19a/miR-19b mimics on luciferase activity, as did transfection of the let-7a over pmiRGlo-Smad4-wt plasmid.
FIG. 6.
Reporter assay for miR-19 binding in Smad4 3′-UTR. Wild-type (pmiR-Glo Smad4-wt) or mutated (pmiR-Glo Smad4-mut) miR-19 predicted binding sites in Smad4 3-UTR was cloned in the pmiR-Glo plasmid. Transient co-transfection of reporter plasmid pmiR-Glo-Smad4-wt with miR-19a/miR-19b mimics significantly reduced luciferase activity, while concomitant anti-miR-19a/anti-miR-19b transfection blocked this effect in normal human thyroid follicular cells, Nthy ori-3.1 cells. The effect of miR-19a/miR-19b mimics was also blocked in cells transfected with a mutated binding site plasmid (pmiR-Glo Smad4-mut). For site binding control, we used pH1-RNA let-7a to overexpress let-7a that does not bind to the Smad4 3′-UTR. Luminescence (a.u.) was normalized by Renilla luciferase activity contained in the pmiR-Glo plasmid. Data are expressed as mean±SD (n=3). *p<0.01 vs. pmiR-Glo Smad4-wt.
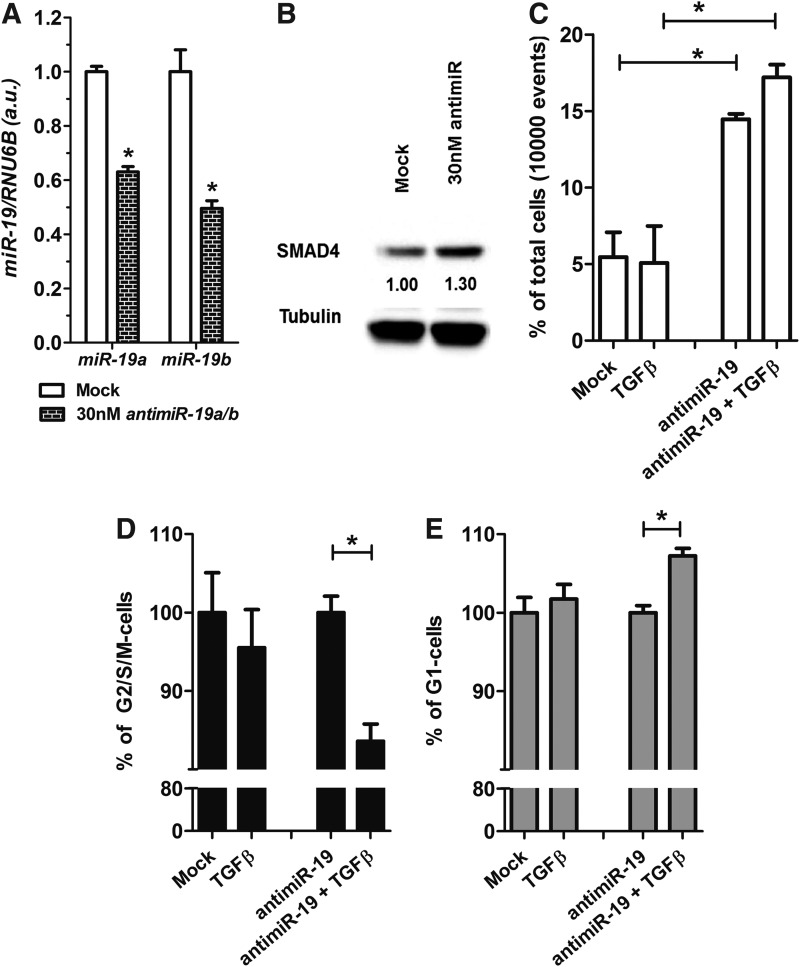
Next, we evaluated the effects of anti-miR-19 treatment in the BCPAP PTC cell line. We observed that anti-miR19 treatment reduced endogenous levels of miR-19a and miR-19b, and increased levels of the SMAD4 protein (Fig. 7A, B). Anti-miR-19 treatment induced BCPAP cell death, evident as an increase in the number of cells with fragmented DNA (three times higher than untreated control cells; Fig. 7C). Moreover, concomitant anti-miR-19 and rTGFβ treatment slightly increased the G1 cell cycle fraction and reduced the G2/S/M fraction, an effect not observed in cells treated with rTGFβ alone (Fig. 7D, E).
