Abstract
Krüppel-like factor 5 (KLF5) is a pro-proliferative transcriptional regulator primarily expressed in the intestinal crypt epithelial cells. Constitutive intestine-specific deletion of Klf5 is neonatal lethal suggesting a crucial role for KLF5 in intestinal development and homeostasis. We have previously shown Klf5 to play an active role regulating intestinal tumorigenesis. Here we examine the effect of inducible intestine-specific deletion of Klf5 in adult mice. Klf5 is lost from the intestine beginning at day 3 after the start of a 5-day treatment with the inducer tamoxifen. Although the mice have no significant weight loss or lethality, the colonic tissue shows signs of epithelial distress starting at day 3 following induction. Accompanying the morphological changes is a significant loss of proliferative crypt epithelial cells as revealed by BrdU or Ki67 staining at days 3 & 5 after start of tamoxifen. We also observed a loss of goblet cells from the colon and Paneth cells from the small intestine upon induced deletion of Klf5. In addition, loss of Klf5 from the colonic epithelium is accompanied by a regenerative response that coincides with an expansion in the zone of Sox9 expression along the crypt axis. At day 11, both proliferation and Sox9 expression return to baseline levels. Microarray and quantitative PCR analyses reveal an upregulation of several regeneration-associated genes (Reg1A, Reg3G and Reg3B) and down-regulation of many Klf5 targets (Ki-67, cyclin B, Cdc2 and cyclin D1). Sox9 and Reg1A protein levels are also increased upon Klf5 loss. Lentiviral-mediated knockdown of KLF5 and exogenous expression of KLF5 in colorectal cancer cell lines confirm that Sox9 expression is negatively regulated by KLF5. Furthermore, ChIP assays reveal a direct association of KLF5 with both the Sox9 and Reg1A promoters. We have shown that disruption of epithelial homeostasis due to Klf5 loss from the adult colon is followed by a regenerative response led by Sox9 and the Reg family of proteins. Our study demonstrates that adult mouse colonic tissue undergoes acute physiological changes to accommodate the loss of Klf5 withstanding epithelial damage further signifying importance of Klf5 in colonic homeostasis.
INTRODUCTION
Krüppel-like factor 5 (KLF5) is a member of the Krüppel-like factor (KLF) family, currently comprises of 17 members (McConnell and Yang, 2010). KLFs are distinct Sp1-like, zincfinger transcription factors that share homology with the Drosophila transcription factor, Krüppel, and regulate myriad biological activities such as development, differentiation, proliferation and cell death (McConnell and Yang, 2010; Nandan and Yang, 2009; Ghaleb et al., 2005; Dang et al., 2000). KLF5, also known as intestine-enriched Krüppel-like factor (IKLF), is predominantly expressed in the proliferating compartment of the intestinal epithelium (Ghaleb et al., 2005; McConnell et al., 2007). KLF5 has a pro-proliferative effect by inducing expression of several cell cycle related genes such as cyclin D1, cyclin B and Cdc2 (Nandan et al., 2005; Nandan et al., 2004). KLF5 is also implicated in intestinal tumorigenesis resulting from both adenomatous polyposis coli gene (Apc) mutation and KRas activation (McConnell et al., 2009; Nandan et al., 2010; Nandan et al., 2008); intestinal tumors derived from Apc mutation or K-Ras activation have increased Klf5 expression. Both tumor number and volume are significantly reduced upon heterozygous deletion of Klf5 in the two mouse models (McConnell et al., 2009; Nandan et al., 2010). Klf5 responds to environmental stresses, including chemical and bacterial insults, in varying manner. Infection of mice with C. rodentium, a mouse intestinal pathogen, resulted in an increase in Klf5 expression and colonic crypt hyperplasia (McConnell et al., 2008). However, heterozygous Klf5 mutant displayed increased susceptibility to Dextran sodium sulfate (DSS)-induced colitis (McConnell et al., 2011). A role for KLF5 in intestinal tumorigenesis and inflammation has been established, however, precise role and importance of KLF5 in normal intestinal homeostasis has still not been well established.
Whole-body homozygous deletion of Klf5 results in embryonic lethality (Shindo et al., 2002). Recently, we developed an intestine-specific mouse model of Klf5 deletion by crossing mice that express homozygously floxed Klf5 alleles with those that express Cre recombinase under the control of the intestine-specific villin promoter (McConnell et al., 2011b). Our results indicate that complete deletion of Klf5 results in neonatal lethality even though mice are born in normal Mendelian ratios. This outcome is attributed to a loss of epithelial proliferation in the intestine. A small percentage of the mice have variegated deletion of Klf5 and survived up to 8 weeks post-partum. These mice display distinct morphological changes including alterations in intestinal epithelial differentiation, migration and barrier function when compared to control mice. Another recent study has reported on the importance of Klf5 in development of the embryonic intestinal epithelium (Bell et al., 2013). Upon deletion of Klf5 using a Sonic hedgehog (Shh)-promoter driven Cre recombinase, the authors observed an phenotype that displayed severe disruption of intestinal maturation and specifically villus morphogenesis. This was evident inspite of no loss in proliferative cells. Furthermore, they also observed an elevated level of FoxA1 and Sox9 expression in the Klf5 deleted embryoes that maintained them in an undifferentiated state. Similarly, we also observed increased expression of a regeneration-associated transcription factor, Sox9, in mice displaying variegated intestine-specific deletion of Klf5 (McConnell et al., 2011b).
Development of the intestinal epithelium is a well-coordinated process. The differentiated cells of the secretory lineage (goblet, enteroendocrine cells and Paneth cells) are dependent on specific expression and repression of Atoh1 (or Math1) or Hes1, respectively (van der Flier and Clevers, 2009; Yeung et al., 2011). Sox9 belongs to the Sexdetermining region Y-box transcription factor family that are high-mobility group proteins and is a Wnt-responsive gene that is predominantly expressed in the crypts and proliferating epithelial cells of both the small intestine and colon (Blache et al., 2004; Formeister et al., 2009). It is also involved in intestinal differentiation, in particular, Paneth cells (Bastide et al., 2007). Intestine-specific deletion of Sox9 results in the disappearance of Paneth cells and a marked reduction of goblet cells and mucin expression (Muc-2) (Bastide et al., 2007). In addition, Bastide et al. also reported an increase in local crypt dysplasia and general hyperplasia with an increase in expression of Wnt-target genes. Sox9 is reported to negatively regulate P-catenin mediated transcription in vivo, thereby reducing c-Myc and cyclin D1 expression upon its induction (Blache et al., 2004; Bastide et al., 2007; Sellak et al., 2012). Lineage tracing of Sox9 expressing cells in the adult intestinal epithelium revealed that these cells could also function as adult progenitor cells (Furuyama et al., 2011).
Due to the extensive neonatal lethality after constitutive intestine-specific Klf5 deletion (McConnell et al., 2011b), it is difficult to gauge the effect of Klf5 deletion from adult intestinal tissues. Hence, we resort to an inducible intestine-specific Cre recombinase system for our current study (el Marjou et al., 2004). Mice with inducible intestine-specific deletion of Klf5 are generated using the Klf5fl/fl mice crossed to Villin-Cre-ERT2 mice, which express an inducible Cre recombinase under the control of a 9 kb mouse villin promoter. This normally inactive Cre recombinase is activated by the addition of a human estrogen receptor (ER) agonist, tamoxifen (el Marjou et al., 2004). Tamoxifen treatment of Villin-Cre-ERT2;Klf5fl/fl mice results in a colonic phenotype marked by epithelial disruption and appearance of a regenerative morphology. Furthermore, Sox9 and several regenerationassociated genes (Reg) are activated upon Klf5 deletion. Based on these observations, we conclude that Klf5 in the adult mouse colonic epithelium regulates both epithelial homeostasis and regeneration.
METHODS
Mice
All studies involving mice were approved by the Stony Brook University Institutional Animal Care and Use Committee (IACUC). C57BL/6 mice carrying Klf5 alleles flanked by loxP sites (Klf5fl/fl) were previously described (Takeda et al., 2010). Also described previously were C57BL/6 mice carrying the inducible Cre recombinase gene under regulation of the villin promoter (Villin-Cre-ERT2 mice) (el Marjou et al., 2004). To establish Villin-Cre-ERT2/Klf5fl/fl (designated as Klf5ERΔ) mice, Klf5fl/fl mice were initially crossed with Villin-Cre-ERT2 mice and its progeny then backcrossed to Klf5fl/fl mice. For all experiments, Klf5fl/fl mice were used as controls. Tamoxifen treatment of mice was per protocol previously described (el Marjou et al., 2004). Eight week old Klf5ERΔ and Klf5fl/fl mice were injected intraperitoneally (I.P.) with 1 mg of tamoxifen (10 mg/ml, dissolved in sterile Corn oil, Sigma, USA) for five consecutive days. Animals were sacrificed on days 3, 5, 7 and 11 after the start of tamoxifen injections.
Cell culture, plasmids and siRNA transfection
HCT116, DLD1 and T84 colon cancer cell lines and HEK293T cells were purchased from ATCC (Manassas, VA, USA) and maintained as previously described (Bialkowska et al., 2011; Yu et al., 2012). HCT116 and DLD1 cells were grown in McCoy’s 5A and RPMI 1640 media (Corning, NY, USA), respectively, supplemented with 10% FBS and 1% Penicillin/Streptomycin (Life Technologies, CA, USA) and maintained at 37 °C in 5% CO2. T84 cells were grown in DMEM/F12 media (Corning, NY, USA) supplemented with 5% FBS and grown under similar conditions. HEK293T cells were grown with DMEM (high glucose) (Corning, NY, USA), also supplemented with 10% FBS and 1% Penicillin/Streptomycin. pCI-Neo-KLF5 was transfected into cells for KLF5 over-expression and protein extracts were collected after 24 hr incubation (McConnell et al., 2009). pCI-Neo (Promega, WI, USA) was used as the empty vector control. Sox9 expression plasmid with the control vector was purchased from Genecopoeia, MD, USA. Short hairpin RNA (shRNA) against KLF5 cloned in pLKO1-puro vector was purchased from Sigma, MO, USA, with the appropriate controls. Klf5-specific silencer siRNA and control silencer siRNA were purchased from Life Technologies, CA. DLD1 cells were grown to 60% confluence and transfected with 50nM silencer siRNA using Lipofectamine RNAimax (Life Technologies, CA) as per protocol. Cells were subsequently used for luciferase reporter assay.
