Catalytic antibody variants with κ and λ light-chain constant domains show differences in their crystal structures which lead to subtle changes in catalytic efficiency and thermodynamic parameters as well as in their affinity for peptide substrates.
Keywords: A17 reactibody, antibodies
Abstract
The engineering of catalytic function in antibodies requires precise information on their structure. Here, results are presented that show how the antibody domain structure affects its functionality. The previously designed organophosphate-metabolizing reactibody A17 has been re-engineered by replacing its constant κ light chain by the λ chain (A17λ), and the X-ray structure of A17λ has been determined at 1.95 Å resolution. It was found that compared with A17κ the active centre of A17λ is displaced, stabilized and made more rigid owing to interdomain interactions involving the CDR loops from the VL and VH domains. These VL/VH domains also have lower mobility, as deduced from the atomic displacement parameters of the crystal structure. The antibody elbow angle is decreased to 126° compared with 138° in A17κ. These structural differences account for the subtle changes in catalytic efficiency and thermodynamic parameters determined with two organophosphate ligands, as well as in the affinity for peptide substrates selected from a combinatorial cyclic peptide library, between the A17κ and A17λ variants. The data presented will be of interest and relevance to researchers dealing with the design of antibodies with tailor-made functions.
1. Introduction
Exquisite specificity and high binding affinity, the hallmarks of the antibody (Ab) response, make Abs excellent tools for biotechnology and biomedical applications. During the past decade, achievements in the field of Ab engineering have markedly expanded the range of these applications. A number of engineering strategies have been applied to modify the functionality of therapeutic monoclonal Abs according to the requirements of the particular biological mechanism to be treated (Kaneko & Niwa, 2011 ▶; Klohn et al., 2013 ▶; Lu et al., 2012 ▶; Vincent & Zurini, 2012 ▶). The development of methods for the humanization and functional expression of Abs and their fragments, together with the emergence of powerful techniques for screening combinatorial libraries and the expansion of structure–function databases aided by refined X-ray analysis, has opened unlimited opportunities for the engineering of Abs with tailor-made properties for specific applications. X-ray crystal structure analysis has played a crucial role in creating novel artificial biocatalysts and engineered Abs (Ekiert et al., 2012 ▶; Golinelli-Pimpaneau et al., 2000 ▶; Guenaga & Wyatt, 2012 ▶; Privett et al., 2012 ▶; Turner et al., 2002 ▶; Zheng et al., 2004 ▶).
Antibody recognition of protein antigens is predominantly mediated by four to six complementarity-determining regions (CDRs), which are variable loops at the tip of each antigen-binding fragment (Fab). Fabs are composed of two polypeptide chains: heavy (H) and light (L). Each chain is folded into two distinct immunoglobulin (Ig) domains, the N-terminal variable domain (VH or VL) and the C-terminal constant domain (CH1 or CL), with the amino-acid residues linking VL to CL and VH to CH1 called the switch residues (J fragment, framework 4). The elbow angle, or elbow bend, defined as the angle between the pseudo-twofold axes relating VL to VH and CL to CH1, is a highly variable parameter in antibodies, and its role in the antigen-binding capacity has been speculated on (Huber et al., 1976 ▶; Landolfi et al., 2001 ▶). It is still unknown how far the changes that a hapten induces in an Ab structure can extend. An analysis of Protein Data Bank (PDB) depositions has shown that in some cases significant differences exist between the elbow angles of liganded and unliganded Fabs (Stanfield, Zemla et al., 2006 ▶; Stanfield, Gorny et al., 2006 ▶). Dramatic changes in the domain structure of NC6.8 (an Ab directed against the compound NC174) caused by small ligand binding were revealed by X-ray structure analysis followed by various molecular-dynamics simulations (Guddat et al., 1994 ▶, 1995 ▶; Sotriffer et al., 2000 ▶). In particular, the elbow angle was shown to change by more than 30°. Mammalian Abs have two types of light chains, κ and λ, which are encoded by different chromosome loci. In general, λ-chain Abs have a less rigid conformation than κ-chain Abs, with the difference being reflected in the values of the elbow angle: about 195° in the former versus 125° in the latter (Stanfield, Zemla et al., 2006 ▶). The apparent hyperflexibility of λ-chain Fabs may be owing to an insertion in their switch region, which usually consists of a glycine residue and hence can also provide more conformational freedom for their molecules.
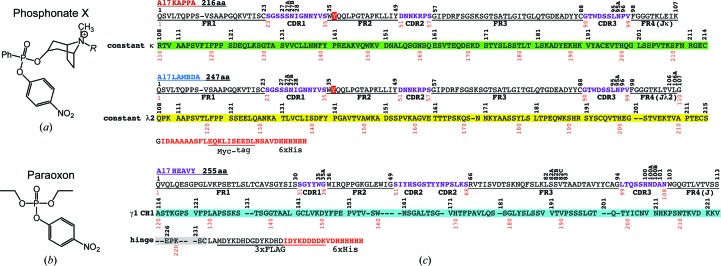
Catalytic function is one of the most sophisticated features of Abs. Recently, the novel ‘reactibody’ approach has been developed, which is based on the chemical selection of biocatalysts from a human semisynthetic Ab variable-fragment library followed by eukaryotic expression in a full-length Ab. This approach has been used to produce a novel organophosphate-metabolizing biocatalyst named the A17 reactibody (Reshetnyak et al., 2007 ▶; Smirnov et al., 2011 ▶). An important task is to develop effective antidotes against very toxic organophosphate compounds, including nerve agents and pesticides. Organophosphate poisoning is a serious clinical problem in rural regions of the developing world, causing the deaths of 200 000 people per year (Eddleston et al., 2008 ▶). It has been shown that the A17 reactibody is capable of irreversibly binding phosphonate X (Fig. 1 ▶ a) and hydrolyzing the organophosphate pesticide paraoxon (Fig. 1 ▶ b) by covalent catalysis with rate-limiting dephosphorylation. The crystal structures of unmodified and phosphonylated Fabs of A17 have previously been solved at 1.5 and 1.36 Å resolution, respectively. Structural analysis combined with kinetic studies has provided an insight into certain mechanistic features of the reaction catalyzed by A17 (Smirnov et al., 2011 ▶). In particular, the catalytic Tyr-L37 in this reactibody proved to be located in a deep active-site cavity, which is not typical of Abs in general. Some differences were observed in the catalytic efficiency of full-length A17 or its Fab fragment compared with its parent single-chain variable-fragment (scFv) molecule, which were possibly owing to structural stabilization of the active centre by additional constant domains. The full-length A17 reactibody contained an artificial light chain in which the λVL domain was fused to κCL through the κ switch region (Fig. 1 ▶ c), which possibly affected the functionality of the Ab. In the case of catalytic Abs, in which the active centre should have a rigid and precise structure to accomplish the catalytic function, cross-domain interaction effects can be extremely important. It has been shown that a change of the heavy-chain isotype (Sapparapu et al., 2012 ▶) or Ab expression in the scFv format (Ponomarenko et al., 2007 ▶) affects the catalytic activity or substrate specificity of Abs. It was reasonable to assume that the κ→λ switch would change the architecture of the binding pocket of the Ab molecule. To study the effects of such a switch, we produced a functionally active catalytic Ab with the constant λ light chain, thereby reconstructing the natural Ab structure.
Figure 1.