FIG. 7.
miR-19 modulation by anti-miR influences SMAD4 and TGFβ signaling responsiveness in BCPAP thyroid cancer cell line. (A) Anti-miR-19a and anti-miR-19b-reduces endogenous levels of miR-19a and miR-19b. (B) Western blotting shows that anti-miR-19 increases SMAD4 protein after 24 hours of transfection. (C) DNA content analysis by flow cytometry shows that anti-miR-19 increases cell death. (D, E) Anti-miR-19 increases G1 cell cycle arrest in the presence of rTGFβ while reducing the G2/S/M fraction. Data are expressed as mean±SE for gene expression and mean±SD for cell cycle (n=3). *p<0.05.
Discussion
Here, we show that the BRAFV600E oncogene stimulates expression of the miR-17-92 cluster and disrupts TGFβ signaling. Maintaining thyroid cells in high iodine-containing medium prevents this effect, blocking miR-17-92 induction and restoring TGFβ responsiveness.
High iodine exerts a protective effect during BRAFV600E oncogene activation by blocking upregulation of miR-17, miR-18, miR-19a, miR-19b, and miR-20. We have previously reported a protective role for high iodine during oncogenic activation of RET/PTC3 in thyroid follicular cells, by blocking pERK phosphorylation (8). A recent study indicated that miR-19 is the main oncogenic component of the cluster, as this miRNA alone is sufficient for promotion of c-Myc-induced lymphoma development via repression of apoptosis and activation of the AKT-mTOR pathway (24). Another study showed that miR-92 is expressed at higher levels in comparison to other miR-17-92 cluster components in colon cancer, serving as the key oncogenic miRNA in this type of cancer through targeting the antiapoptotic gene BIM (25). In thyroid cancer, selective inhibition of miR-19a or miR-17-5p resulted in strong growth reduction and restored expression of PTEN and RB, respectively, in an ATC cell line (10). Moreover, our present study indicates the importance of miR-19 in BRAF-mediated thyroid oncogenesis.
We observed distinct levels of induction of miR-17-92 components in response to BRAFV600E expression, indicating that intrinsic mechanisms of miRNA processing are associated with unequal expression arising from a single primary transcript. A recent study showed that the primary miR-17-92 transcript is differentially processed, depending on the position of the miRNAs within the tertiary structure of the pri-mi-17-92 polycistron, which generates different expression levels (26). This observation could explain the differing basal levels of miR-17, miR-18, and miR-19, which are substantially lower than those of miR-20 and miR-92 in rat thyroid follicular cells. Moreover, treatment with iodine alone stimulates punctually miR-20 and miR-92, indicating the existence of a complex and controlled mechanism of primary-miR-17-92 processing or an independent precursor miRNA induction mechanism. Interestingly, a recent study revealed that the presence of the RNA-binding protein, hnRNP1 A1, is important for the processing of miR-18a (27), pointing to the existence of a finely tuned balance of independent expression of these miRNAs.
One of the known stimulators of miR-17-92 is the proto-oncogene c-Myc. The human, mouse, and rat miR-17-92 promoter regions contain multiple consensus binding sequences for c-Myc, indicating possible regulation by this transcription factor. O'Donnell et al. showed that c-Myc binds to the promoter region and induces miR-17-92 expression in HeLa cells and that Myc deletion in rat cells reduces miR-17-92 levels, indicating an important influence of c-Myc on transcriptional regulation of the cluster (28). However, in our study, we observed that BRAFV600E-induced miR-17-92 expression is not associated with enhanced Myc binding in thyroid follicular cells (data not shown), indicating that miR-17-92 upregulation could be associated with a Myc-independent mechanism. Indeed, bioinformatic analyses of the rat miR-17-92 upstream region revealed putative binding sites for Hes1 (induced by Notch signaling) and NFκB transcription factors, in addition to c-Myc, which could influence the transcription of miR-17-92. NFκB (29) and Notch (17) signaling are among the pathways activated by BRAFV600E in thyroid follicular cells. Here, we show that high iodine blocks BRAF-induced activation of Notch signaling and that Notch1 knockdown inhibits expression of miR-19a/b. Moreover, hyperactivation of Notch signaling induced a robust increase in miR-19a and miR-19b expression, consistent with the participation of the Notch pathway in BRAF-induced miR-17-92 enhancement.