Lentiviral transduction
Lentivirus production and transduction were performed as previously described with modifications (Kutner et al., 2009). The envelope and helper plasmids (pLTR-G and pCD/NL-BH*ΔΔΔ) were a gift from Dr. Jakob Reiser (FDA, Bethesda, MD). HEK293T cells were transfected with helper, envelope and KLF5-specific or control shRNA-containing plasmids. The KLF5-specific (Mission clone# TRCN 0000013635) and control shRNA plasmids (Mission pLKO.1-puro #SHC001) were both purchased from Sigma, MO, USA. The lentivirus-containing supernatant was harvested 2 days post-transfection. T84 cells were transduced with lentivirus for 24 hr and then selected with puromycin (4 µg/ml) (Santa Cruz Biotechnology, CA, USA) for 7 days to establish stable T84 cells with KLF5 knockdown.
Antibodies
Antibodies used in the experiments were previously described (McConnell et al., 2011b; Nandan et al., 2010). For Western blot analysis, antibodies against KLF5 (Strategic Diagnostics, Newark, DE) were synthesized and used at 1:4,000 dilution. For immunofluorescence analysis, KLF5 polyclonal antibodies (Santa Cruz Biotechnologies, CA, USA; Cat# sc-22797) were used at 1:200 dilution. Anti-Ki67 antibodies were purchased from Novocastra (Leica Microsystems, IL, USA; Cat# KI67P-CE) and used at 1:500 dilutions.
Sox9 antibodies (used at 1:100 dilution; Cat# sc-20095) were obtained from Santa Cruz Biotechnologies were used for both immunohistochemistry (IHC) and immunoblots. Sox9 antibody purchased from Millipore (Billerica, MA; Cat# AB5535) was used for immunofluorescence staining at 1:100 dilution. Monoclonal antibody against Reg1 alpha was purchased from Abcam, MA, USA (Cat# ab86374) and used at 1:100 dilution. Antibodies derived against Chromogranin A (Epitomics, CA, USA; Cat# 1773-1) and Carbonic Anhydrase (Santa Cruz Biotechnologies; Cat# sc-50896) were used at 1:500 dilution. Monoclonal P-actin antibodies used as loading control on immunoblots were purchased from Sigma Aldrich, MO, USA (Cat# A1978) and used at 1:2000 dilution. Cleaved caspase-3 antibody was purchased from Cell Signaling Technology, MA, USA(Cat# 9664S) used at 1: 200 dilution. Lysozyme antibody used at 1:2000 dilution was acquired from Dako North America, CA, USA (Cat# A0099).
Alcian blue staining
Alcian blue (AB) staining was performed as previously described (McConnell et al., 2011b). Paraffin-embedded sections were de-paraffinized and stained with AB to identify goblet cells. This was carried out using the Alcian Blue Staining Kit (Biocare Medical, CA, USA) per manufacturer′s instructions.
Microarray and statistical analyses
Total RNA was extracted from scraped colonic musosa of Klf5ERΔ and Klf5fl/fl mice (n=3, each), after 3 day of tamoxifen treatment, using Trizol reagent (Life Technologies, CA, USA). The extracted RNA was purified using RNeasy columns (Qiagen, CA, USA) upon DNase digestion (Life Technologies, CA, USA). RNA samples were visualized on gels to ascertain proper integrity and prepared for microarray analysis on an Illumina MouseWG-6 v2.0 Expression BeadChip Kit (Illumina, CA, USA). Microarray and statistical analysis were performed as described earlier (Hagos E, 2011). The arrays were scanned using the BeadStation 500 Instrument (Illumina, San Diego, CA) and data were normalized using the GenomeStudio v1.0.2 and Illumina Beadchip software (Illumina, San Diego, CA). Significant genes were identified using significance analysis of microarrays (SAM) with a FDR of 1%. The differentially expressed genes were then annotated and analysed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (david.abcc.ncifcrf.gov). Fold change of > 1.5 or <−1.5 and P<0.05 were used as criteria to parse through the data. Pathway analysis was performed using Ingenuity Pathway Analysis (IPA) where genes were sorted into groups based on function and ordered according to their P-values.
Quantitative polymerase chain reaction
RNA from scraped colonic musosa of Klf5ERΔ and Klf5 fl/fl mice (n=3, each), prepared for microarray analysis, was used for quantitative PCR. Specific primers against mouse Klf5, Ki-67, cyclin B1, Cdk1, Reg3G, Reg1A, Reg3B, Cyp4B1 and Sox9 were purchased from Qiagen (CA, USA). Their respective catalog numbers are QT01057756, QT00247667, QT00152040, QT00167734, QT00147455, QT00178514, QT00239302, QT00107450, QT00163765 Quantitative PCR was performed using the Power SYBR Green RNA-to-CT 1-Step kit (Life Technology, CA, USA) as per protocol. Observed CT levels were then used to calculate fold change using the 2-ΔΔCt method of relative quantification (Livak and Schmittgen, 2001).
Immunohistochemistry
Immunohistochemical analysis was modified from a previously described protocol (Nandan et al., 2010). Intestinal tissues dissected from mice were fixed overnight in 10% formalin buffer (Fisher Scientific, Fair Lawn, PA, USA). The tissues were then paraffin-embedded using an automated processor and sectioned at 5 µm. The sections were then dried onto charged slides and used for staining. The slides were baked in a 65 °C oven for 1 hr and subsequent were deparaffinized in xylene. Sections were incubated in a 3% hydrogen peroxide bath to block endogenous tissue peroxidases and were then rehydrated by incubation in a decreasing alcohol bath series (100%, 95%, 70%) followed by antigen retrieval in citrate buffer solution (10mM Sodium citrate, 0.05% Tween-20, pH 6.0) at 125 °C for 10 min using a decloaking chamber (Biocare Medical, CA, USA). Tissue sections were first incubated with blocking buffer (Biocare Medical, CA, USA) for 30 min and then incubated with primary antibody at 4°C overnight with shaking. Sections were washed and incubated with secondary antibodies (HRP-conjugated or fluorescent-tagged) at the appropriate concentration for 30 min at 37 °C. Betazoid DAB (Biocare Medical, CA, USA) was used to reveal IHC staining in tissues. The sections were then incubated in Gill’s Hematoxylin (Vector Labs, CA, USA), dehydrated and cover-slipped for observation for IHC sections. For fluorescent sections, slides were washed after secondary antibody treatment and then stained with DAPI (Fisher Scientific, PA, USA) and mounted with Prolong gold antifade (Life Technologies, CA, USA). Slides were observed under a Nikon Eclipse 90i microscope (Nikon, NY, USA) and representative pictures taken. Morphology of sections was observed upon staining 5 µm sections with Hematoxylin and Eosin (H&E) (Vector Labs, CA, USA). Slides were subsequently analysed by our pathologist, Dr. Kenneth Shroyer at Stony Brook University. All cell count and staining intensity measurements were performed using ImageJ software (http://imagej.nih.gov/).
5-Bromo-2-deoxyuridine (BrdU) labelling
Mice were injected intraperitoneally (IP) with BrdU (Sigma) at 50 µg/g body weight, then sacrificed at 4 hr post-injection. Immunostaining for BrdU was performed using monoclonal BrdU antibodies (BD Biosciences, CA, USA) at 1:50 dilution. Following immunostaining, the number of BrdU-positive cells was counted from at least 50 crypts per mouse per time point.
Western blot analyses
Western blot analyses were performed as previously described (Nandan et al., 2010). Proteins were extracted from 20 µm paraffin embedded tissue sections using a previously established protocol (Shi et al., 2006). Tissue sections were deparaffinized using xylene with the addition of 7.5% methanol and were then centrifuged and the pellet dried in a fume hood for 3 min. The pellets were then resuspended in 20 mM Tris-HCl (pH 7.5) containing 2% SDS and the suspension heated in a 100 °C heat block for 20 min. Subsequently, the samples were incubated in a 60 °C oven for 2 hr. Protein content was measured and equal amounts of samples were loaded onto Tris-Glycine gels (Life Technologies, CA, USA). Proteins were transferred to nitrocellulose membranes (BioRad, CA, USA) and probed with appropriate primary antibodies. Blots were then washed and secondary antibodies applied at appropriate concentrations. Protein bands were subsequently visualized on film upon chemiluminescent detection.
Chromatin Immunoprecipitation (ChIP)
Chromatin Immunoprecipitation assays were performed as per protocol (SimpleChIP Enzymatic Chromatin IP kit, Cell Signaling Technologies, MA, USA). Proteins are initially cross-linked to DNA and nuclei are pelleted and sonicated to shear DNA. The cross-linked DNA was immunoprecipitated with appropriate antibodies overnight at 4 °C with rotation, DNA-Antibody complexes were bound to ChIP beads, pulled down, washed and then eluted from beads. Following reversal of cross-linkage purified DNA was used for Quantitative PCR using ChIP PCR primers against Sox9, Reg1a, Reg3g, Ccnd1 that were purchased from Qiagen, CA, USA. Putative KLF5 and Sox9 binding sites were identified using TESS (Transcription Element Search System) database at http://www.cbil.upenn.edu/tess. Primers were directed to the area spanning the 1Kb upstream of the transcription start site, where the putative KLF5/Sox9 binding sequences were localized (Qiagen catalog#s GPH1005991(−)01A, GPH1007558(−)01A, GPH1007557(−)01A, GPH1002541(−)01A respectively). Immunoprecipitation efficiency was calculated by normalizing sample CT values against control IgG values and calculating ratios of sample CT values relative to Input values.
