Upon transplant, functional HSC clones preferentially expand in certain skeletal locations, exhibiting only limited migration toward other niches.
Abstract
Hematopoietic stem cells (HSCs) are able to migrate through the blood stream and engraft bone marrow (BM) niches. These features are key factors for successful stem cell transplantations that are used in cancer patients and in gene therapy protocols. It is unknown to what extent transplanted HSCs distribute throughout different anatomical niches in the BM and whether this changes with age. Here we determine the degree of hematopoietic migration at a clonal level by transplanting individual young and aged mouse HSCs labeled with barcoded viral vector, followed by assessing the skeletal distribution of hundreds of HSC clones. We detected highly skewed representation of individual clones in different bones at least 11 mo after transplantation. Importantly, a single challenge with the clinically relevant mobilizing agent granulocyte colony-stimulating factor (G-CSF) caused rapid redistribution of HSCs across the skeletal compartments. Old and young HSC clones showed a similar level of migratory behavior. Clonal make-up of blood of secondary recipients recapitulates the barcode composition of HSCs in the bone of origin. These data demonstrate a previously unanticipated high skeletal disequilibrium of the clonal composition of HSC pool long-term after transplantation. Our findings have important implications for experimental and clinical and stem cell transplantation protocols.
Continuous generation and regeneration of all blood and immune cells over the lifespan of an organism is ensured by a limited number of hematopoietic stem cells (HSCs). The vast majority of HSCs reside in the BM, whereas a small fraction of functional HSCs can be found in the blood circulation, both in mice and humans (Goodman and Hodgson, 1962; Richman et al., 1976; Dorie et al., 1979; Körbling et al., 1981). In early development, the ability of HSCs to migrate and engraft niches is important at the stage when HSCs exit the fetal liver and populate the BM (Orkin and Zon, 2008). In adults, HSCs have been shown to move toward the site of injury or inflammation and participate in tissue repair (Lapid et al., 2012). The migrating ability of HSCs is routinely used in clinical transplantation and gene therapy protocols, which are applied in the treatment of an increasing number of hematopoietic and nonhematopoietic diseases. Thus far, it is unknown how individual HSC clones migrate and distribute among skeletal niches after transplantation and how this is affected by mobilization-inducing cytokines.
Our limited knowledge of HSC migration is primarily based on results from parabiotic rodents sharing a common circulation (Warren et al., 1960; Dorie et al., 1979; Wright et al., 2001; Abkowitz et al., 2003). These studies suggest that egress of HSCs into blood is continuous. Migrating cells are capable of reengrafting the BM and further contributing to hematopoiesis (Wright et al., 2001). Based on approximate calculations, it was claimed that 1–5% of all HSCs are circulating daily (Bhattacharya et al., 2009). If this claim was correct, HSC distribution within the same mouse or across parabiotic mice would approach equilibrium within a few months. However, direct measurements of chimerism in parabiotic mice demonstrated relatively slow rates of equilibration (Wright et al., 2001). Although this rate was dramatically increased upon administration of G-CSF, it did not result in full equilibration of HSCs between parabiotic mice (Abkowitz et al., 2003).
G-CSF–induced mobilization is routinely used in clinical BM transplantation and gene therapy protocols, allowing harvest of the HSC-enriched fraction from the donors’ blood (To et al., 1997). Stem cell mobilization in patients has been claimed to decline with age (Morris et al., 2003; Pozotrigo et al., 2013); however, experimental data underlying this phenomenon are limited and contradictory. Although multiple studies found a homing defect of old mouse HSCs (Liang et al., 2005; Dykstra et al., 2011), another study suggested that G-CSF–induced mobilization in aged mice was more efficient than in young (Xing et al., 2006).
In this study, we analyzed posttransplantation skeletal localization of hundreds of young and old hematopoietic clones. To track individual stem cell clones, we labeled highly purified HSCs with a viral barcode label before transplantation (Gerrits et al., 2010; Verovskaya et al., 2013). We questioned whether old and young HSCs would respond differently to mobilizing stimuli. Our data demonstrate that migration of clones under steady-state conditions is very limited, such that clonal distribution does not reach equilibrium up to 11 mo after transplantation. However, migration was strongly activated and led to complete clonal equilibration upon a single mobilizing challenge. Clonal differences in HSC composition of specific skeletal sites were inherited upon secondary transplantations from those particular bones and also resulted in different functional activity in secondary recipients.