Chemical compounds and antibody chains used in this study. (a) R = H, p-nitrophenyl 8-methyl-8-azabicyclo[3.2.1]octylphenylphosphonate (phosphonate X); R = biotin, biotinylated phosphonate X (BtX). (b) O,O-Diethyl O-(4-nitrophenyl)phosphate (paraoxon). (c) Amino-acid sequences of A17 κ and λ light chains and the heavy chain. Residues are numbered using the Kabat system (superlinear) and sequential numbering (interlinear). Frameworks (FR1–FR4) are underlined; switch residues of the J segment are designated as FR4 according to Kabat; CDRs are coloured magenta; constant domains κCL, λCL and CH1 are coloured green, yellow and blue, respectively.
In this study, we present data on the effect of a κ→λ light-chain switch on the structure and function of the A17 reactibody. The available high-resolution structural data allowed us to trace the changes caused by this switch in the active centre and antigen-binding site, and to observe how the interdomain interactions of the CDR loops narrow the cavity entrance, thereby forming a more rigid structure. A comparative structural analysis of the A17λ and A17κ variants showed that the κ→λ switch results in a decrease in the Ab elbow angle, in contrast to previously published data. It was somewhat unexpected that A17λ proved to have a rigid structure compared with A17κ, with its catalytic activity and affinity changing only slightly despite major structural modifications. Hopefully, the data presented below will contribute to the advancement of research on the design of antibodies with tailor-made functions.
2. Materials and methods
2.1. Protein expression and purification
Recombinant FabA17 containing κ or λ light-chain constant regions was produced in the methylotrophic yeast Pichia pastoris GS115 using the modified expression vector pPICZαA/Jk1 (Zakharov et al., 2011 ▶) based on pPICZαA (Invitrogen, USA) (Supplementary Fig. S11).
Expression constructs for HCH (human Ab heavy chain containing the CH1 constant domain with hinge region) and A17 κ light chain (Zakharov et al., 2011 ▶) and the constant region of human λ2 light chain with the J2 (joining) segment (Gabibov et al., 2011 ▶) were prepared as described in the respective studies. To construct pPICZαA/Jλ2, the PCR-amplified DNA fragment corresponding to J2λ2 was digested with KpnI and SacII and ligated into pPICZαA/Jk1 at the appropriate restriction sites. VLA17 was amplified by PCR, digested with BspMI and SpeI, and cloned into pPICZαA/Jλ2 at the BsmBI and AvrII restriction sites. All constructs were verified by DNA sequencing.
Procedures of electrocompetent cell preparation, electroporation of P. pastoris GS115 cells, Mut+ or Muts phenotype determination and selection on zeocin followed Invitrogen protocols. Analytical or large-scale expression of recombinant FabA17 was performed in cultures of BMGY and BMMY media according to the Invitrogen protocol. Methanol was added every 24 h after induction (up to 0.5%).
The culture medium was concentrated by ultrafiltration, equilibrated with 50 mM sodium phosphate buffer pH 8.0 containing 300 mM NaCl and purified on a Talon resin column (Clontech, USA). The eluted fraction was desalted against 50 mM sodium phosphate buffer pH 7.4 and separated by anion-exchange chromatography on a Mono Q column (Sigma) with salt-gradient elution (0–1 M NaCl in 50 mM sodium phosphate buffer pH 7.4). Fractions corresponding to Fabs were then purified on a Superdex 75 column (GE Healthcare, United States) equilibrated with 50 mM sodium phosphate buffer or 50 mM Tris–HCl buffer pH 7.4. The purity and identity of the eluted Fabs were tested by 12% SDS–PAGE with Coomassie staining and Western blot analysis. Horseradish peroxidase-conjugated anti-FLAG and anti-human light chain Abs (Sigma, USA) were used for detecting HCH and CL, respectively.
2.2. Crystallization and data collection
Crystals of FabA17λ were grown using the hanging-drop vapour-diffusion method by mixing equal volumes of protein (7 mg ml−1 in 50 mM Tris–HCl buffer pH 7.4) and precipitant solution [0.25 M ammonium sulfate and 20%(w/v) polyethylene glycol 5000 monomethyl ether in 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES) pH 6.5]. Rod-shaped crystals of approximately 0.4 × 0.1 × 0.1 mm in size were obtained after 3–4 d and X-ray data were collected on EMBL beamline P14 at the PETRA III storage ring (DESY, Hamburg, Germany) at a wavelength of 1.2234 Å using a MAR CCD 225 mm detector. The data were collected at a cryogenic temperature of 100 K, and the mother-liquor solution supplemented with 20%(v/v) PEG 400 was used as a cryoprotectant. Images of 0.25° oscillation were collected over a total rotation of 85° using 2 s exposure per image. The diffraction data were indexed and integrated with XDS (Kabsch, 2010 ▶) and scaled using SCALA (Evans, 2006 ▶). The values of I/σ(I) and CC1/2 (Karplus & Diederichs, 2012 ▶) were used as a guide to determine the resolution cutoff (Table 1 ▶).
Table 1. Data-collection and refinement statistics.
| Data-collection statistics | |
| Source | MX beamline P14, PETRA III, EMBL/DESY |
| No. of images | 380 |
| Oscillation range () | 0.25 |
| Space group | P41212 |
| Unit-cell parameters () | a = b = 60.63, c = 279.64 |
| Wavelength () | 1.2234 |
| Resolution () | 25.01.95 (2.061.95) |
| R merge (%) | 9.6 (85.4) |
| R r.i.m. (%) | 10.4 (92.0) |
| R p.i.m. (%) | 3.8 (33.9) |
| CC1/2 | 99.7 (79.3) |
| I/(I) | 15.6 (2.7) |
| Completeness (%) | 99.9 (100) |
| Multiplicity | 7.3 (7.3) |
| Estimated B factor from Wilson plot (2) | 26.9 |
| Refinement statistics | |
| Resolution range () | 25.01.95 |
| No. of reflections used for R free calculation | 39334/1971 |
| R work/R free (%) | 20.5/25.5 |
| No. of atoms | |
| Protein | 3326 |
| Ligands | 12 |
| Solvent | 310 |
| B factors (2) | |
| Protein | 24.8 |
| VH | 18.4 |
| CH | 34.6 |
| VL | 20.6 |
| CL | 26.1 |
| Ligands | 19.1 |
| Solvent | 27.8 |
| Root-mean-square deviations | |
| Bond lengths () | 0.01 |
| Bond angles () | 1.12 |
| Ramachandran plot, residues in | |
| Most favoured regions | 427 [96.8%] |
| Favoured regions | 20 [3.2%] |
| Disallowed regions | 0 |
2.3. Structure solution and refinement
The A17λ structure was solved by molecular replacement using MOLREP (Vagin & Teplyakov, 2010 ▶) with the heavy chain of the FabA17κ structure (PDB entry 2xza; Smirnov et al., 2011 ▶) as a search model. Attempts to use the whole Fab structure as a model were unsuccessful, probably owing to the difference in the overall shape of the molecule.
The partial structure solution was followed by location of the VL domain, which is identical in the λ and κ variants, and building of the rest of the Fab molecule was achieved using the ARP/wARP program (Langer et al., 2008 ▶). A β-sheet of the CL domain could not be built automatically and was added using several manual interventions with Coot (Emsley & Cowtan, 2004 ▶) and PHENIX (Afonine et al., 2012 ▶), including simulated annealing. After completion of model building, refinement was carried out with PHENIX and REFMAC5 (Murshudov et al., 2011 ▶).