Luciferase reporter assay
Sox9 and Reg1A luciferase promoter reporter plasmids (Cat# S705795 & S700990) were purchased from Switchgear Genomics, CA, USA. DLD1 colon cancer cells that were knockdown with Klf5-specific or control silencer siRNA for 24 hrs were transfected with Sox9, Reg1A or empty reporter plasmids using Lipofectamine 2000 (Life Technologies, CA) as per protocol. Lysates were collected and analysed using LightSwitch Luciferase assay kit (Switchgear Genomics, CA) as per protocol.
RESULTS
Klf5 deletion in adult mouse intestine results in epithelial damage and a regenerative morphology
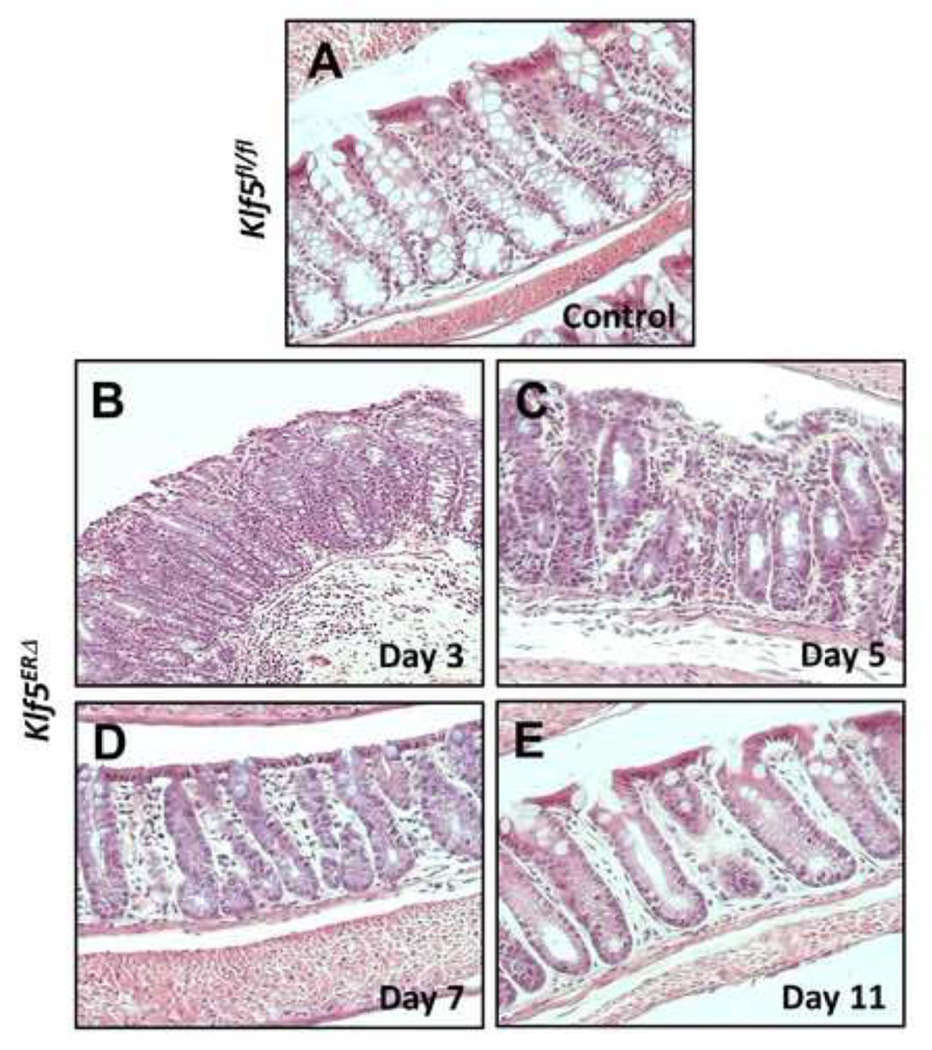
To better understand the physiologic functions of Klf5 in the adult mouse intestine, we generated mice with inducible intestine-specific deletion of Klf5. Here, we induced Cre recombinase expression in the intestinal tissues by treating 8-week old Vil-Cre-ERT2 /Klf5fl/fl (designated as Klf5ERΔ) and control Klf5fl/fl mice with tamoxifen for five consecutive days (Suppl. Fig. 1). Intestinal tissue samples were collected at days 3, 5, 7 and 11 from the start of tamoxifen treatment (Suppl. Fig. 1). Histological sections from day 3 tamoxifen-treated Klf5ERΔ colon (Fig. 1B & Suppl. Fig. 3B) display focal acute inflammation involving crypts and adjacent lamina propria compared to control (day 5 data shown in Fig. 1A & Suppl. Fig. 3A, data from other days not shown). In areas of inflammation, the crypts showed mild reactive atypia, including nuclear enlargement with prominent nucleoli, with a decrease in the number of goblet cells. The adjacent submucosa exhibited acute inflammation, including neutrophils and plasma cells. Reactive/regenerative changes persist in day 5 tamoxifen-treated Klf5ERΔ colon (Fig. 1C & Suppl. Fig. 3C) reflected by decreased goblet cell lineage and mild nuclear enlargement but mucosal inflammation appeared to subside. Day 7 tamoxifen-treated Klf5ERΔ colon (Fig. 1D & Suppl. Fig. 3D) showed shortened crypts but there was restoration of goblet cells and no evidence of active inflammation. By day 11, the tamoxifen-treated Klf5ERΔ colon epithelium (Fig. 1E & Suppl. Fig. 3E) appeared to be restored to normal. Analogous changes were observed in the small intestinal tissues of the Klf5ERΔ mice with a shortening of crypt heights compared to the control Klf5fl/fl mice (data not shown). We observed a complete loss of Klf5 expression from the colonic crypt cells of the Klf5ERΔ mice (Figs. 2E, H, K & N) compared to tamoxifen-treated control Klf5fl/fl mice (Fig. 2B). There was not a significant difference in the change in weights between the Klf5ERΔ and Klf5fl/fl mice up to 8 days after the start of tamoxifen treatment (Suppl. Fig. 2).
Figure 1. Acute short-term inflammation of colonic epithelium upon Klf5 deletion from Klf5ERΔ mice.
Representative H&E images are presented at 10X magnification both from colons of day 5 tamoxifen-treated Klf5fl/fl controls (Panel A) and days 3, 5, 7 & 11 after the start of tamoxifen treatment in Klf5ERΔ mice (Panels B, C, D and E, respectively). Acute focal inflammation with mild nuclear enlargement, prominent nucleoli and loss of goblet was evident in days 3 and 5. Day 7 & 11 panels (D & E) show a moderate resolution of inflammation and restoration of goblet cells.
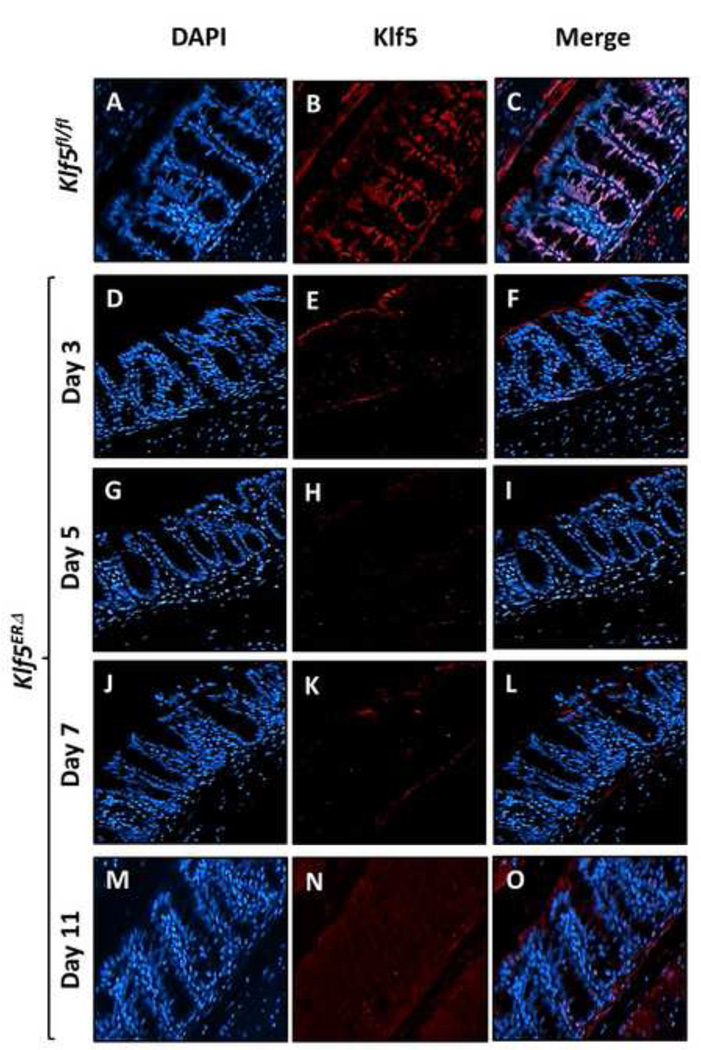
Figure 2. Tamoxifen treatment of Klf5ERΔ mice results in deletion of Klf5 from colonic tissues.