RESULTS
Quantification of HSC migration by clonal analysis of distinct skeletal locations
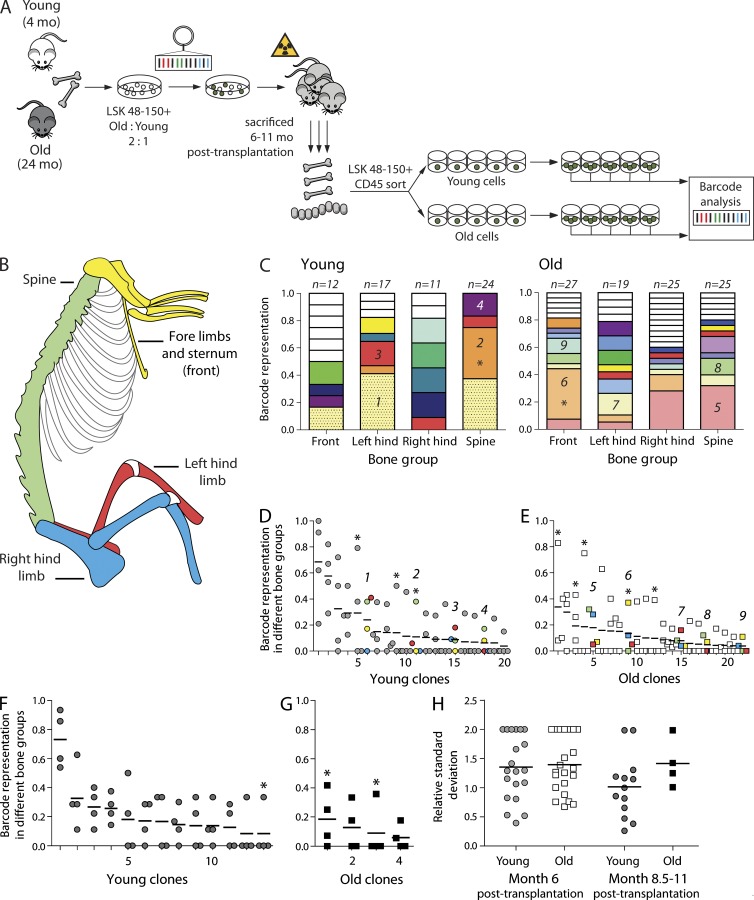
Experiments with parabiotic animals demonstrated quite a slow exchange of HSC pools during an observation time up to 22 wk (Wright et al., 2001; Abkowitz et al., 2003). Other authors, however, claimed a continuously high level of HSC migration (Bhattacharya et al., 2009). Based on these observations, one would predict the clonal composition of the HSC compartment in all skeletal sites to equilibrate within a few months after transplantation. To address the extent of skeletal distribution of old and young HSC clones, we compared the barcode composition in four skeletal sites in recipients of barcoded BM cells (Fig. 1, A and B). Purified young (4 mo) and aged (24 mo) lineage− Sca1+ c-Kit+ CD48− CD150+ (LSK48−150+) cells were mixed and transduced with a barcoded vector library and transplanted into irradiated hosts. To achieve a statistically sufficient number of young and old clones, several spatially closely related bones were pooled (Fig. 1 B). Young and old stem cells were isolated from four distinct skeletal regions: (1) spine; (2) sternum and bones of fore limbs; (3) left femur, left tibia, and left half of pelvic bone; and (4) right counterparts of these bones (Fig. 1 B). Single GFP+ LSK48−150+ cells from young and old origin were sorted from primary recipients and clonally expanded in the presence of cytokines to confirm the high proliferative capacity of these cells and to generate enough cells for robust barcode analysis (Fig. 1 A).
Figure 1.