Solvent molecules were located automatically using PHENIX and were confirmed by visual inspection; all of them were well defined in density. All located water molecules were refined with unit occupancy. The final model consisted of one Fab molecule (Fig. 2 ▶) with 445 residues and 310 water molecules. The positions of two N-terminal residues of the light chain could not be located in the electron density. Close to the active centre, there was a well resolved residual density for a MES molecule that was present in the crystallization condition and refined with occupancy value of 0.8 (Fig. 3 ▶). A stereochemical analysis of the structure using PROCHECK (Laskowski et al., 1993 ▶) showed that 96.8% of the residues were in the most favoured regions of the Ramachandran plot and 3.2% were in favoured regions. The data were nominally collected to 1.89 Å resolution, but during processing the resolution was reduced to 1.95 Å. This moderate data truncation slightly improved the regions of poor electron density as well as the refinement statistics. The refinement was deemed to have converged at an R factor of 20.5% and an R free of 25.5%. Data-collection and refinement statistics are presented in Table 1 ▶.
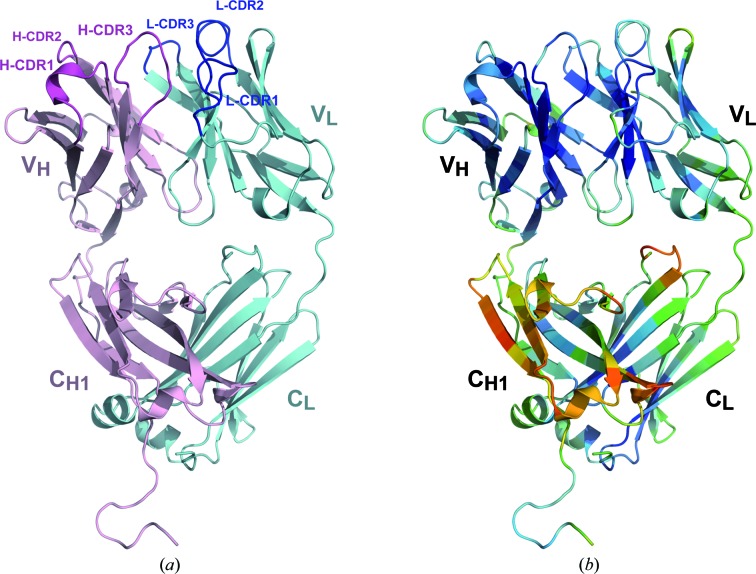
Figure 2.
(a) The overall structure of the A17λ antibody. Heavy (VH/CH1) and light chains are shown in magenta and cyan, respectively. (b) The A17λ structure coloured according to the Cα atomic displacement parameters (ADP), with a colour transition from blue to red indicating increasing ADP values.
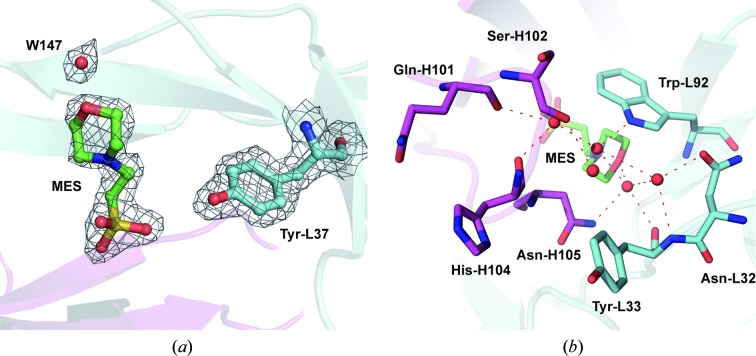
Figure 3.
(a) 2F o − F c electron-density map contoured at the 1.5σ level above the mean (0.4 e Å−3) showing the MES molecule, water 147 and the catalytic Tyr-L36. Heavy and light chains are shown in magenta and cyan, respectively. (b) An extended hydrogen-bonding pattern involving the MES molecule and five intact water molecules located close to the A17λ active centre (red dashed lines).
To check whether different search models or algorithms could affect the results of structure solution by molecular replacement, we also tried to use as search models individual domains from the A17κ structure or other reported Ab structures that share high homology with A17λ (e.g. PDB entries 3mly and 4evn; Jiang et al., 2010 ▶; Lingwood et al., 2012 ▶). Thus, solving the A17κ structure with MOLREP, we first located the VH domain using the VH domain of the 2xza or the 3mly structure as a model and then the CH1 domain of the same structures. Thereafter, the VL and CL domains were located using as models the homologous domains from the 2xza and 4evn structures, respectively. Similar results were obtained by first locating the domains of the light chain and then those of the heavy chain or first the two variable domains and then the constant domains of the Ab. By carrying out molecular replacement with Phaser (McCoy et al., 2007 ▶), it was also possible to locate the four domains of the Fab molecule by using individual domains of 2xza or 3mly for VH and CH1 and of 4evn for VL and CL as search ensembles. The same results were also obtained by altering the order of the search ensembles. Finally, using the BALBES pipeline (Long et al., 2008 ▶) it was possible to individually locate the VH, CH1 and VL domains of A17λ and finally to use ARP/wARP to build most of the CL domain. All of the solutions were very similar, with r.m.s.d. values of less than 1 Å for Cα atoms after rigid-body refinement.
2.4. Molecular-dynamics simulation
The GPU-accelerated GROMACS 4.6.3 software package (Pronk et al., 2013 ▶) was used for the simulation and analysis of MD trajectories using the Amber ff99SB-ILDN force field (Lindorff-Larsen et al., 2010 ▶). Explicit solvent simulations were performed at a temperature of 300 K with a time constant for coupling of 0.1 ps under the control of a velocity rescaling thermostat and isotropic constant-pressure boundary conditions under the control of the Berendsen algorithm of pressure coupling with a time constant of 5 ps and application of the particle mesh Ewald method for electrostatic interactions (PME). A triclinic box of TIP3P water molecules was added around the protein to a depth of 20 Å on each side of the solute. Charges were neutralized by the addition of chloride ions. Additional NaCl was added to the systems to a final concentration of 0.14 M. In each of the simulations, there were two temperature-coupling groups, the first consisting of protein and the second consisting of water with Na+ and Cl− ions. The time step for integration in all simulations was 2 fs. Coordinates were written to output as a trajectory file every 10 ps and the total time of simulation was 250 ns. All simulations were performed on a Lomonosov supercomputer provided by the SRCC of Moscow State University. Analysis of the trajectories was also performed using the GROMACS 4.6 software package.
2.5. Evaluation of kinetic and thermodynamic parameters for the reactibody reactions
Kinetic measurements were made as described by Reshetnyak et al. (2007 ▶) and Smirnov et al. (2011 ▶). Briefly, reactions of FabA17λ and FabA17κ (3–32 µM) with phosphonate X or paraoxon (Figs. 1 ▶ a and 1 ▶ b) over a concentration range of 10–500 µM were carried out in 0.1 M sodium phosphate buffer pH 7.4 at different temperatures. Reaction rates were determined from the changes in absorbance at 405 nm owing to p-nitrophenol formation, and the rate constants were calculated using a p-nitrophenol extinction coefficient ∊ of 12 300 M −1 cm−1. Active Ab concentrations were extrapolated from the A max at 405 nm in the presence of excess phosphonate X. Modifications of rate constants k 1 were estimated by Kitz–Wilson analysis (for details, see Supporting Information §S1).