Immunofluorescence analysis of Klf5 expression in colonic tissues from both Klf5ERΔ and control Klf5fl/fl mice after tamoxifen treatment. Images are stained with DAPI (blue), Klf5 (red) and merged (DAPI + Klf5). Panels A through C show colonic tissues from day 5 tamoxifen-treated control Klf5fl/fl and Panels D through O, those from Klf5ERΔ mice. Panels D, E & F are representative of 3 day tamoxifen treatment of Klf5ERΔ mice. Panels G-I, J-L and M-O represent 5, 7, and 11 day post-tamoxifen treatment respectively. Klf5ERΔ mice show a complete loss of Klf5 (Panels E, H, K & N) from the nuclei of colonic epithelial cells when compared to Klf5fl/fl colon (Panel B).
The numbers of proliferative crypt cells are reduced upon Klf5 deletion in Klf5ERΔ mouse colons
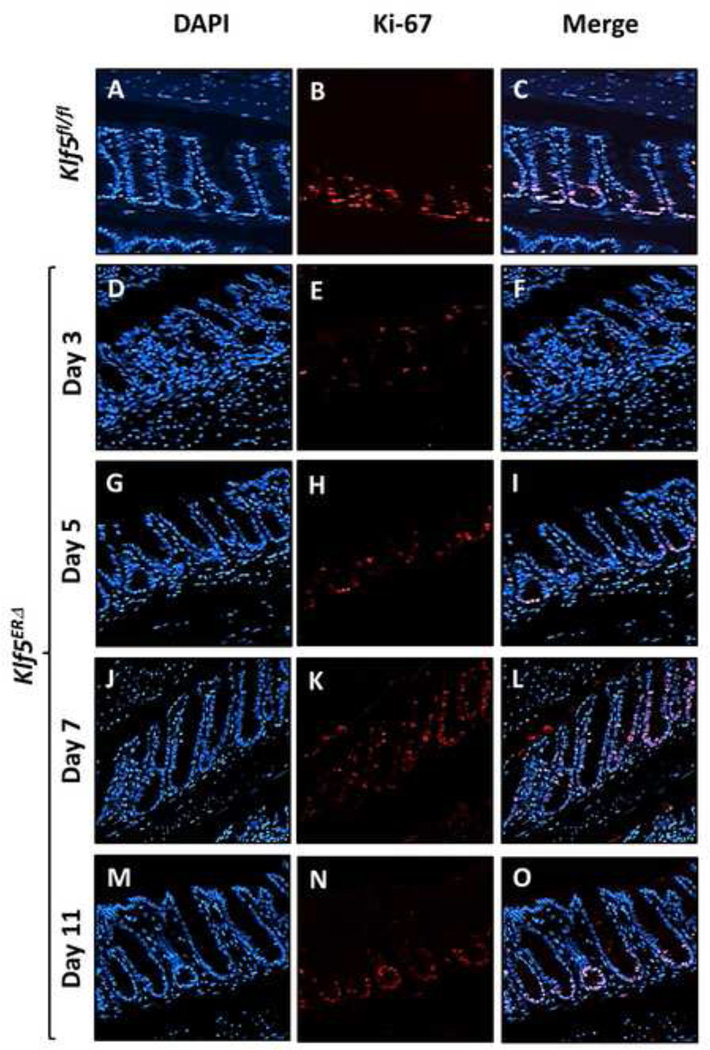
We performed immunofluorescence analyses to determine the proliferative capacity of the crypts in the colon of mice following Klf5 deletion. Expression of Ki-67, a marker for proliferation, was restricted to the bottom half of the colonic crypts in control Klf5fl/fl mice after 5 days of tamoxifen treatment (Fig. 3B) in the location that correlates with Klf5 expression (Fig. 2B). This finding is consistent with our previous studies demonstrating that KLF5 functions as a mitogenic factor (Nandan et al., 2010; Nandan et al., 2008). At days 3 and 5 of tamoxifen treatment, we observed a significant decrease in the number of Ki-67 positive cells in the colonic crypts of the Klf5ERΔ mice (Figs. 3E & H and Suppl. Figs. 4B, 4C & 5B) when compared to control Klf5fl/fl mice (Fig. 3B and Suppl. Figs. 4A & 5B). The number of Ki-67 positive cells on day 7 following start of tamoxifen treatment was higher than days 3 and 5 and extended to almost the top of the colonic crypts (Fig. 3K & Suppl. Fig. 4D). By day 11, Ki-67 was restored to baseline levels and its localization returned to the bottom of the colonic crypts (Fig. 3N & Suppl. Fig. 4E). The decrease in the number of proliferative cells at days 3 and 5 was confirmed by BrdU labelling of tamoxifen treated mice. There was an immediate loss of proliferative cells at day 2 after start of tamoxifen treatment compared to the untreated mice. The decrease in proliferative cells continued up to day 5 (Suppl. Fig. 5A). We did not find significant differences in apoptosis as denoted by cleaved caspase 3 staining between the two genotypes (Suppl. Figs. 8A & B). These results suggest that loss of Klf5 results in an immediate reduction in proliferative cells from the colonic crypts.
Figure 3. The numbers of Ki67-positive proliferative crypt cells are reduced upon Klf5 deletion in Klf5ERΔ mouse colon.
Proliferation was monitored in Klf5ERΔ and control Klf5fl/fl colon by staining sections for Ki-67. Images are stained with DAPI (blue), Ki-67 (red) and merged (DAPI + Ki-67). Panels A, B and C show colonic tissues from day 5 tamoxifen-treated control Klf5fl/fl and Panels D through O, those from Klf5ERΔ mice. Panels D-F, G-I, J-L and M-O are representative of 3, 5, 7, and 11 day post-tamoxifen treatment respectively. In the control Klf5fl/fl mice, a strong Ki-67 staining pattern was located at the bottom of the colonic crypts (Panel B). Panels E and H show a decrease in Ki-67 staining at days 3 and 5 after the start of tamoxifen treatment, respectively. Ki-67 expression gradually returns to baseline by days 7 and 11 (Panels K and N, respectively).
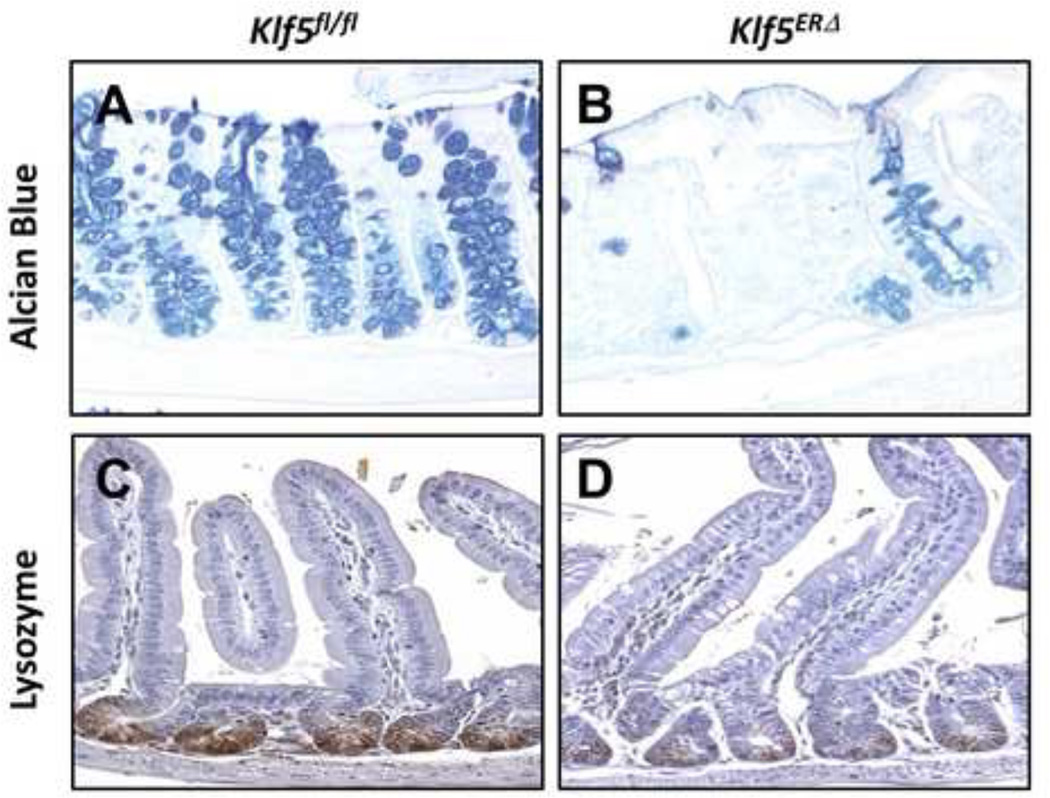
Differentiated epithelial cells of the secretory lineage, particularly goblet and Paneth cells, are attenuated in Klf5ERΔ mouse colon and small intestine, respectively
An interesting observation arose while analyzing the differentiated epithelial cell compartments of the Klf5ERΔ and Klf5fl/fl mice (Fig. 4 and Suppl. Fig. 8). Day 5 Klf5ERΔ colonic mucosa displayed a decreased number of goblet cells (Fig. 4B) when compared to Klf5fl/fl mouse colons (Fig. 4A). However, Chromogranin A positive epithelial cells, representative of enteroendocrine cells, were not significantly changed in the day 5 Klf5ERΔ colons (Suppl. Fig. 8D) compared to Klf5fl/fl mice (Suppl. Fig. 8C). Similarly, there was no change in epithelial cells expressing carbonic anhydrase, representative of absorptive colonocytes, between both day 5 Klf5ERΔ and Klf5fl/fl mouse colons (Suppl. Figs. 8E & F). In contrast, we observed a marked decrease in the number of lysozyme-stained Paneth cells in the day 5 Klf5ERΔ small intestines compared to Klf5fl/fl (Figs. 4C & D). These results suggest that Klf5 deletion affects epithelial differentiation of cells belonging to certain secretory lineages but not of the absorptive lineage.