Old and young HSCs are asymmetrically distributed among various skeletal niches and migrate at a similar rate. (A) Experimental setup. BM cells were isolated from young and aged donors, and LSK48−150+ cells were separately purified by cell sorting and subsequently pooled. After transduction with a barcoded vector library, cells were transplanted into lethally irradiated animals. Recipient mice were sacrificed 6–11 mo after transplant. BM cells were isolated from four skeletal locations, and LSK48−150+ cells of young and old origin were single-cell sorted into cytokine-rich medium. Colonies expanded to ∼30,000 cells were harvested for barcode analysis. (B) Schematic representation of the four bone groups that were analyzed. Bones analyzed in the same group are shown in the same color. (C) Skeletal distribution of barcoded clones in young (left) and old (right) LSK48−150+ cells in a single mouse, analyzed 6 mo after transplantation. Clones that are colored white are unique, i.e., were found only once. The asterisks identify clones with distributions that are significantly different from random (P < 0.05 based on Monte Carlo simulations). The number of barcoded colonies analyzed for every skeletal location is indicated above the column. Clones found in four or more colonies and further used for distribution analysis are numbered. (D) Summary of all clones of young origin found in four or more colonies 6 mo after transplantation (n = 6; data were derived from two cohorts of mice). X axis shows the rank of clone size, based on mean value over all four locations, ranked from most frequent to the least frequent. Y axis shows the contribution of a certain clone to the total pool in one location. Dots on the same vertical line reflect the size of a particular clone in each of the four different locations. The mean contribution for a given clone is shown with a horizontal line. As in C, clones significantly deviating from equal distribution (P < 0.05 based on Monte Carlo simulations) are marked with an asterisk. For clarity, four numbered clones from C are colored according to the bone group colors as in B. (E) Same as D, but shown for old clones (summary of two independent cohorts of mice; n = 6). (F) Same as D, but for young clones from mice sacrificed 8.5–11 mo after transplantation (one cohort; n = 4). (G) Same as F, but for old clones (n = 4). (H) Summary of the RSDs for all clones shown in D–G. Horizontal lines indicate mean value.
Both old and young HSC clones demonstrate a highly skewed skeletal distribution for at least 11 mo after transplantation
Mice were sacrificed 6–11 mo after transplantation to assess the dynamics of the migration process. Six mice were analyzed 6-mo after transplant, whereas additional groups of two mice each were analyzed 10 and 20 wk later (at 8.5 and 11 mo, respectively). Approximately 100 colonies per mouse (mean 98 ± 35; ranging from 38 to 160) were obtained and barcodes successfully analyzed. Detailed results of such an analysis for one of the mice are shown in Fig. 1 C. The distribution of different barcoded clones was highly diverse: although some clones were detected in all four analyzed locations (e.g., bottom [pink, #5] clone in panel on old cells), others were found only in one site. Such heterogeneity of spatial distribution was observed both for old and young HSC clones, and it was not related to the total number of clones found in an animal. The observed differences in clonal distribution were of two kinds. Some clones were present in some bones and absent in others. Other clones were present in all observed bones, yet their sizes were very different. As differences in the occurrence of barcode frequencies might be purely random or related to differences in the total number of clones analyzed (i.e., minor error of measurement caused by a relatively small colony number), we tested whether the observed skewed distributions were statistically significant or rather happening by chance. To this end the dataset was tested against the null hypotheses of equal skeletal distribution of HSCs. Only relatively abundant clones represented by at least four colonies were taken for analysis (smaller clones per definition cannot be found in four locations). The data for all mice sacrificed 6 mo after transplantation are summarized in Fig. 1 D (clones derived from young HSCs) and Fig. 1 E (clones derived from old HSCs). For individual clones, the contribution to the local stem cell compartment size at different locations could vary as much as from 8 to 83% or 0 to 79% (Fig. 1, D and E). It appeared that 8 out of 42 clones were significantly deviating from the expected mean (P < 0.05, marked with asterisks). To test whether skeletal distribution was equalized in time, four additional mice were analyzed 10 and 20 wk later (Fig. 1, F and G). Similarly to clones analyzed 6 mo after transplantation, we detected skewed clonal representation among the four skeletal sites. For 3 of 17 clones, this skewing was statistically significant, confirming that local dominance of HSC clones in certain skeletal regions is retained for at least 11 mo after transplantation.
Because previous studies demonstrated a decreased homing or binding to stroma of old HSCs (Liang et al., 2005; Xing et al., 2006; Dykstra et al., 2011), we questioned whether the extent of migration/spatial distribution of old HSCs across the skeleton was different from their young counterparts. To address this issue, we calculated the relative SD (RSD) for each of the clones found four or more times. The RSD allows us to compare variations in clones with different mean contributions. An RSD < 1 demonstrates low-level variance of distribution, whereas an RSD > 1 indicates high-level variance. Most young and old clones have RSD values >1, but the mean RSD for both age groups is very similar (Fig. 1 H), demonstrating once more that both populations have highly unequal skeletal distributions.