Stopped-flow measurements with fluorescence detection were made using an SX.18MV stopped-flow spectrometer (Applied Photophysics, England). All experiments were carried out in 20 mM sodium phosphate buffer pH 7.4 with 150 mM NaCl at different temperatures (280–293 K). The Trp fluorescence was excited at 290 nm and monitored at >320 nm. Each trace in the diagrams is the average of no fewer than four individual recordings. The concentration of Fabs in all experiments was 10 µM and the concentrations of phosphonate and paraoxon were varied from 5 to 300 µM. Kinetic parameters were calculated as described by Smirnov et al. (2011 ▶) using the DynaFit software (Kuzmic, 1996 ▶) (for details, see Supporting Information §S1).
Thermodynamic parameters (rate constants k 2 and equilibrium constants K d; Supplementary Table S1) for the interaction of phosphonate X with A17 were determined.
2.6. Determination of denaturation temperature
The denaturation temperatures (T d) of A17κ and A17λ were measured in a VP-DSC microcalorimeter (MicroCal, USA) in 0.5 ml cells at a heating rate of 1 K min−1 as described in Mitkevich et al. (2003 ▶). Test solutions contained 0.6–1.5 mg ml−1 protein in 50 mM sodium phosphate buffer pH 7.4. The accuracy of the measurements was ±0.1 K. To analyze functions of excess heat capacity, the Origin-DSC program package was used.
2.7. Phage library selection for binding the single-chain antibody scFvA17
The random cyclic heptapeptide phage library CX 7C was used (Koivunen et al., 1994 ▶). The solid-phase selection procedures were performed according to Yribarren et al. (2003 ▶), with some modifications (for details, see Supporting Information §S2). The pool of phage-bound peptides selected after each round was tested for specificity toward scFvA17 by means of phage ELISA (Supplementary Fig. S2). DNA fragments encoding the peptides from 20 clones randomly taken after the fifth round were amplified by PCR and sequenced. As a result of amino-acid sequence alignment, two consensus sequences were identified: CRNPWGLTC (pep50) and CPNPWGLLC (pep54).
2.8. Peptide synthesis
The peptides were synthesized by standard solid-phase Nα-Fmoc chemistry (for details, see Supporting Information §S3). Two selected peptides were obtained in cyclic (50C or 54C) and linear (50L or 54L) forms, each consisting of 18 amino acids. The two residues at the N-terminus belonged to the bacteriophage pIII protein and were followed by a peptide sequence flanked by two cysteines. Lysine in the C-terminal part (GAAGAEK), which is also found in the bacteriophage pIII protein, was conjugated with a biotin molecule. The final sequences of the peptides were as follows: NH2-GACRNPWGLTCGAAGAEK(Biot)NH2 (50) and NH2-GACPNPWGLLCGAAGAEK(Biot)NH2 (54).
2.9. Surface plasmon resonance (SPR) analysis
The surface plasmon resonance experiments were performed using a Biacore T200 system (GE Healthcare) equipped with a research-grade SA sensor chip. Chemically synthesized peptides (oxidized pep50C and pep54C, reduced pep50L and pep54L and a control peptide) were immobilized according to the manufacturer’s protocol. Flow cell 1 was left blank as a reference surface. To collect kinetic binding data, A17κ, A17λ and control Abs were injected at a range of concentrations into the four flow cells at a flow rate of 10 µl min−1 at 298 K. Ab–peptide association and dissociation were each monitored for 300 s. The surfaces were regenerated by a 100 s injection of 100 mM glycine–HCl pH 2.0. Data were collected at a rate of 1 Hz and fitted with a 1:1 binding model using the affinity-analysis option available within the BIAevaluation software.
3. Results and discussion
3.1. Molecular organization of the A17 reactibody
Organophosphate-metabolizing A17scFv with variable domains corresponding to IGHV4-b*/IGLV1-51* germline genes was selected from a human semisynthetic Ab variable fragment library using a covalent capture selection strategy (Reshetnyak et al., 2007 ▶). To express full-length A17 reactibody in CHO cells, we used vectors permitting the production of a corresponding Ab with human constant domains of subclass IgG1/κ (Smirnov et al., 2011 ▶). Some differences were observed in the catalytic efficiency of full-length A17 or its Fab fragment compared with the parent scFv molecule (Zakharov et al., 2011 ▶). On one hand, these differences could result from structural stabilization of the active centre owing to the presence of additional constant domains. Crystallographic studies show that there is a close association between VL and VH and between CL and CH1 in the Fab (Padlan, 1994 ▶). Typically, Fv shares similar antigen-binding properties with Fab. However, the relative orientation of VL and VH in Fv is obviously not necessarily the same as in the Fab because the stabilizing effect of the CL–CH1 module (observed in Fab) is absent in Fv (Narciso et al., 2011 ▶). On the other hand, the full-length A17 reactibody contained the artificial light chain with the λVL domain fused to κCL through the κ switch region (Fig. 1 ▶ c) and this non-native domain could also affect the properties of Ab.
To solve this question, the natural λ light chain was constructed (Fig. 1 ▶ c) and recombinant FabA17 containing a κ or λ light-chain constant region was produced in the methylotrophic yeast P. pastoris.
3.2. Structure of the A17λ Fab reactibody and its comparison with the A17κ variant
3.2.1. Quality of the final model
The A17λ crystals belonged to the tetragonal space group P41212 and the model was refined to an R factor and R free of 20.5 and 25.5%, respectively. The results showed that the asymmetric unit of the crystal contained one Fab molecule and four domains (VL, CL, VH and CH1) with the canonical β-sandwich immunoglobulin fold (Fig. 2 ▶). The final model comprises residues 1–231 of the heavy chain and 3–216 of the light chain. Most of the structure, including the CDR regions, shows well defined electron density, except for the loops H 136–138, H 160–164 and L 170–173 (sequential numbering; for numbering according to Kabat, see Fig. 1 ▶ c).
3.2.2. Comparison of the A17λ and A17κ structures
As calculated by the PISA program (Krissinel & Henrick, 2007 ▶), the interface area between the heavy and light chains in A17λ is 1785 Å2 and there are 18 hydrogen bonds, one salt bridge and one disulfide bond (Supplementary Table S2). This interface is comparable to others reported for Fab structures, but is more extended than that in the A17κ structure (PDB entry 2xza; 1527 Å2 with 12 hydrogen bonds and one salt bridge).