Figure 4. Differentiation of epithelial cells is repressed in tamoxifen-treated Klf5ERΔ mouse colon and small intestine compared with controls.
Day 5 tamoxifen-treated colon tissue sections stained with Alcian blue (as blue stain in panels A and B) and similarly treated small intestinal tissues stained with lysozyme (as brown stain in panels C and D). Panels A & C represent control Klf5fl/fl mice, while Panels B & D represents Klf5ERΔ tissues. There was a significant reduction in Alcian blue and lysozyme staining in Klf5ERΔ tissues (Panels B & D) compared to controls (Panels A & C).
Klf5 deletion from the mouse colon results in an increase in regeneration-associated gene products and a concomitant decrease in proliferation-associated products
KLF5 regulates several important regulatory pathways including proliferation, differentiation and apoptosis (McConnell and Yang, 2010). Recently, we reported that constitutive intestinespecific deletion of Klf5 leads to down-regulation of several proteins including β-catenin, cyclin D1, dishevelled 2, Wnt 2, Wnt 4 and Sox17 (McConnell et al., 2011b). In contrast, Sry (sex determining region of Y)-box 9 (Sox9) protein was found to be up regulated (up to 4-fold increase) upon Klf5 deletion.
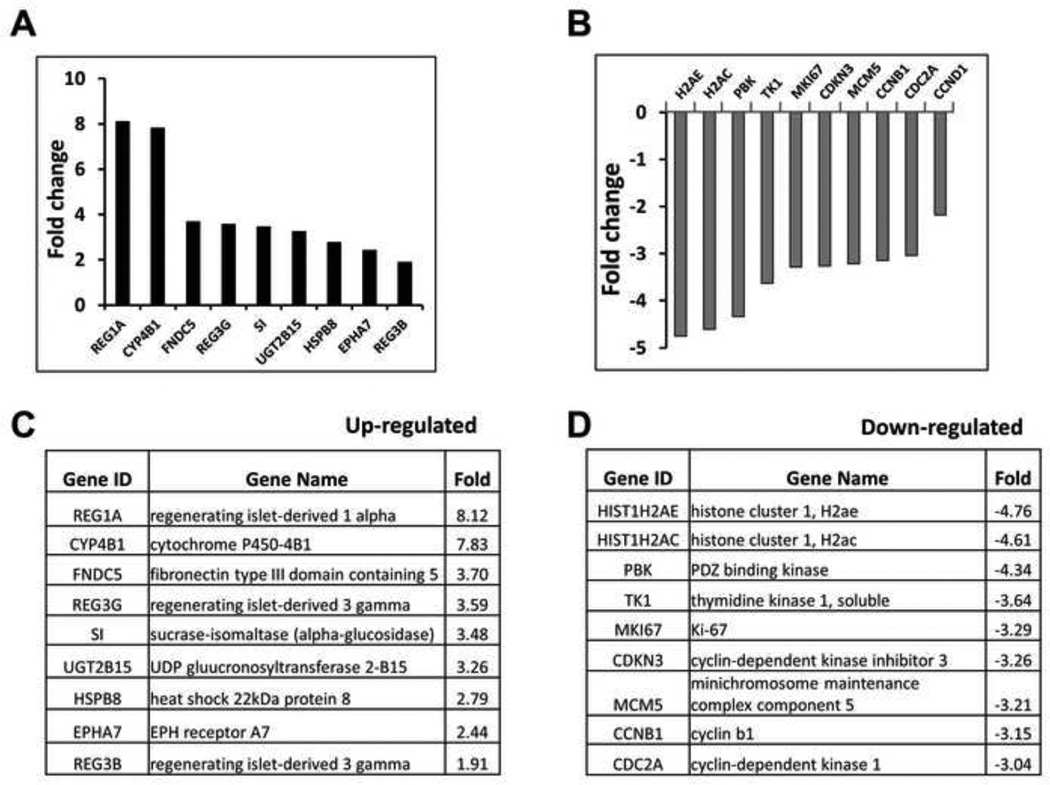
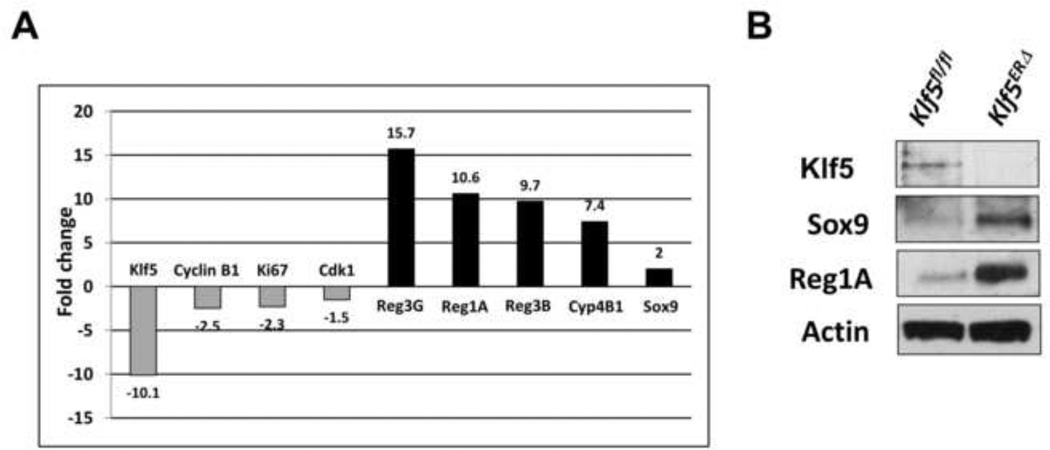
To determine the effect of Klf5 deletion in adult mouse colon, we performed gene microarray analysis of epithelial cells extracted from day 3 tamoxifen-treated Klf5ERΔ and Klf5fl/fl colons. Some of the most highly expressed transcripts upon deletion of Klf5 were regeneration-associated genes including Reg1A, Reg3G and Reg3B (Figs. 5A & C). Among genes that were decreased upon Klf5 deletion were common targets of KLF5 including Ki-67, cyclin D1, cyclin B and Cdc2 (Figs. 5B & D). We confirmed the results of the microarray experiment by performing quantitative PCR reactions with primers for selected genes (Fig. 6A). We observed that expression of Reg1A, Reg3G, Reg3B and Sox9 was increased upon Klf5 deletion while that of cyclin B1, Ki-67 and Cdk1 was decreased (Fig. 6A). We also extracted protein from day 3 tamoxifen-treated Klf5ERΔ and Klf5fl/fl mouse colons and performed immunoblots against Klf5, Sox9, Reg1A and actin (as a loading control). As expected, Klf5 was absent from the colon of Klf5ERΔ mice upon tamoxifen treatment (Fig. 6B). Consistent with the microarray and quantitative PCR analyses, the levels of Sox9 and Reg1A proteins were higher in the colon of Klf5ERΔ mice when compared to control (Fig. 6B).
Figure 5. Microarray analyses from Klf5ERΔ and Klf5fl/fl tissues reveal up-regulation of several regeneration associated genes and down-regulation of known Klf5 targets.
Panels A-D represent microarray data from day 3 tamoxifen-treated Klf5ERΔ colonic tissues normalized to control Klf5fl/fl tissues. RNA extracted from mice was analyzed using an Illumina Mouse Array to specify gene expression patterns. Panel A and C display genes that were highly up-regulated in Klf5ERΔ colonic tissues when compared to controls. Presence of several regeneration associated (Reg) genes were noted. Panels B and D display genes that were significantly down-regulated in Klf5ERΔ colonic tissues when compared to controls. We observed many previously known targets of Klf5 to be down-regulated.
Figure 6. Quantitative PCR and Western blot analyses confirm the up/down-regulation of microarray-selected Klf5 targets.
Quantitative PCR was performed on day 3 tamoxifen-treated RNAs as discussed in Methods. The results contained in Panel A show that Klf5, Ki-67, cyclin B1 & Cdk1 were several folds down-regulated when compared to the controls, while Reg3G, Reg1A, Reg3B, Cyp4B1 and Sox9 were up-regulated. Panel B represents immunoblot analyses of Klf5, Sox9, Reg1A and actin. Lane 1 represents control Klf5fl/fl colon lysates and lane 2 represents Klf5ERΔ samples. Actin was used as a loading control.