G-CSF–induced mobilization leads to redistribution of HSC clones
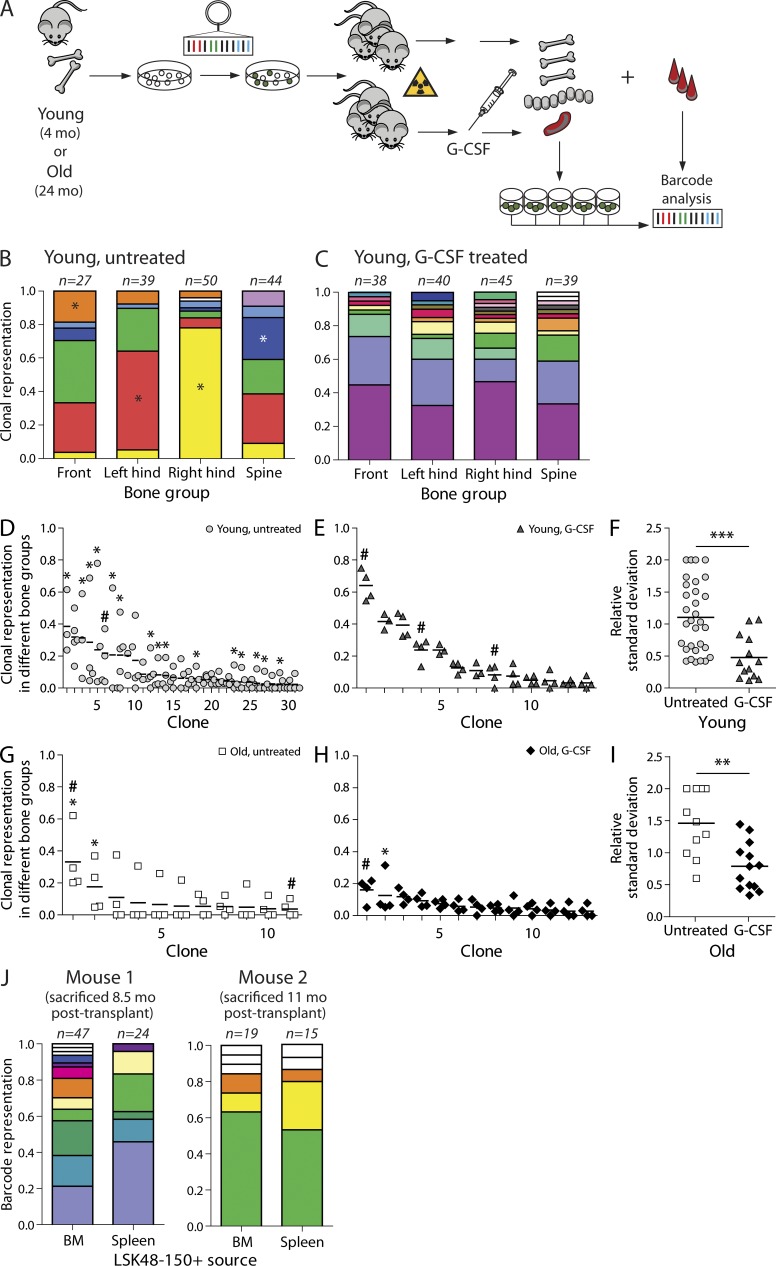
G-CSF administration mobilizes HSCs and progenitors into the circulation. G-CSF has also been shown to increase partner chimerism when administered to a parabiotic animal (Abkowitz et al., 2003). To assess whether G-CSF affects clonal skeletal distribution, we analyzed a cohort of seven animals transplanted with young (4 mo old) barcoded LSK48−150+ cells. As only one cell type was transplanted, it was possible to generate more in vitro colonies per clone and increase the accuracy of the measurements. Three mice were mobilized 7 mo after transplantation, whereas four mice were left untreated. 10–13 wk after G-CSF treatment, mice were sacrificed, and the barcode composition of the LSK48−150+ pool was analyzed in different locations (Fig. 2 A). Additionally, LSK48−150+ cells were sorted from the spleen. Similar to our results described above, skeletal distribution of HSC clones in all four untreated animals was highly asymmetrical (Fig. 2, B and D), and 15 of 31 clones were significantly skewed across bone sites. Strikingly, a single G-CSF challenge led to complete randomization of HSC clones across the four studied locations (Fig. 2, C and E). Skewing completely disappeared in this group (Fig. 2 E). This indicates that HSC clones are readily mobilized and reenter the niches after mobilization. Colony-forming efficiency of LSK48−150+ cells in treated and untreated animals was similar. We have also been able to retrieve up to three barcoded LSK48−150+ colonies in spleen per mouse. The clones present in spleen are marked with a hashtag in Fig. 2.