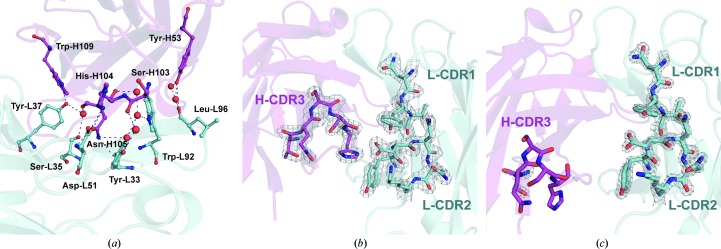
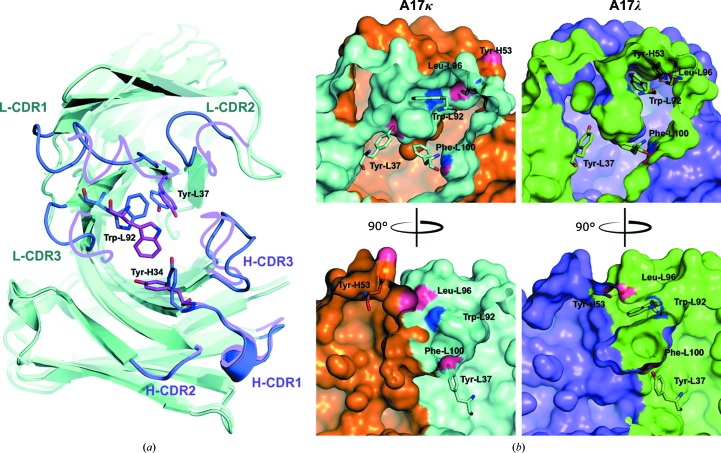
An important feature of the A17λ structure and the key difference between the two variants is a large interface between the two variable domains of the Fab molecule, where interactions take place between the CDR loops located close to the A17λ active centre, as well as Tyr-H53 from H-CDR2 (Fig. 4 ▶ a). They include strong direct interactions between residues of the H-CDR3 loop with L-CDR1 and L-CDR2, between H-CDR2 and L-CDR3 (Supplementary Table S2), as well as contacts via water molecules and a bound MES molecule. These interactions result in the displacement of these CDR loops, apart from H-CDR2, as follows from their structural alignment with the A17κ structure (r.m.s.d. of 2.4 Å for Cα atoms using LSQKAB; Kabsch et al., 1976 ▶), and in the formation of a fairly rigid ensemble by the two variable domains, unlike in A17κ, where these domains do not interact directly. In contrast, the H-CDR1 loop (which does not take part in this interface) and the H-CDR2 loop, which lies farther away from the active centre (similar to H-CDR1), are well aligned in the two structures (r.m.s.d. of 0.9 Å using LSQKAB). The presence of a large interdomain interface in A17λ appears to provide stabilization of the CDR loops, as can be deduced from a comparison of their atomic displacement parameters (ADPs). Although the average ADP value for all protein atoms in the A17λ structure is higher than in A17κ (25.1 versus 21.5 Å2), the ADPs of the A17λ CDR loops are much lower (17.0 versus 25.4 Å2) (Supplementary Table S3). Furthermore, the electron density in this region is very well defined (Fig. 4 ▶ b), in contrast to the A17κ structure, in which the electron density of the CDR loops is relatively poor (Fig. 4 ▶ c). Comparison of the normalized ADP for residues of CDR loops for A17κ (PDB entry 2xza), phosphonylated A17κ (PDB entry 2xzc) and A17λ (PDB entry 3zl4) demonstrated that the ADP values for all CDR loops are generally higher for the two A17κ structures compared with A17λ, with no considerable dependence on the crystal contacts taking place in this loop (Supplementary Table S4 and Fig. S3). This in turn means that the active centre of the A17λ variant is more rigid than that of A17κ.
Figure 4.
(a) Interdomain interactions between the CDR loops of A17λ and interactions involving catalytic Tyr-L37. Protein residues involved in these interactions are shown in ball-and-stick representation, water molecules are shown as spheres and hydrogen-bond interactions are shown as blue dashed lines. Here and in (b) and (c), heavy and light chains are shown in magenta and cyan, respectively. Tyr-L33 and Ser-L35 belong to L-CDR1, Asp-L51 to L-CDR2, Trp-L92 and LeuL96 to L-CDR3, Tyr-H53 to H-CDR2, and Ser-H103, His-H104 and Asn-H105 to H-CDR3. (b) 2F o − F c electron-density map contoured at the 2σ level (0.5 e Å−3) above the mean for the CDR loop region of A17λ. (c) 2F o − F c electron-density map contoured at the 2σ level (0.5 e Å−3) above the mean for the CDR loop region of A17κ.
The reported A17λ structure is characterized by marked differences in the ADPs between the variable and constant domains of the chains. The average ADPs for all of the protein atoms in the VH and VL domains are 18.4 and 20.6 Å2, compared with 34.6 and 26.1 Å2 in the CH1 and CL domains, respectively (Fig. 2 ▶ b). This is presumably owing to an extended interaction between the variable domains, with the constant domains being more mobile. In contrast, the A17κ variant shows an even distribution of ADP values, which average 19.2 and 19.2 Å2 for VH and VL and 17.3 and 19.3 Å2 for CH1 and CL, respectively. Certain differences in the ADPs between the variable and constant domains (with a slightly higher mobility for the constant domains) are not rare. In a survey of 200 high-resolution structures of unliganded Fab molecules deposited in the PDB, we found that the average ADPs for the Cα atoms of variable domains are almost identical, 29 Å2 for VH and 28 Å2 for VL, with this parameter being slightly higher for the constant domains: 31 Å2 for both CH1 and CL (Supplementary Fig. S4). Obviously, the corresponding differences in the case of A17λ are much greater.
The active centre of A17λ structurally deviates from that of A17κ. The major difference is that the upper part of the A17λ active centre is shifted away from the light chain. This shifting of the L-CDR3 loop, which is mainly caused by the strong interaction between Tyr-H53 and Leu-L96, leads to the displacement of Trp-L92 by about 4 Å, its flipping and rotation (Fig. 5 ▶). This displacement of L-CDR3, and Trp-L92 in particular, which is probably facilitated by the crystal contacts or the presence of the MES molecule (Fig. 3 ▶ b), results in the enlargement of the hydrophobic pocket surface and the formation of a lid above the cavity entrance (Fig. 5 ▶ b). To estimate the level of involvement of the crystal contacts in the active-centre architecture, we performed an MD simulation of the behaviour of A17κ and A17λ in a dilute solution environment and without any neighbouring molecules, including MES for A17λ. In A17κ, displacement of the symmetry-related molecules by water leads to nonsignificant relaxation of the structure (protein backbone r.m.s.d. of 1.2 and 1.3 Å for the heavy and light chains, respectively). The same estimation performed for A17λ revealed noticeable changes in the structure (protein backbone r.m.s.d. of 2.2 and 2.7 Å for the heavy and light chains, respectively) and movement of L-CDR3 and H-CDR3 followed by hydrogen-bond formation between the side chain of Trp-L92 and the main chain of Asn-H105 (Supplementary Fig. S5). It should be noted that this is not observed in the case of the A17κ MD structure during the entire simulation time; therefore, the rearrangement of A17λ mainly occurred owing to the removal of the MES molecule from the active site. The observations described above allow us to suggest that crystal contacts do not play a significant role in the structure of the A17κ active centre, but the impact of packing interactions for A17λ cannot be ruled out.
Figure 5.
(a) Superposition of the A17λ and A17κ variable domains. A17λ CDR loops are shown in magenta and A17κ CDR loops are shown in blue. The flip of the Trp-L92 side chain is indicated. (b) The flipping of Trp-L92 provides the enlargement of the hydrophobic pocket surface (Trp-L92–Phe-L100) and the formation of a kind of lid above the cavity entrance. The light chain of A17κ is shown in cyan and the heavy chain is shown in brown; the light chain of A17λ is shown in green and the heavy chain in shown in magenta.