KLF5 regulates expression of Sox9 and Reg genes
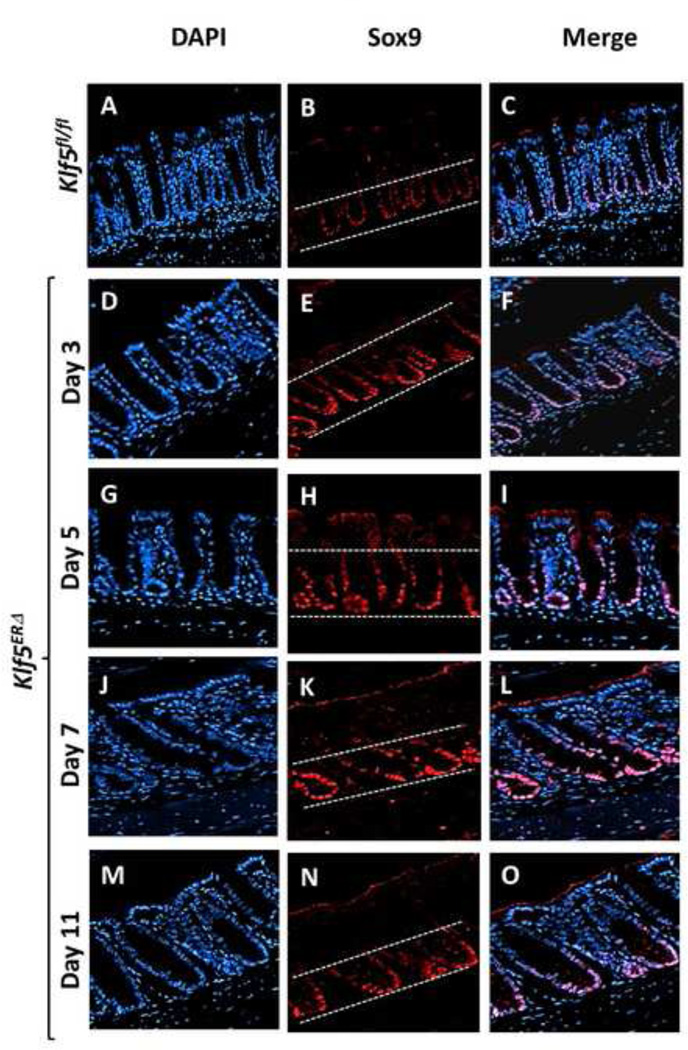
Based on the results of previous (McConnell et al., 2011b) and current studies, we consistently found an increase in Sox9 transcript and protein levels upon deletion of Klf5 from the colonic epithelium. We therefore performed immunofluorescence staining to confirm this increase in Sox9 levels. We observed that in day 5 tamoxifen-treated control Klf5fl/fl colons, Sox9 staining was restricted to the bottom proliferative compartment of the colonic epithelium (Fig. 7B & Suppl. Fig. 6A). During the immediate period following deletion of Klf5 (days 3 and 5), the Sox9-positive zone was considerably expanded in tamoxifen-treated Klf5ERΔ mice (Figs. 7E & H and Suppl. Figs. 6B, 6C & 7). We also observed a quantitative increase in the number of epithelial cells staining positive for Sox9 in the Klf5ERΔ mouse colons in this period when compared to controls (Suppl. Fig. 7). A similar Sox9 staining pattern was observed at day 7 after start of tamoxifen treatment (Fig. 7E & Suppl. Figs. 6D & 7). Subsequently, by day 11 the Sox9 staining returned to a pattern similar to control mice (Figs. 7N & B and Suppl. Figs. 6E, 6A & 7). Of note, results of our previous study also showed a similar expansion of the Sox9 positive zone upon constitutive intestinespecific Klf5 deletion (McConnell et al., 2011b).
Figure 7. Temporal enlargement of Sox9 expression zone in mouse colons following induced deletion of Klf5.
Images are stained with DAPI (blue), Sox9 (red) and merged (DAPI + Sox9). Panels A, B and C show colonic tissues from day 5 tamoxifen-treated control Klf5fl/fl and Panels D through O, those from Klf5ERΔ mice. Panels D-F, G-I, J-L and M-O are representative of 3, 5, 7, and 11 day post-tamoxifen treatment respectively. White dashed lines (Panels B, E, H, K and N) mark the zone of epithelial cells staining positive for Sox9. Expansion of Sox9-positive zone was observed in Panels E, H & K compared to control (Panel B). Restoration of Sox9 expansion to the bottom of crypts was observed in Panel N similar to control (Panel B).
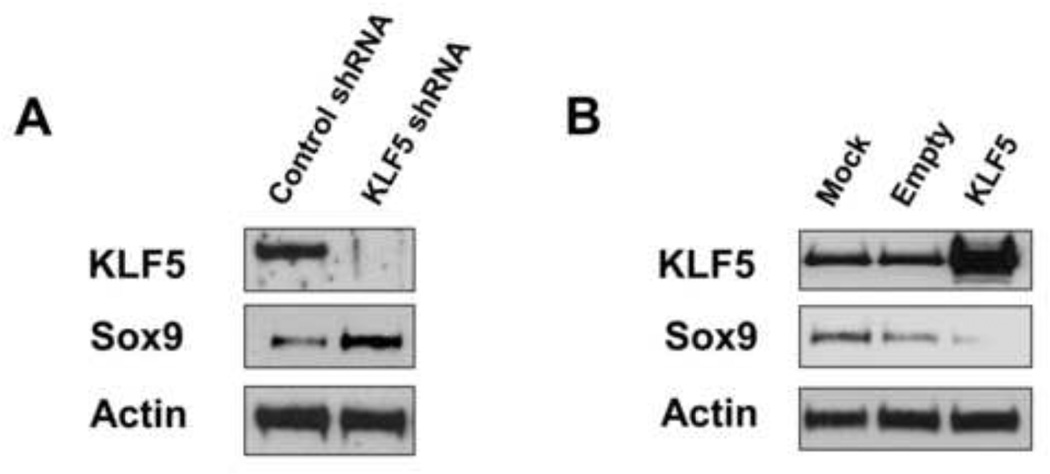
To examine the mechanism by which KLF5 modulates Sox9 expression, we performed in vitro experiments in two colon cancer cell lines, T84 and HCT116, in which KLF5 was knocked down or over-expressed, respectively. Lentiviral-mediated short-hairpin RNA-mediated knockdown of endogenous KLF5 expression in T84 cells resulted in increased Sox9 protein levels when compared to control (Fig. 8A). Conversely, exogenous expression of KLF5 in HCT116 cells resulted in a reduction of Sox9 expression levels when compared to both the empty vector and mock-treated controls (Fig. 8B). Furthermore, we performed chromatin immunoprecipitation (ChIP) experiments in HCT116 cells with expression of exogenous KLF5 or Sox9 (empty vector used as control). Both KLF5 and Sox9 directly bound to Sox9 and Reg1A promoters (Fig. 9). In contrast, KLF5, but not Sox9, bound to the cyclin D1 promoter (Fig. 9) similar to previous observations (Nandan et al., 2004). We confirmed the negative regulation of Sox9 promoter activity by KLF5 by performing luciferase reporter assay in DLD1 colon cancer cells (Suppl. Fig. 9). Sox9 promoter activity and expression was significantly increased by KLF5 knockdown when compared to controls. Reg1A promoter did not show significant activity with KLF5 reduction (Suppl. Fig. 9A).
Figure 8. Modulation of KLF5 expression regulates Sox9 expression in colorectal cancer cell lines.
T84 and HCT116 colon cancer cell lines were used in Panels A and B, respectively. T84 cells were transduced with lentiviruses containing either short hairpin RNA (shRNA) sequence against KLF5 or control shRNA. The lysates were collected from stable T84 knockdown or control cell lines and immunoblots performed for KLF5, Sox9 and actin (loading control) (Panel A). Lane 1 represents lysates from control shRNA transduction and lane 2 from KLF5-specific shRNA transduction. In Panel B, HCT116 cells were either mock-transfected (Lane 1), transfected with empty vector (Lane 2) or with pCI-Neo-KLF5 (Lane 3). Sox9 expression is inversely correlated to KLF5 expression in both T84 and HCT116 experiments (Panels A and B).
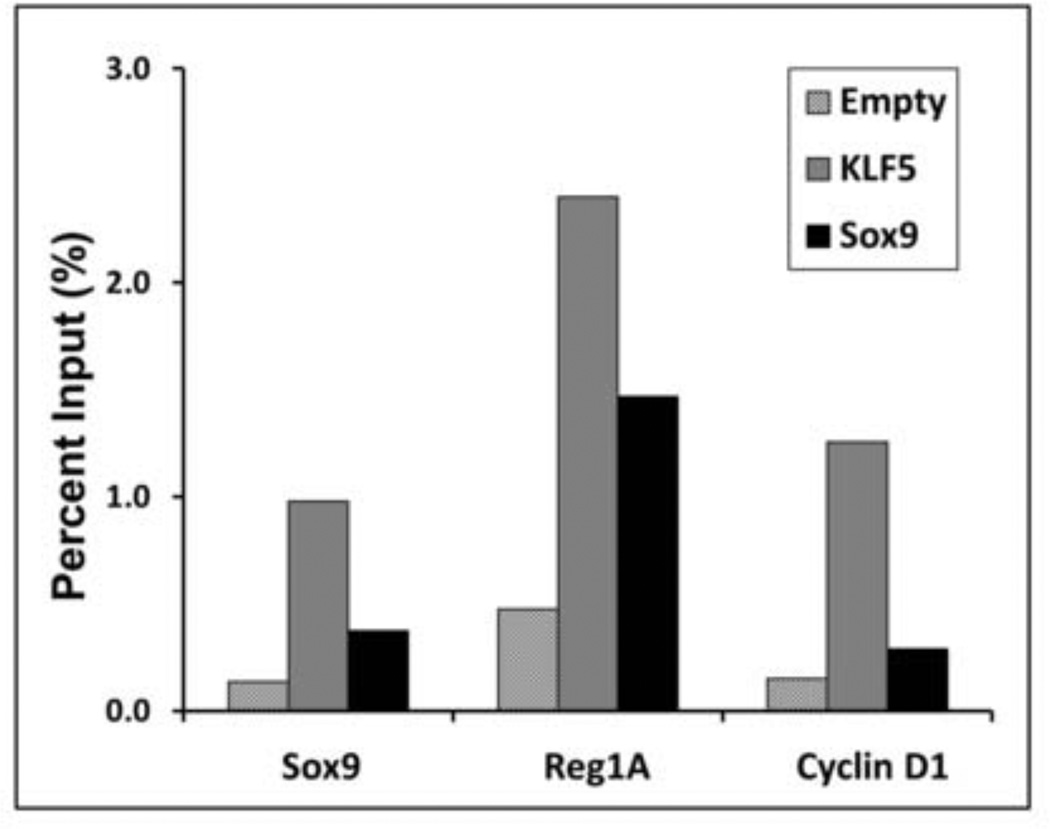
Figure 9. Chromatin Immunoprecipitation (ChIP) assay show strong binding of KLF5 to Sox9, Reg1A and Cyclin D1 promoters.