Figure 2.
G-CSF–induced mobilization leads to homogenization of skeletal HSC distribution of both young and old clones. (A) Experimental setup. Recipient mice were transplanted with young or old transduced LSK48−150+ cells. The groups of mice were challenged by G-CSF 7 (young mice) or 8 mo (old mice) after transplantation. 10–18 wk after treatment, all mice (both treated and untreated groups) were sacrificed. Clonal composition of different skeletal locations, spleen, and blood was investigated by barcode analysis. Recipients of young cells belong to one cohort of seven animals. Recipients of old LSK48−150+ cells relate to two cohorts (n = 4; one of the untreated animals originated from a separate cohort). (B) Example of clonal barcode representation in an untreated mouse transplanted with young cells. As in Fig. 1, identical colors relate to the same clone, and clones found only once are shown as white. (C) Same as B, but for a G-CSF–treated mouse. (D) Summary of skeletal clonal distributions in four recipient mice transplanted with young stem cells. The asterisks identify clones with distributions that are significantly different from random (P < 0.05 based on Monte Carlo simulations). (E) Same as D, but for G-CSF–treated recipients (n = 3). (F) Summary of RSDs for all clones found in four or more colonies for the groups shown in D and E. The difference between the groups was tested by two-tailed Mann–Whitney test (***, P = 0.0004). (G) Analysis of clonal skeletal distribution in two recipient animals transplanted with old LSK48−150+ cells. Statistical analysis was performed as in D and E. (H) Same as G for two G-CSF–treated recipients transplanted with old cells. (I) Summary of RSDs for all clones found in four or more colonies for the groups shown in G and H. The difference between the groups was assessed by two-tailed Mann–Whitney test (**, P = 0.0037). (D–I) Hashtags indicate the clones that were found in the spleen. Horizontal lines indicate mean contributions of a corresponding clone. (J) Comparison of clonal composition of LSK48−150+ cells in BM and spleen/blood 7 d after G-CSF treatment (n = 2). BM refers to pooled data from all four skeletal sites. Correlation between clone sizes in spleen and BM was significant for young cells in both mice (Pearson correlation, r = 0.62, P = 0.032; and r = 0.91, P < 0.001, respectively). Data for old cells are not shown because of low old cell chimerism in one of the analyzed mice.
It has been reported that the mobilization potential of HSCs might be affected by the age of the patient. For instance, older myeloma patients have been shown to be poorer mobilizers compared with young counterparts (Morris et al., 2003; Pozotrigo et al., 2013). However, mobilization in old mice was shown to be more efficient than in young individuals (Xing et al., 2006). We addressed this controversy at the clonal level: we studied the extent of skeletal allocation of old barcoded HSC clones (24 mo at time of transduction and transplantation; Fig. 2, G and H). Transplantations were performed as for recipients of young cells, and G-CSF was administered to half of the animals 8 mo after transplant. One of G-CSF–treated mice suffered from a spleen rupture and died 1 wk after injection. The two remaining mice were sacrificed 10 wk after mobilization. The response was similar to young clones (Fig. 2, F and I), with the exception of 1 out of 13 clones (clone 2; Fig. 2 H). Although most of the clones equilibrated after G-CSF challenge, clone 2 was dominant in spine (11 of 35 colonies carried this barcode). Although all young clones seem to be equally prone to mobilization, within the old cell population we cannot exclude the presence of both good and bad mobilizers.
To test this in a more direct manner, we studied clonal representation of the LSK48−150+ cells in BM and spleen at the peak of mobilization (1 wk after G-CSF administration). Recipients of cotransplanted young and old cells (as shown at Fig. 1 A) were used to study the response within the same animal. The animals were mobilized 8.5 or 11 mo after transplantation, and LSK48−150+ clones were harvested from BM and spleen (Fig. 2 J). The clonal composition of young LSK48−150+ cells in BM and spleen was very similar (r = 0.62, P = 0.032; and r = 0.91, P < 0.001 for mouse 1 and 2, respectively; Fig. 2 J). Data for aged cells are not shown because of low cell chimerism.