miR-17-92 cluster components share targets in the TGFβ pathway, such as the miR-17/20 target TGFBR2 and the miR-18 targets SMAD2/SMAD4 (30), which when downregulated could ablate TGFβ inhibitory signaling and contribute to tumorigenesis. Reduced SMAD4 expression is observed in PTC cell lines, which limits the response to TGFβ antiproliferative signaling, as overexpression of SMAD4 blocks cell proliferation and reduces invasion (31). Here, we show that BRAFV600E-induced expression of miR-19a/miR-19b targets Smad4 and impairs TGFβ-induced G1-arrest, which can be reversed by treatment with iodine. Moreover, inhibition of miR-19 in a thyroid cancer cell line increases SMAD4 expression and activates cell cycle arrest in response to TGFβ. Interestingly, Smad4 is also controlled by miR-146b (32), which is overexpressed in PTC (33), and whose expression is driven by the BRAFV600E and RET/PTC oncogenes. These complex interactions of miRNAs targeting the same mRNA indicate an additive effect of miR-19, miR-18, and miR-146b for the ablation of TGFβ inhibitory signaling during thyroid oncogenesis.
miR-17-92 is also overexpressed in aggressive lung cancer (11), and combined expression of miR-17-3p and miR-19b-1 interacts with c-Myc expression to accelerate tumor development in a mouse B-cell lymphoma model (12). ATC, the most aggressive histotype, exhibits miR-17-92 overexpression, while antisense inhibition of miR-17-92 reduces proliferation and induces apoptosis and cell senescence (10). Our results show that inhibition of BRAFV600E by the specific inhibitor PLX4032 in the KTC-2 ATC cell line reduces expression of the miR-17-92 cluster and that knockdown of NOTCH1 in cells also inhibits miR-19a/b, indicating ATC cells exhibit BRAFV600E/NOTCH addiction to maintain miR-17-92 upregulation.
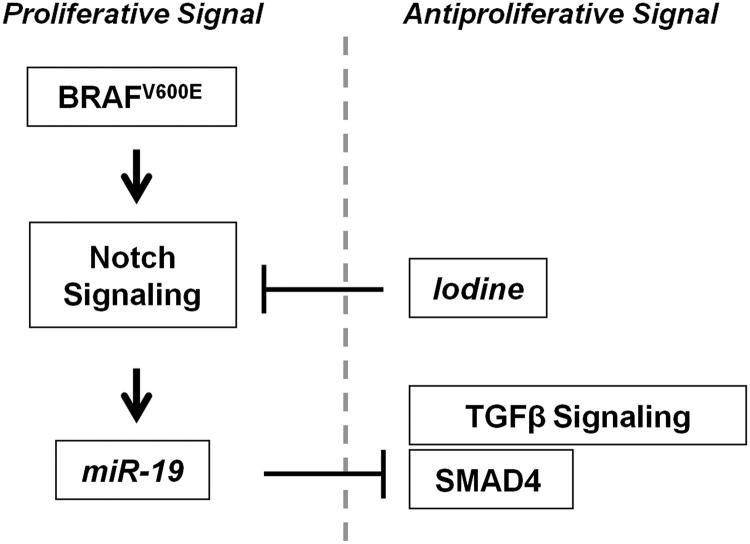
In this study, we show that high iodine exerts a protective influence over BRAF-activated thyroid cells. Iodine attenuates acute BRAF oncogene induction of miR-19, a newly identified regulator of Smad4, and blocks loss of responsiveness to the TGFβ pathway in a mechanism dependent on Notch signaling in thyroid follicular cells (Fig. 8). Detailed study of the functions of individual miR-17-92 cluster components is important for understanding the overall effect of BRAFV600E-induced miR-17-92 expression in thyroid follicular cells. However, the precise molecular mechanism by which iodine interferes with BRAF oncogene-induced effects and miR-17-92 cluster expression remains to be elucidated.
FIG. 8.