Chromatin immunoprecipitation (ChIP) assays were performed as per Methods. HCT116 cells were transfected with KLF5, Sox9 or empty vector before collecting lysates for immunoprecipitations (IP) with Sox9, Reg1Aand cyclin D1 antibodies. DNA purified from the IP samples were then analysed with quantitative PCR using primers against Sox9, Reg1A and cyclin D1 respectively. Promoter binding affinity was plotted in the Y-Axis as Percent Input (%) against different promoter sequences in the X-axis. These results indicate that KLF5 strongly binds to Sox9, Reg1A and cyclin D1 promoters. Sox9 displays strong binding to Reg1A promoter and weak interactions with cyclin D1 and its own promoter.
DISCUSSION
In our previous study we generated mice with intestine-specific deletion of Klf5 regulated by Villin gene promoter (McConnell et al., 2011b). One major impediment in that study was the death of about 80% of neonates with complete Klf5 deletion, with the remaining 20% surviving due to incomplete deletion (McConnell et al., 2011b). Our current study describes the phenotype of inducible intestine-specific deletion of Klf5 in 8-week old adult mice. We had hypothesized that Klf5 loss in adult mice would lead to severe inflammation and epithelial loss followed by morbidity or mortality. Contrary to our expectations, mice with induced deletion of Klf5 have no grossly untoward effect up to 11 days following the start of tamoxifen treatment. However, the most noticeable effect of Klf5 deletion occurred in the days during and immediately following tamoxifen treatment, days 3 and 5 (Figs. 1B & C). These changes included acute focal inflammation, loss of proliferation and emergence of a regenerative phenotype. By day 11 following start of tamoxifen treatment, the effects of Klf5 loss were significantly reduced. These suggest that in spite of dramatic changes in the mucosal epithelia due to loss of Klf5, certain intrinsic processes retain the ability to repair and rebuild the epithelium.
KLF5 has a well-established pro-proliferative role in the intestinal epithelium (Ghaleb et al., 2005; McConnell et al., 2007; McConnell and Yang, 2010). As we had previously described, induced Klf5 deletion was accompanied by a reduction in the number of proliferative cells in the colonic mucosa, as manifested by the loss of BrdU and Ki-67 staining (Fig. 3 & Suppl. Fig. 5). This effect is most apparent during the immediate phase of Klf5 deletion (days 3 and 5). However, by day 11 after the start of tamoxifen treatment, Ki67 staining has returned to a pattern that is similar to control (Fig. 3N). This is also accompanied by the relative restoration of the epithelial architecture at this time. We predict that the early loss of proliferative cells and their later appearance allude to multiple levels of proliferative control. Immediate Klf5 loss could entail an initial proliferative loss but other pathways, some compensatory in nature, could contribute to reappearance of proliferative cells.
We did not observe any significant changes in survival or weight in adult mice upon loss of Klf5 (Suppl. Fig. 2), suggesting that Klf5 is dispensable during adulthood but not in embryonic development (McConnell and Yang, 2010). A recent study by Bell et al. (Bell et al., 2013) observed that the embryonic intestinal-specific deletion of Klf5 resulted in severe disruption in intestinal villus morphogenesis but without any significant loss in proliferation. The authors also observed loss of goblet cells and enteroendocrine cells signifying a role for Klf5 in early embryonic intestinal development and differentiation (Bell et al., 2013)
Our results indicate that the loss of Klf5 resulted in a decrease in secretory lineage, goblet cells in the colon and Paneth cells in the small bowel (Fig. 4). This correlated with the expansion of Sox9 expression zone in the colon (Fig. 7 & Suppl. Figs. 6 & 7). It is interesting that previously, to the contrary, Sox9 inactivation in the intestine has been reported to have deleterious effects on Paneth cells and reduce goblet cells (Bastide et al., 2007). Our results suggest that Klf5 could directly control secretory lineage differentiation and/or localization in lieu of Sox9 expression.
The established link between loss of epithelial integrity and Klf5 deletion influenced our decision to perform microarray analysis of samples at day 3 tamoxifen treatment. Our analysis revealed that immediately upon Klf5 loss several regeneration-associated genes (Reg1A, Reg3G & Reg3B) were up-regulated (Figs. 5A & C). The genes that were down-regulated included several known targets of KLF5 (Ki-67, cyclin B1, Cdc2 & cyclin D1) (Figs 5B & D). Quantitative PCR performed at the same time point supported the evidence obtained from microarray analysis (Fig. 6A). These observations indicate that deletion of Klf5 results in changes in the profiles of gene expression pro in at least two major programs, proliferation and regeneration, which could either be concurrently or sequentially connected to each other. Upon immediate loss of Klf5, we found an increase in Sox9 and Reg1 transcript & protein (Fig. 6A & B). We propose that the induction of the Sox9 and Reg genes immediately following the loss of Klf5 denotes the start of a regenerative phase.
We confirmed the in vitro regulation of Sox9 by KLF5 both upon lentiviral-mediated knock-down of KLF5 in T84 colorectal cancer cells and over-expression in HCT116 colorectal cancer cells (Fig. 8) and through luciferase reporter assays (Suppl. Fig. 9). ChIP assay in HCT116 cells also demonstrated direct binding of KLF5 to both Sox9 and Reg1A promoters and binding of Sox9 to the Reg1A promoter (Fig. 9). Thus, KLF5 could putatively control Reg1A expression either directly or through Sox9 in colon epithelial cells both in vitro and in vivo.
We now have evidence that connects loss of Klf5 with a regenerative phenotype and induction of several regeneration-associated genes. However the absence of catastrophic epithelial destruction in the Klf5-deleted adult mice in contrast to constitutive Klf5 deletion (McConnell et al., 2011b) could have two potential explanations. One is a putative redundancy in Klf5’s role in governing cell proliferation in adult mouse intestinal epithelium. Regeneration-associated genes could compensate for the loss of Klf5 and facilitate downstream proliferative processes. Another potential explanation is that there could be an incomplete deletion of Klf5 in intestinal crypt stem cells in the Villin-Cre-ERT2 mouse model. We have observed Klf5 expression in the Lgr5-positive intestinal stem cell compartment (unpublished results) suggesting that the damaged intestinal epithelium could recover from stem cells based at the bottom of colonic crypts that may not have lost Klf5.
Previous studies have reported a protective role for Klf5 by promoting epithelial repair in DSS-mediated intestinal inflammation (McConnell et al., 2011a; Tetreault et al., 2012). Based on the current study, Klf5 deletion in adult mouse intestines is better tolerated than its deletion in embryonic tissues. Our results show that inducible Klf5 loss initiates epithelial damage and disruption combined with decrease in proliferation in the colonic epithelium. Furthermore, we have demonstrated that Klf5 loss also activates various regeneration-associated genes, including Sox9. The presence of a regenerative intestinal morphology preceded by epithelial disruption suggests the importance for Klf5 in maintaining homeostasis. The precise role of KLF5 in the regenerative response controlled by Sox9 and the family of Reg genes remains to be determined.
Supplementary Material
HIGHLIGHTS.
Mice with inducible, intestine-specific deletion of Klf5 are viable.
The colonic epithelium of mutant mice shows signs of epithelial distress soon after Klf5 deletion.
Later, colonic epithelium is restored by a regenerative response involving Sox9.
Several regeneration-associated genes (Reg) also contribute to epithelial repair.
Klf5 binds to the Sox9 and Reg1A promoters and modulates Sox9 promoter activity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Mandayam O. Nandan, Email: Mandayam.Nandan@stonybrookmedicine.edu.
Amr M. Ghaleb, Email: Amr.Ghaleb@stonybrookmedicine.edu.
Yang Liu, Email: yangliu5@ic.sunysb.edu.
Agnieszka B. Bialkowska, Email: Agnieszka.Bialkowska@stonybrookmedicine.edu.
Beth B. McConnell, Email: Beth.McConnell@data2impact.com.
Kenneth R. Shroyer, Email: Kenneth.Shroyer@stonybrookmedicine.edu.
Sylvie Robine, Email: Sylvie.Robine@curie.fr.
Vincent W. Yang, Email: Vincent.Yang@stonybrookmedicine.edu.