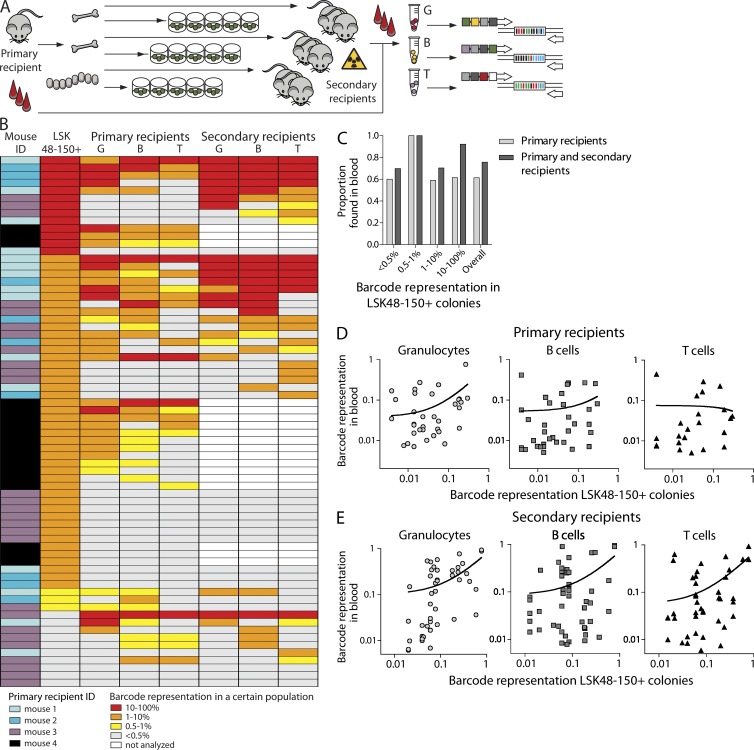
LSK48−150+ cells represent the functional HSC population with highly variable developmental potential
In naive mice, the LSK48−150+ phenotype allows us to purify a highly enriched HSC fraction (Kiel et al., 2005). However, ex vivo culture, transduction, and transplantation could in theory affect the phenotype of primitive cells. To confirm that LSK48−150+ colonies originated from functional HSCs, we compared barcode spectra in mature blood populations with those from single-cell expanded clones. Barcode fluctuations in blood samples were followed for a period of 10–11 mo in primary recipients and for an additional 6 mo in secondary recipients (Fig. 3 A). Granulocytes and T and B lymphocytes were purified from blood of primary and secondary transplant recipients by FACS, and barcodes were analyzed by high-throughput Solexa sequencing. Previously, we showed that clonal representation of the hematopoietic system becomes stable 3 mo after transplantation (Verovskaya et al., 2013); therefore, here we focused on maximum representation of a particular barcode in blood 12 wk and later after transplant. Results of such analysis for 4 primary recipients of young LSK48−150+ cells and 13 secondary recipients are summarized in Fig. 3 B. We considered a barcode present if it was detected at a level of at least 0.5% in a relevant population (Verovskaya et al., 2013). Of 70 barcodes detected in LSK48−150+ cells, 43 were found in primary recipients, 17 of them were found in both primary and secondary recipients, and 10 of the remaining 27 barcodes were detected only in secondary animals. Surprisingly, the probability to detect a particular barcode in blood of primary recipients was not proportional to the corresponding clone size in LSK48−150+ cells (Fig. 3 B). There seems to be no apparent relation between clone size of a particular stem cell and its repopulating activity measured in blood samples.
Figure 3.
Developmental potential of migrating HSC clones in primary and secondary recipients. (A) Experimental setup. Mature cells were purified from blood of primary and secondary transplant recipients by FACS, and barcodes were analyzed by high-throughput sequencing. Clonal composition in blood was compared with barcodes from expanded single stem cell–initiated colonies. (B) Barcode representation in primary LSK48−150+ colonies and in three blood populations in primary or secondary recipients is plotted. The graph summarizes data from 70 clones found in 4 primary recipients of young cells and 13 secondary recipients transplanted with cells from 3 of these primary recipients. For one of the primary recipients (corresponding to 18 barcodes), no secondary transplantations were performed. The colored legend identifies each of the four primary recipients from which the barcode data originate. Each row reflects a single clone/barcode and shows the maximum frequency with which that particular clone contributes to a particular lineage, measured over a period of observation starting at 12 wk after transplantation. (C) Proportion of clones contributing to blood formation in primary and secondary recipients as a function of the clone size in the LSK48−150+ pool. Note that overall ∼40% and ∼20% of the clones detected in BM LSK48−150+ cells remains undetected in blood in primary or secondary recipients, respectively. (D) Correlation between barcode representation in BM LSK48−150+ colonies and three mature blood populations in primary recipients (in one cohort of four mice). The best-fit line of linear regression analysis is shown (for granulocytes, slope [±CI95%] = 0.70 ± 0.26, P = 0.0125; for B cells, slope = 0.24 ± 0.21, P = 0.2776; and for T cells, slope = −0.07, P = 0.8). (E) Same as in D, but now in secondary recipients (13 mice). The slopes of the best-fit lines and p-values are as follows: for granulocytes, slope = 0.70 ± 0.16, P < 0.0001; for B cells, slope = 0.63 ± 0.18, P = 0.001; for T cells, slope = 0.63 ± 0.15, P = 0.0001.