Summary of key findings.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Norisato Mitsutake (Nagasaki University Graduate School of Biomedical Sciences, Nagasaki, Japan) for kindly donating PC-BRAFV600E-6 and KTC2 cells; Dr. S. Diane Hayward (The Johns Hopkins University School of Medicine, Baltimore, MD) for the 4xCBF1Luc and 4xmCBF1Luc reporter plasmids; Dr. Takashi Takahashi (Nagoya University Graduate School of Medicine, Nagoya, Japan) for the pH1-RNA-puro-let7a plasmid; and Dr. Sissy M. Jhiang (The Ohio State University, Columbus, OH) for donating the NIS antibody.
This study was supported by grants #2010/51704-0 and #2011/50732-2 from the São Paulo Research Foundation (FAPESP), a grant from National Council for Scientific and Technological Development (CNPq), and a grant from University of Sao Paulo, NapMiR Research Support Center.
Author Disclosure Statement
The authors declare that there are no conflicts of interest that would prejudice the impartiality of this study.
References
- 1.Eng PH, Cardona GR, Fang SL, Previti M, Alex S, Carrasco N, Chin WW, Braverman LE.1999Escape from the acute Wolff–Chaikoff effect is associated with a decrease in thyroid sodium/iodide symporter messenger ribonucleic acid and protein. Endocrinology 140:3404–3410 [DOI] [PubMed] [Google Scholar]
- 2.Leoni SG, Galante PA, Ricarte-Filho JC, Kimura ET.2008Differential gene expression analysis of iodide-treated rat thyroid follicular cell line PCCl3. Genomics 91:356–366 [DOI] [PubMed] [Google Scholar]
- 3.Yamazaki K, Tanigawa K, Suzuki K, Yamada E, Yamada T, Takano K, Obara T, Sato K.2010Iodide-induced chemokines and genes related to immunological function in cultured human thyroid follicles in the presence of thyrotropin. Thyroid 20:67–76 [DOI] [PubMed] [Google Scholar]
- 4.Yuasa R, Eggo MC, Meinkoth J, Dillmann WH, Burrow GN.1992Iodide induces transforming growth factor beta 1 (TGF-beta 1) mRNA in sheep thyroid cells. Thyroid 2:141–145 [DOI] [PubMed] [Google Scholar]
- 5.Matsuo SE, Fiore AP, Siguematu SM, Ebina KN, Friguglietti CU, Ferro MC, Kulcsar MA, Kimura ET.2010Expression of SMAD proteins, TGF-beta/activin signaling mediators, in human thyroid tissues. Arq Bras Endocrinol Metabol 54:406–412 [DOI] [PubMed] [Google Scholar]
- 6.Eloy C, Santos J, Cameselle-Teijeiro J, Soares P, Sobrinho-Simoes M.2012TGF-beta/Smad pathway and BRAF mutation play different roles in circumscribed and infiltrative papillary thyroid carcinoma. Virchows Arch 460:587–600 [DOI] [PubMed] [Google Scholar]
- 7.Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, Solari A, Bobisse S, Rondina MB, Guzzardo V, Parenti AR, Rosato A, Balmain A, Piccolo S.2009A mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell 137:87–98 [DOI] [PubMed] [Google Scholar]
- 8.Fiore AP, Fuziwara CS, Kimura ET. 2009High iodine concentration attenuates RET/PTC3 oncogene activation in thyroid follicular cells. Thyroid 19:1249–1256 [DOI] [PubMed] [Google Scholar]
- 9.Petrocca F, Vecchione A, Croce CM.2008Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res 68:8191–8194 [DOI] [PubMed] [Google Scholar]
- 10.Takakura S, Mitsutake N, Nakashima M, Namba H, Saenko VA, Rogounovitch TI, Nakazawa Y, Hayashi T, Ohtsuru A, Yamashita S.2008Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci 99:1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T.2005A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 65:9628–9632 [DOI] [PubMed] [Google Scholar]
- 12.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM.2005A microRNA polycistron as a potential human oncogene. Nature 435:828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM.2006A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103:2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iorio MV, Croce CM.2009MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol 27:5848–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA.2003High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res 63:1454–1457 [PubMed] [Google Scholar]