REFERENCES
- Abe M, Nata K, Akiyama T, Shervani NJ, Kobayashi S, Tomioka-Kumagai T, Ito S, Takasawa S, Okamoto H. Identification of a novel Reg family gene, Reg IIIdelta, and mapping of all three types of Reg family gene in a 75 kilobase mouse genomic region. Gene. 2000;246:111–122. doi: 10.1016/s0378-1119(00)00059-7. [DOI] [PubMed] [Google Scholar]
- Bartoli C, Baeza N, Figarella C, Pellegrini I, Figarella-Branger D. Expression of peptide-23/pancreatitis-associated protein and Reg genes in human pituitary and adenomas: comparison with other fetal and adult human tissues. The Journal of clinical endocrinology and metabolism. 1998;83:4041–4046. doi: 10.1210/jcem.83.11.5217. [DOI] [PubMed] [Google Scholar]
- Bastide P, Darido C, Pannequin J, Kist R, Robine S, Marty-Double C, Bibeau F, Scherer G, Joubert D, Hollande F, Blache P, Jay P. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. The Journal of cell biology. 2007;178:635–648. doi: 10.1083/jcb.200704152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialkowska AB, Crisp M, Bannister T, He Y, Chowdhury S, Schurer S, Chase P, Spicer T, Madoux F, Tian C, Hodder P, Zaharevitz D, Yang VW. Identification of small-molecule inhibitors of the colorectal cancer oncogene Kruppel-like factor 5 expression by ultrahigh-throughput screening. Molecular cancer therapeutics. 2011;10:2043–2051. doi: 10.1158/1535-7163.MCT-11-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blache P, van de Wetering M, Duluc I, Domon C, Berta P, Freund JN, Clevers H, Jay P. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. The Journal of cell biology. 2004;166:37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Regulation of Krupple-like factor 5 by targeted protein degradation. Methods Mol Biol. 2010;647:267–277. doi: 10.1007/978-1-60761-738-9_16. [DOI] [PubMed] [Google Scholar]
- Dang DT, Pevsner J, Yang VW. The biology of the mammalian Kruppel-like family of transcription factors. The international journal of biochemistry & cell biology. 2000;32:1103–1121. doi: 10.1016/s1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Caro A, Lohse J, Sarles H. Characterization of a protein isolated from pancreatic calculi of men suffering from chronic calcifying pancreatitis. Biochemical and biophysical research communications. 1979;87:1176–1182. doi: 10.1016/s0006-291x(79)80031-5. [DOI] [PubMed] [Google Scholar]
- el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- Formeister EJ, Sionas AL, Lorance DK, Barkley CL, Lee GH, Magness ST. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. American journal of physiology. Gastrointestinal and liver physiology. 2009;296:G1108–G1118. doi: 10.1152/ajpgi.00004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, Masui T, Kawaguchi M, Takaori K, Doi R, Nishi E, Kakinoki R, Deng JM, Behringer RR, Nakamura T, Uemoto S. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nature genetics. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell research. 2005;15:92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracz AD, Magness ST. Sry-box (Sox) transcription factors in gastrointestinal physiology and disease. American journal of physiology. Gastrointestinal and liver physiology. 2011;300:G503–G515. doi: 10.1152/ajpgi.00489.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf R, Schiesser M, Lussi A, Went P, Scheele GA, Bimmler D. Coordinate regulation of secretory stress proteins (PSP/reg, PAP I, PAP II, and PAP III) in the rat exocrine pancreas during experimental acute pancreatitis. The Journal of surgical research. 2002;105:136–144. doi: 10.1006/jsre.2002.6387. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Akiyama T, Nata K, Abe M, Tajima M, Shervani NJ, Unno M, Matsuno S, Sasaki H, Takasawa S, Okamoto H. Identification of a receptor for reg (regenerating gene) protein, a pancreatic beta-cell regeneration factor. The Journal of biological chemistry. 2000;275:10723–10726. doi: 10.1074/jbc.275.15.10723. [DOI] [PubMed] [Google Scholar]
- Kutner RH, Zhang XY, Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nature protocols. 2009;4:495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- McConnell BB, Bialkowska AB, Nandan MO, Ghaleb AM, Gordon FJ, Yang VW. Haploinsufficiency of Kruppel-like factor 5 rescues the tumor-initiating effect of the Apc(Min) mutation in the intestine. Cancer research. 2009;69:4125–4133. doi: 10.1158/0008-5472.CAN-08-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BB, Ghaleb AM, Nandan MO, Yang VW. The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. BioEssays : news and reviews in molecular, cellular and developmental biology. 2007;29:549–557. doi: 10.1002/bies.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BB, Kim SS, Bialkowska AB, Yu K, Sitaraman SV, Yang VW. Kruppel-like factor 5 protects against dextran sulfate sodium-induced colonic injury in mice by promoting epithelial repair. Gastroenterology. 2011a;140:540–549. doi: 10.1053/j.gastro.2010.10.061. e542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BB, Kim SS, Yu K, Ghaleb AM, Takeda N, Manabe I, Nusrat A, Nagai R, Yang VW. Kruppel-like factor 5 is important for maintenance of crypt architecture and barrier function in mouse intestine. Gastroenterology. 2011b;141:1302–1313. doi: 10.1053/j.gastro.2011.06.086. 1313 e1301–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiological reviews. 2010;90:1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandan MO, Chanchevalap S, Dalton WB, Yang VW. Kruppel-like factor 5 promotes mitosis by activating the cyclin B1/Cdc2 complex during oncogenic Ras-mediated transformation. FEBS letters. 2005;579:4757–4762. doi: 10.1016/j.febslet.2005.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandan MO, Ghaleb AM, McConnell BB, Patel NV, Robine S, Yang VW. Kruppel-like factor 5 is a crucial mediator of intestinal tumorigenesis in mice harboring combined ApcMin and KRASV12 mutations. Molecular cancer. 2010;9:63. doi: 10.1186/1476-4598-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandan MO, McConnell BB, Ghaleb AM, Bialkowska AB, Sheng H, Shao J, Babbin BA, Robine S, Yang VW. Kruppel-like factor 5 mediates cellular transformation during oncogenic KRAS-induced intestinal tumorigenesis. Gastroenterology. 2008;134:120–130. doi: 10.1053/j.gastro.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandan MO, Yang VW. The role of Kruppel-like factors in the reprogramming of somatic cells to induced pluripotent stem cells. Histology and histopathology. 2009;24:1343–1355. doi: 10.14670/hh-24.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandan MO, Yoon HS, Zhao W, Ouko LA, Chanchevalap S, Yang VW. Kruppel-like factor 5 mediates the transforming activity of oncogenic H-Ras. Oncogene. 2004;23:3404–3413. doi: 10.1038/sj.onc.1207397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H. The Reg gene family and Reg proteins: with special attention to the regeneration of pancreatic beta-cells. Journal of hepato-biliary-pancreatic surgery. 1999;6:254–262. doi: 10.1007/s005340050115. [DOI] [PubMed] [Google Scholar]
- Parikh A, Stephan AF, Tzanakakis ES. Regenerating proteins and their expression, regulation and signaling. Biomolecular concepts. 2012;3:57–70. doi: 10.1515/bmc.2011.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechreche H, Montalto G, Mallo GV, Vasseur S, Marasa L, Soubeyran P, Dagorn JC, Iovanna JL. pap, reg Ialpha and reg Ibeta mRNAs are concomitantly up-regulated during human colorectal carcinogenesis. International journal of cancer. Journal international du cancer. 1999;81:688–694. doi: 10.1002/(sici)1097-0215(19990531)81:5<688::aid-ijc3>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Sarles H, Dagorn JC, Giorgi D, Bernard JP. Renaming pancreatic stone protein as 'lithostathine'. Gastroenterology. 1990;99:900–901. doi: 10.1016/0016-5085(90)90999-h. [DOI] [PubMed] [Google Scholar]
- Sellak H, Wu S, Lincoln TM. KLF4 and SOX9 transcription factors antagonize beta-catenin and inhibit TCF-activity in cancer cells. Biochimica et biophysica acta. 2012;1823:1666–1675. doi: 10.1016/j.bbamcr.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SR, Liu C, Balgley BM, Lee C, Taylor CR. Protein extraction from formalin-fixed, paraffin-embedded tissue sections: quality evaluation by mass spectrometry. Journal of Histochemistry and Cytochemistry. 2006;54:739–43. doi: 10.1369/jhc.5B6851.2006. [DOI] [PubMed] [Google Scholar]
- Shindo T, Manabe I, Fukushima Y, Tobe K, Aizawa K, Miyamoto S, Kawai-Kowase K, Moriyama N, Imai Y, Kawakami H, Nishimatsu H, Ishikawa T, Suzuki T, Morita H, Maemura K, Sata M, Hirata Y, Komukai M, Kagechika H, Kadowaki T, Kurabayashi M, Nagai R. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nature medicine. 2002;8:856–863. doi: 10.1038/nm738. [DOI] [PubMed] [Google Scholar]
- Takeda N, Manabe I, Uchino Y, Eguchi K, Matsumoto S, Nishimura S, Shindo T, Sano M, Otsu K, Snider P, Conway SJ, Nagai R. Cardiac fibroblasts are essential for the adaptive response of the murine heart to pressure overload. The Journal of clinical investigation. 2010;120:254–265. doi: 10.1172/JCI40295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetreault MP, Alrabaa R, McGeehan M, Katz JP. Kruppel-like factor 5 protects against murine colitis and activates JAK-STAT signaling in vivo. PloS one. 2012;7:e38338. doi: 10.1371/journal.pone.0038338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno M, Nata K, Noguchi N, Narushima Y, Akiyama T, Ikeda T, Nakagawa K, Takasawa S, Okamoto H. Production and characterization of Reg knockout mice: reduced proliferation of pancreatic beta-cells in Reg knockout mice. Diabetes. 2002;51(Suppl 3):S478–S483. doi: 10.2337/diabetes.51.2007.s478. [DOI] [PubMed] [Google Scholar]
- van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annual review of physiology. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- Yeung TM, Chia LA, Kosinski CM, Kuo CJ. Regulation of self-renewal and differentiation by the intestinal stem cell niche. Cellular and molecular life sciences : CMLS. 2011;68:2513–2523. doi: 10.1007/s00018-011-0687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Dalton WB, Yang VW. CDK1 regulates mediator of DNA damage checkpoint 1 during mitotic DNA damage. Cancer research. 2012;72:5448–5453. doi: 10.1158/0008-5472.CAN-12-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenilman ME, Kim S, Levine BA, Lee C, Steinberg JJ. Ectopic expression of reg protein: A marker of colorectal mucosa at risk for neoplasia. Journal of gastrointestinal surgery : official journal of the Society for Surgery of the Alimentary Tract. 1997;1:194–201. doi: 10.1016/s1091-255x(97)80109-6. discussion 201-192. [DOI] [PubMed] [Google Scholar]
- Ziemer LT, Pennica D, Levine AJ. Identification of a mouse homolog of the human BTEB2 transcription factor as a beta-catenin-independent Wnt-1-responsive gene. Molecular and cellular biology. 2001;21:562–574. doi: 10.1128/MCB.21.2.562-574.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.