We decided to study this issue of clone size representation in more detail. For the barcodes found in both blood and HSCs (reflecting active stem cells), we next asked whether the frequency of a barcode in LSK48−150+ pool was correlated to frequencies in granulocytes and B and T lymphocytes. Fig. 3 C summarizes such analysis in primary recipients. It is apparent that the barcode representation data are quite disparate, and no obvious trend can be seen. To test whether the barcode representation in blood and LSK48−150+ colonies from BM showed a linear relationship, we calculated the relation between these two parameters by testing whether the slope of the regression line was significantly different from zero (Fig. 3 D). For granulocytes, the slope was significantly different from the null hypothesis (P = 0.0125). For T and B cells, the difference was insignificant, indicating no linear relationship between two variables (P = 0.2776 and P = 0.8, respectively). We also tested Spearman correlation between different cell subsets: only LSK48−150+ and B cells significantly correlated, although the degree of correlation was low (ρ = 0.3725, P = 0.0275).
Several factors could contribute to the lack of linearity between barcode size in lymphocytes and LSK48−150+. First, lymphocytes are long-lived blood cells that persist in the circulation for months, in contrast to granulocytes that are replenished every 2 d. Thus, detected lymphocytes can originate from stem cells that have exhausted long before analysis took place. Second, as only a fraction of bones was taken for analysis, several clones (contributing to B and T cell production) could be missed in the analysis. As these factors are irrelevant in secondary transplantations, we compared barcodes in blood of 13 secondary recipients to the LSK48−150+ pool composition in primary animals (Fig. 3 E). For all three populations (granulocytes and B and T cells), clonal distribution in blood was proportional to the corresponding LSK48−150+ pool of the donor, and the slope was significantly different from zero (P < 0.0001, P = 0.001, and P = 0.0001, respectively). Although in the primary animals only B cells and BM composition were significantly correlating, in secondary recipients clonality of both granulocytes and T cells significantly correlated with that of primitive cells originating from primary recipient (for granulocytes, ρ = 0.6704, P < 0.0001; for T cells, ρ = 0.3303, P = 0.0349). Note, however, that this is a mean regression for all clones. Individual data are scattered, and the degree of variation is substantial. It strongly argues against a simple relation between clone size in BM and blood. Obviously, there are other factors than clone size in the BM that affect clonal representation in the blood.