- 16.Nikiforova MN, Kimura ET, Gandhi M, Biddinger PW, Knauf JA, Basolo F, Zhu Z, Giannini R, Salvatore G, Fusco A, Santoro M, Fagin JA, Nikiforov YE.2003BRAF mutations in thyroid tumors are restricted to papillary carcinomas and anaplastic or poorly differentiated carcinomas arising from papillary carcinomas. J Clin Endocrinol Metab 88:5399–5404 [DOI] [PubMed] [Google Scholar]
- 17.Yamashita AS, Geraldo MV, Fuziwara CS, Kulcsar MA, Friguglietti CU, da Costa RB, Baia GS, Kimura ET.2013Notch pathway is activated by MAPK signaling and influences papillary thyroid cancer proliferation. Transl Oncol 6:197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sethi N, Kang Y.2011Notch signalling in cancer progression and bone metastasis. Br J Cancer 105:1805–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitsutake N, Knauf JA, Mitsutake S, Mesa C, Jr, Zhang L, Fagin JA.2005Conditional BRAFV600E expression induces DNA synthesis, apoptosis, dedifferentiation, and chromosomal instability in thyroid PCCL3 cells. Cancer Res 65:2465–2473 [DOI] [PubMed] [Google Scholar]
- 20.Simon P.2003Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics 19:1439–1440 [DOI] [PubMed] [Google Scholar]
- 21.Hsieh JJ, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD.1996Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol 16:952–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Massachusetts Institute of Technology Target Scan Human. Available at: http://targetscan.org (accessed January2013)
- 23.Schug J, Overton GC.1997TESS: Transcription Element Search Software on the WWW. Technical Report CBIL-TR-1997-1001-v0.0
- 24.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L.2009miR-19 is a key oncogenic component of mir-17-92. Genes Dev 23:2839–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuchida A, Ohno S, Wu W, Borjigin N, Fujita K, Aoki T, Ueda S, Takanashi M, Kuroda M.2011miR-92 is a key oncogenic component of the miR-17-92 cluster in colon cancer. Cancer Sci 102:2264–2271 [DOI] [PubMed] [Google Scholar]
- 26.Chaulk SG, Thede GL, Kent OA, Xu Z, Gesner EM, Veldhoen RA, Khanna SK, Goping IS, MacMillan AM, Mendell JT, Young HS, Fahlman RP, Glover JN.2011Role of pri-miRNA tertiary structure in miR-17∼92 miRNA biogenesis. RNA Biol 8:1105–1114 [DOI] [PubMed] [Google Scholar]
- 27.Guil S, Caceres JF.2007The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol 14:591–596 [DOI] [PubMed] [Google Scholar]
- 28.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT.2005c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435:839–843 [DOI] [PubMed] [Google Scholar]
- 29.Palona I, Namba H, Mitsutake N, Starenki D, Podtcheko A, Sedliarou I, Ohtsuru A, Saenko V, Nagayama Y, Umezawa K, Yamashita S.2006BRAFV600E promotes invasiveness of thyroid cancer cells through nuclear factor kappaB activation. Endocrinology 147:5699–5707 [DOI] [PubMed] [Google Scholar]
- 30.Mestdagh P, Bostrom AK, Impens F, Fredlund E, Van Peer G, De Antonellis P, von Stedingk K, Ghesquière B, Schulte S, Dews M, Thomas-Tikhonenko A, Schulte JH, Zollo M, Schramm A, Gevaert K, Axelson H, Speleman F, Vandesompele J.2010The miR-17-92 microRNA cluster regulates multiple components of the TGF-beta pathway in neuroblastoma. Mol Cell 40:762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D'Inzeo S, Nicolussi A, Ricci A, Mancini P, Porcellini A, Nardi F, Coppa A.2010Role of reduced expression of SMAD4 in papillary thyroid carcinoma. J Mol Endocrinol. 45:229–244 [DOI] [PubMed] [Google Scholar]
- 32.Geraldo MV, Yamashita AS, Kimura ET.2012MicroRNA miR-146b-5p regulates signal transduction of TGF-beta by repressing SMAD4 in thyroid cancer. Oncogene 31:1910–1922 [DOI] [PubMed] [Google Scholar]
- 33.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, de la Chapelle A.2005The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA 102:19075–19080 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.