In the reported A17λ structure, the catalytic Tyr-L37 forms a hydrogen bond to Trp-H109, while a water molecule bridges it to Ser-L35 and Asn-H105, thereby forming a rigid entity (Fig. 4 ▶ a). In A17κ, Tyr-L37 does not directly interact with neighbouring residues but forms an extended hydrogen-bonding network through solvent molecules (Smirnov et al., 2011 ▶). In the presence of co-crystallized MES, Tyr-H34 in A17λ is not directed towards Tyr-L37 but turns to form a hydrogen bond with Ser-H51. The positions of Tyr-L33 and Asn-H105 deviate from those in A17κ owing to a hydrogen-bonding pattern that is formed via one MES and five water molecules located close to the active centre. We think that this fairly extended hydrogen bonding (Fig. 3 ▶ b) stabilizes the protein and at the same time prevents the modification of A17λ by phosphonate X in crystals containing MES, in contrast to the crystal structure of A17κ, in which this modification is possible.
3.2.3. Stability of the antibody variants
To determine whether this extended hydrogen-bonding pattern at the active centre of A17λ and interdomain interaction stabilize the Fab molecule, we measured the denaturation temperatures (T d) of κ and λ Fabs and their phosphonylated forms. The denaturation curves for the proteins had only one peak, indicating that the Fab molecules formed a single structural ensemble in all variants. The T d values of the two λ variants were higher than those of the κ variants (339.8 versus 335.5 K for unmodified proteins and 347.1 versus 344.9 K for phosphonylated proteins, respectively).
These results indicate that the κ→λ switch leads to stabilization of the antibody molecule, possibly owing to the aforementioned interaction between the CDR loops. Furthermore, higher T d values in the phosphonylated variants compared with the unmodified antibodies indicate that the phosphonylation stabilizes the antibody molecule.
3.2.4. Structural alignment
Structure-based sequence alignment of λ and κ light chains revealed a high similarity between them. The variable domains of the light chain are identical (Supplementary Fig. S6), while the CL domains and J regions connecting the VL and CL domains show only 42% sequence identity and 83% secondary-structure matching as calculated by the PDBeFold algorithm (Krissinel & Henrick, 2004 ▶). Alignment of the Cα atoms of the two structures using the LSQKAB algorithm resulted in r.m.s.d. estimates of 1.1 Å for the heavy chains and 4.1 Å for the light chains. Such a large structural deviation between κ and λ chains mainly appears to be owing to the J segment, especially to the Gly-L110 insertion in the λ variant, which changes the orientation of the CL domain relative to VL so that it forms a rotation angle of 29° with the latter (Fig. 6 ▶). In addition to the Gly-L110 insertion, the substitution of the positively charged Arg-L111 in A17κ by Gln-L111 in A17λ also appears to play a role in differentiation between the two light chains, since Gln-L111 strongly interacts with Tyr-L143, Val-L108 and Glu-L84. In A17κ, Arg-L111 is turned away from Tyr-L143, possibly because of steric effects, and forms a hydrogen bond with Asp-L172.
Figure 6.
Structural superposition of the A17κ (blue) and A17λ (magenta) light chains on the VL domain. The Gly-L110 insertion in A17λ accounts for the change in the VL–CL orientation compared with that in A17κ.
3.2.5. Comparison of the elbow angles
The elbow angle of the FabA17λ molecule is 126°. This value is at the lower limit of the range previously recorded in Fab molecules (125–225°) and differs considerably from that characteristic of λ chain-type Fabs, which tend to adopt large elbow angles (Stanfield, Zemla et al., 2006 ▶). Structural comparisons of A17λ with A17κ (PDB entry 2xza) and A17κ phosphonylated by phosphonate X (PDB entry 2xzc) allowed us to conclude that neither the κ→λ light-chain switch nor phosphonylation have any significant effect on the elbow angle (Fig. 7 ▶). As for the overall structure of the two constant domains (CH1 and CL) in the Fab molecule, the change in the CL domain of A17λ resulted in some differences in its orientation relative to CH1, as follows from the comparison of the twist/tilt angles between the two domains (176/97° in A17λ versus 184/104° in A17κ) and in the alignment of these modules in the two structures (r.m.s.d. of 3.2 Å when aligned using the LSQKAB algorithm).
Figure 7.
Elbow angles in A17 reactibody variants. A17κ-P is a phosphonylated variant.
3.3. Functional characteristics of FabA17 reactibody variants with κ and λ light chains
The newly obtained FabA17λ is a fully functional reactibody with a reactivity comparable to that of the κ variant. Comparison between A17 reactibodies with κ and λ light chains with respect to the steady-state kinetic parameters of their interaction with the phosphonate X molecule showed that K d and k 2 for A17λ were similar to those for A17k (Table 2 ▶). The slight difference in K d indicates a certain divergence between these variants at the first (rapid) stage of ligand binding (see Scheme 1 in Supporting Information S§1).
Table 2. Kinetic parameters for the interaction of phosphonate X with and variants of A17.
| Phosphonate X | Paraoxon | |||||||
|---|---|---|---|---|---|---|---|---|
| k 2 (min1) | K d (M) | k 2/K d (M 1min1) | G (kcalmol1) | H (kcalmol1) | S (calmol1 K1) | E a † (kcalmol1) | k 2/K d (M 1min1) | |
| A17 | 0.24 0.03 | (120 15) 106 | 2000 500 | 6.5 0.1 | 4.1 0.3 | 4.0 1.2 | 12.5 1.3 | 1.2 0.5 |
| A17 | 0.28 0.02 | (130 15) 106 | 2200 400 | 6.5 0.1 | 3.0 0.4 | 7.6 1.8 | 11.4 0.5 | 1.6 0.6 |
E a is the activation-energy parameter.
To reveal the details of the reaction process, thermodynamic and pre-steady-state kinetic parameters were evaluated for A17κ and A17λ.
Thermodynamic parameters were calculated from the values of the K d and k 2 constants as a function of temperature (Supplementary Table S1 and Fig. S7). The energies of A17κ and A17λ modification by phosphonate X are comparable in terms of enthalpy and free energy; however, the reaction of A17λ with phosphonate X is more entropically favourable (Table 2 ▶).
The pre-steady-state kinetic analysis of interaction of A17λ with phosphonate X revealed two noncovalent binding stages, with the first rapid stage corresponding to the bimolecular interaction of the reactibody with the phosphonate ligand and the second stage involving induced-fit conformational changes of the reactibody. This is in agreement with previously reported data on the A17κ reactibody (Smirnov et al., 2011 ▶). Comparison of kinetic parameters for A17κ and A17λ shows that the induced-fit stage is more rapid in the κ variant (Table 3 ▶). This may be owing to the less rigid structure of A17κ, with the entrance to its active centre being wider than in A17λ, where it is significantly narrowed because of interactions between the CDR loops. These conformational peculiarities provide more rapid and precise fitting of this reactibody to the phosphonate molecule.
Table 3. Stopped-flow kinetic parameters of the interaction of phosphonate X with and variants of A17.
The errors indicated are 1 SD.
| k 1 (M 1s1) | k 1 (s1) | k 2 (s1) | k 2 (s1) | k obs1 | k obs2 | |
|---|---|---|---|---|---|---|
| A17 | (6.6 0.5) 106 | 460 35 | 46 7 | 150 20 | 530 90 | 130 30 |
| A17 | (17.5 0.8) 106 | 480 35 | 65 10 | 90 12 | 580 90 | 80 20 |
An X-ray analysis of both A17 variants has shown that they have a well developed, deep active centre with the nucleophilic Tyr-L37 at the bottom of the cavity (15 Å from the surface of the molecule). A similar architecture of the active centre with Tyr-L37 has previously been described for the mouse catalytic Ab 13G5 (Heine et al., 1998 ▶). This part of the Ab molecule is highly conserved and belongs to its structural core, which usually does not interact with antigens (Narciso et al., 2012 ▶). This fact may account for the absence of marked differences between A17κ and A17λ in the kinetic parameters of their interaction with organophosphates. The observed structural divergence of CDRs loops can lead to changes in the antigen-binding properties of Ab molecules. Since A17 was selected as a biocatalyst using artificial chemical substrates, it was relevant to find out whether there is an epitope to which A17 is naturally intended to bind. As we failed to detect such an antigen among nucleic acids, lipids and polysaccharides (data not shown), we focused on epitopes of a protein nature. To address the question of the antigen specificity of A17κ and A17λ, we used a combinatorial approach. ‘Epitope mapping’ was performed by screening a phage-displayed cyclic heptapeptide library. To facilitate this procedure, scFvA17 was used for peptide-epitope selection. The scFv molecule consists of only the variable domains of the light and heavy chains connected through an (SG4)2SGGSAL linker; therefore, any effect from the constant domains was excluded. The scFvA17 proved to specifically bind two phage-displayed peptides, pep50 and pep54. It was found that the recombinant Fab reactibodies were also capable of binding the selected phage-displayed peptides pep50 and pep54, retaining this capacity after modification by phosphonate X (Supplementary Fig. S8). However, phosphonylation of the Fabs reduced the level of binding (Supplementary Fig. S8a), whereas such a modification of scFv significantly enhanced the ELISA signals (Supplementary Fig. S8b). The phosphonate ligand is completely buried in the active centre, being inaccessible for direct interaction with the peptides. Therefore, we suggest that covalent modification may have an indirect effect on the interaction of the antibody with the peptides.
To make a more accurate comparison of antigen-binding properties between the two A17 variants, we performed SPR analysis of reactibody binding with synthetic cyclic (pep50C and pep54C) and linear (pep50L and pep54L) peptides.
The affinity parameters determined from the sensorgrams are shown in Table 4 ▶ and Supplementary Fig. S9. Both A17κ and A17λ proved to have high affinity for the pep50L and pep50C peptides. However, A17κ was more active in binding either the linear or the cyclic form of pep50, which could be owing to its less rigid structure. In the case of pep54C, A17λ was more active than A17κ. A probable explanation is that the cyclic structure of pep54C fits better to the rigid A17λ antigen-binding site.
Table 4. BiaCore affinity constants for interactions of A17 and A17 reactibodies with linear (50L and 54L) and cyclic (50C and 54C) peptides.
| K d(50L) (106 M) | K d(54L) (106 M) | K d(50C) (106 M) | K d(54C) (106 M) | |
|---|---|---|---|---|
| A17 | 1.3 0.1 | 25.0 5.0 | 0.9 0.1 | 25.7 0.5 |
| A17 | 2.3 0.3 | 18.0 4.0 | 2.20 0.08 | 4.8 0.6 |
4. Conclusions
Determination of the structure of the A17λ reactibody variant and its comparison with the previously reported structure of A17κ has provided an insight into changes in the structure–function relationship upon the κ→λ switch. It should be emphasized that the results presented above are based on direct comparison between the two light-chain variants of the same antibody A17, rather than on a statistical survey of structures reported for different antibodies. They show that an exchange of the light-chain constant domain produces an effect on the active-centre architecture, altering the overall shape of the light chain and consequently its orientation relative to the heavy chain. This alteration is also reflected in the interaction between the variable domains via their CDR loops, which leads to deformation of the cavity entrance and makes the binding pocket of A17λ different from that of A17κ; however, the impact of packing interactions cannot be formally ruled out. The elbow angle differs slightly between the two variants and is slightly reduced in A17λ compared with A17κ, in contrast to the data reported previously for antibodies with κ and λ light chains. As suggested previously (Stanfield, Zemla et al., 2006 ▶), changes in elbow angles may simply serve to increase the flexibility of the Fab. Such changes could affect the reaction mechanism of the biocatalyst. Overall, the physicochemical (crystallographic, kinetic and thermodynamic) data and the results of artificial epitope mapping for both light-chain variants of A17 show that the replacement of the light-chain constant domain has an effect on the stability and antigen-binding properties of the reactibody, but not on its reactivity. In our opinion, the domain structure of an antibody molecule should be taken into account in the design of novel artificial biocatalysts.
Supplementary Material
PDB reference: A17λ, 3zl4
Supporting Information.. DOI: 10.1107/S1399004713032446/wd5216sup1.pdf
Acknowledgments
This study, performed within the framework of RFBR–EMBL program ‘Structure–Function Interrelation in Artificial Enzymes’ (project no. 12-04-92428), was supported by the Skolkovo program, RFBR grants (projects nos. 11-04-02016-a, 12-04-00474-a, 12-04-01609-a, 12-04-33258, 13-04-40277, 14-04-31223, 14-04-31207, 13-04-40279-N KOMFI and 13-04-40280-N KOMFI); programs of the Presidium of Russian Academy of Sciences ‘Fundamental Sciences for Medicine’, ‘Molecular and Cellular Biology’ (AAM, OSF, AG) and ?Nanotechnologies and Nanomaterials? (no. 24); the Scientific School Support Program ‘Chemical Basis of Biocatalysis’ (project no. 2046.2014.4); the Ministry of Education and Science of the Russian Federation (project no. 8283); grant no. 4238 from PICS Centre National de la Recherche Scientifique France–?Russia. Computer resources were provided by the Research Computing Center of Moscow State University. The supercomputer ‘Lomonosov’ was used for all modelling studies. The authors acknowledge the access to the macromolecular crystallography beamline P14 (EMBL/DESY, Hamburg) and are grateful to N. A. Gorgolyuk for his help in preparing the manuscript.
Footnotes
Supporting information has been deposited in the IUCr electronic archive (Reference: WD5216).
References
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Eddleston, M., Buckley, N. A., Eyer, P. & Dawson, A. H. (2008). Lancet, 371, 597–607. [DOI] [PMC free article] [PubMed]
- Ekiert, D. C. et al. (2012). Nature (London), 489, 526–532.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Gabibov, A. G. et al. (2011). FASEB J. 25, 4211–4221. [DOI] [PubMed]
- Golinelli-Pimpaneau, B., Goncalves, O., Dintinger, T., Blanchard, D., Knossow, M. & Tellier, C. (2000). Proc. Natl Acad. Sci. USA, 97, 9892–9895. [DOI] [PMC free article] [PubMed]
- Guddat, L. W., Shan, L., Anchin, J. M., Linthicum, D. S. & Edmundson, A. B. (1994). J. Mol. Biol. 236, 247–274. [DOI] [PubMed]
- Guddat, L. W., Shan, L., Fan, Z.-C., Andersen, K. N., Rosauer, R., Linthicum, D. S. & Edmundson, A. B. (1995). FASEB J. 9, 101–106. [DOI] [PubMed]
- Guenaga, J. & Wyatt, R. T. (2012). PLoS Pathog. 8, e1002806. [DOI] [PMC free article] [PubMed]
- Heine, A., Stura, E. A., Yli-Kauhaluoma, J. T., Gao, C., Deng, Q., Beno, B. R., Houk, K. N., Janda, K. D. & Wilson, I. A. (1998). Science, 279, 1934–1940. [DOI] [PubMed]
- Huber, R., Deisenhofer, J., Colman, P. M., Matsushima, M. & Palm, W. (1976). Nature (London), 264, 415–420. [DOI] [PubMed]
- Jiang, X., Burke, V., Totrov, M., Williams, C., Cardozo, T., Gorny, M. K., Zolla-Pazner, S. & Kong, X.-P. (2010). Nature Struct. Mol. Biol. 17, 955–961. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Kabsch, W., Kabsch, H. & Eisenberg, D. (1976). J. Mol. Biol. 100, 283–291. [DOI] [PubMed]
- Kaneko, E. & Niwa, R. (2011). BioDrugs, 25, 1–11. [DOI] [PubMed]
- Karplus, P. A. & Diederichs, K. (2012). Science, 336, 1030–1033. [DOI] [PMC free article] [PubMed]
- Klohn, P. C., Wuellner, U., Zizlsperger, N., Zhou, Y., Tavares, D., Berger, S., Zettlitz, K. A., Proetzel, G., Yong, M., Begent, R. H. & Reichert, J. M. (2013). MAbs, 5, 178–201. [DOI] [PMC free article] [PubMed]
- Koivunen, E., Wang, B. & Ruoslahti, E. (1994). J. Cell Biol. 124, 373–380. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Kuzmic, P. (1996). Anal. Biochem. 237, 260–273. [DOI] [PubMed]
- Landolfi, N. F., Thakur, A. B., Fu, H., Vásquez, M., Queen, C. & Tsurushita, N. (2001). J. Immunol. 166, 1748–1754. [DOI] [PubMed]
- Langer, G. G., Cohen, S. X., Perrakis, A. & Lamzin, V. S. (2008). Nature Protoc. 3, 1171–1179. [DOI] [PMC free article] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- Lindorff-Larsen, K., Piana, S., Palmo, K., Maragakis, P., Klepeis, J. L., Dror, R. O. & Shaw, D. E. (2010). Proteins, 78, 1950–1958. [DOI] [PMC free article] [PubMed]
- Lingwood, D., McTamney, P. M., Yassine, H. M., Whittle, J. R. R., Guo, X., Boyington, J. C., Wei, C.-J. & Nabel, G. J. (2012). Nature (London), 489, 566–570. [DOI] [PMC free article] [PubMed]
- Long, F., Vagin, A. A., Young, P. & Murshudov, G. N. (2008). Acta Cryst. D64, 125–132. [DOI] [PMC free article] [PubMed]
- Lu, Z.-J., Deng, S.-J., Huang, D.-G., He, Y., Lei, M., Zhou, L. & Jin, P. (2012). World J. Biol. Chem. 3, 187–196. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Mitkevich, V. A., Schulga, A. A., Ermolyuk, Y. S., Lobachov, V. M., Chekhov, V. O., Yakovlev, G. I., Hartley, R. W., Pace, C. N., Kirpichnikov, M. P. & Makarov, A. A. (2003). Biophys. Chem. 105, 383–390. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Narciso, J. E. T., Uy, I. D. C., Cabang, A. B., Chavez, J. F. C., Pablo, J. L. B., Padilla-Concepcion, G. P. & Padlan, E. A. (2011). New Biotechnol. 28, 435–447. [DOI] [PubMed]
- Narciso, J. E. T., Uy, I. D. C., Cabang, A. B., Chavez, J. F. C., Pablo, J. L. B., Padilla-Concepcion, G. P. & Padlan, E. A. (2012). Philipp. Sci. Lett. 5, 63–89.
- Padlan, E. A. (1994). Mol. Immunol. 31, 169–217. [DOI] [PubMed]
- Parmley, S. F. & Smith, G. P. (1988). Gene, 73, 305–318. [DOI] [PubMed]
- Ponomarenko, N. A., Pillet, D., Paon, M., Vorobiev, I. I., Smirnov, I. V., Adenier, H., Avalle, B., Kolesnikov, A. V., Kozyr, A. V., Thomas, D., Gabibov, A. G. & Friboulet, A. (2007). Biochemistry, 46, 14598–14609. [DOI] [PubMed]
- Privett, H. K., Kiss, G., Lee, T. M., Blomberg, R., Chica, R. A., Thomas, L. M., Hilvert, D., Houk, K. N. & Mayo, S. L. (2012). Proc. Natl Acad. Sci. USA, 109, 3790–3795. [DOI] [PMC free article] [PubMed]
- Pronk, S., Páll, S., Schulz, R., Larsson, P., Bjelkmar, P., Apostolov, R., Shirts, M. R., Smith, J. C., Kasson, P. M., van der Spoel, D., Hess, B. & Lindahl, E. (2013). Bioinformatics, 29, 845–854. [DOI] [PMC free article] [PubMed]
- Reshetnyak, A. V., Armentano, M. F., Ponomarenko, N. A., Vizzuso, D., Durova, O. M., Ziganshin, R., Serebryakova, M., Govorun, V., Gololobov, G., Morse, H. C. III, Friboulet, A., Makker, S. P., Gabibov, A. G. & Tramontano, A. (2007). J. Am. Chem. Soc. 129, 16175–16182. [DOI] [PMC free article] [PubMed]
- Sapparapu, G., Planque, S., Mitsuda, Y., McLean, G., Nishiyama, Y. & Paul, S. (2012). J. Biol. Chem. 287, 36096–36104. [DOI] [PMC free article] [PubMed]
- Smirnov, I. et al. (2011). Proc. Natl Acad. Sci. USA, 108, 15954–15959.
- Sotriffer, C. A., Rode, B. M., Varga, J. M. & Liedl, K. R. (2000). Biophys. J. 79, 614–628. [DOI] [PMC free article] [PubMed]
- Stanfield, R. L., Gorny, M. K., Zolla-Pazner, S. & Wilson, I. A. (2006). J. Virol. 80, 6093–6105. [DOI] [PMC free article] [PubMed]
- Stanfield, R. L., Zemla, A., Wilson, I. A. & Rupp, B. (2006). J. Mol. Biol. 357, 1566–1574. [DOI] [PubMed]
- Turner, J. M., Larsen, N. A., Basran, A., Barbas, C. F. III, Bruce, N. C., Wilson, I. A. & Lerner, R. A. (2002). Biochemistry, 41, 12297–12307. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Vincent, K. J. & Zurini, M. (2012). Biotechnol. J. 7, 1444–1450. [DOI] [PubMed]
- Yribarren, A. S., Thomas, D., Friboulet, A. & Avalle, B. (2003). Eur. J. Biochem. 270, 2789–2795. [DOI] [PubMed]
- Zakharov, A. V., Smirnov, I. V., Serebryakova, M. V., Dronina, M. A., Kaznacheeva, A. V., Kurkova, I. N., Belogurov, A. A., Friboulet, A., Ponomarenko, N. A., Gabibov, A. G. & Bobik, T. V. (2011). Mol. Biol. 45, 74–81. [PubMed]
- Zheng, L., Goddard, J. P., Baumann, U. & Reymond, J. L. (2004). J. Mol. Biol. 341, 807–814. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: A17λ, 3zl4
Supporting Information.. DOI: 10.1107/S1399004713032446/wd5216sup1.pdf