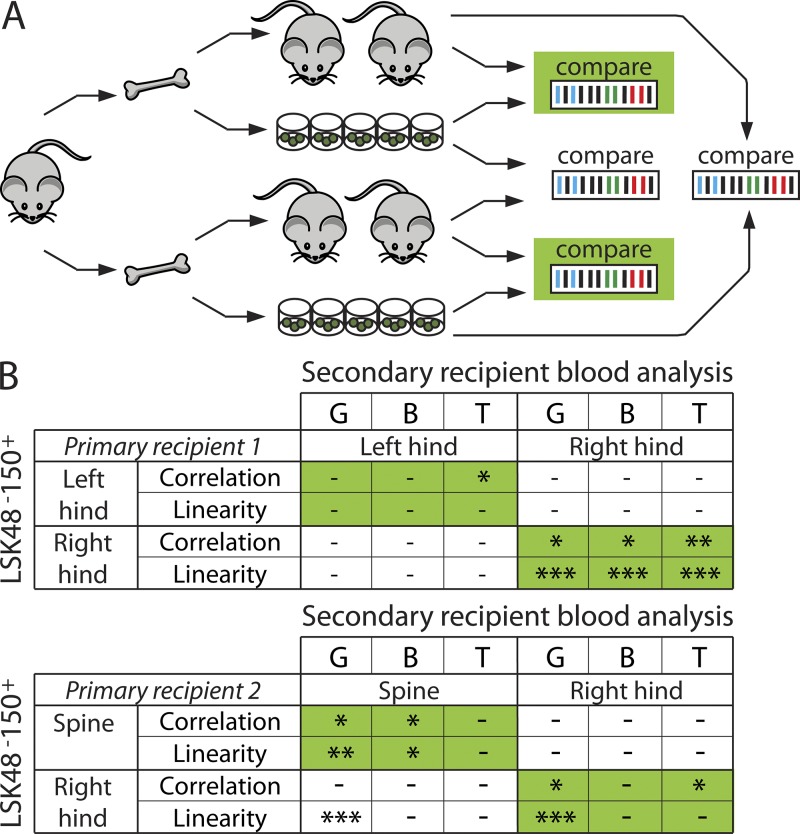
Clonal composition of secondary transplant recipients depends on donor bone of origin and resembles the clonal composition of donor LSK48−150+
As the clonal composition of the HSC pool differs in different skeletal locations, this could have ramifications on the interpretation of BM biopsy composition or on the clonality of secondary recipients, resulting in an underestimated clonal representation compared with the primary recipient. We tested whether the clonal composition of blood in secondary recipients, transplanted with BM from a single skeletal site, was similar to the HSC (LSK48−150+) pool of the corresponding donor bone. As a negative control, we compared the data with those obtained from another skeletal site from the very same donor mouse. As some clones found in the LSK48−150+ cell population were not contributing to blood (Fig. 3), these could not be taken for similarity analysis using correlation estimates. Therefore, we focused only on the barcodes found in blood at levels of >1% in either primary or secondary animals that correspond to the active stem cells and excluded barcodes that occurred less frequently. We further analyzed correlations and tested linear relationships between barcode make-up of the LSK48−150+ pool in a certain skeletal site and matched (marked in green) or unmatched (marked in white) secondary recipients (Fig. 4). Despite large variation in the amount of output of mature blood cells per LSK48−150+ clone (Fig. 3), both linearity and correlation were higher in the matched samples (Fig. 4 A, green). In 8 out of 12 cases of blood versus BM comparisons, barcode makeup of blood in secondary recipients was significantly similar to the barcode composition of LSK48−150+ cells in the donor bone of origin (P < 0.05). In contrast, correlations were insignificant in all unmatched samples. Similarly, we observed good linearity between clone sizes in blood and HSCs in six groups (P < 0.05). Among unmatched groups, linear dependence was observed in one case (where the granulocytic population showed good fit to the bones of right hind leg, while originating from spine), further underscoring restricted interdependence between primitive and mature cells. This example demonstrates that although Pearson correlation and linear regression both are testing linear relationship between two sets of data, the results might be different, and conclusions should be drawn with care.
Figure 4.
Clonal blood composition in secondary recipients reflects the clonal HSC makeup of bone of origin. (A) Experimental setup. BM cells were isolated from two primary recipients (left and right hind legs from recipient 1 and spine and right hind leg from recipient 2) and were transplanted in four secondary recipients each. In parallel with secondary transplant, barcode frequencies were assessed in donor LSK48−150+ cells. Granulocytes (G) and B and T cells were isolated from blood of secondary recipients 12–24 wk after transplant, and clonal analysis was performed by high-throughput sequencing. The relation between barcode representation in LSK48−150+ colonies and blood of secondary transplant recipients was evaluated in matched (green) and unmatched (unmarked) samples. (B) The clonal composition of HSCs in the LSK48−150+ pool in a certain skeletal site was compared with blood composition of related and unrelated secondary recipients using Spearman correlation and linear regression analysis. The tables summarize conditions in which correlation was significant and the slope was significantly different from zero. Significance levels are as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
Understanding the kinetics and molecular events underlying trafficking of HSCs is important both in basic hematology research and for implementation and interpretation of experimental and clinical BM transplantation protocols. Here, we studied posttransplantation skeletal distribution of hundreds of HSC clones to address the extent of migration in steady-state conditions and upon G-CSF–induced mobilization. Genetic barcoding of highly purified hematopoietic cells was used to quantitatively analyze HSC clone sizes in different bones and contributions of these clones to blood.
Our findings demonstrate that at very extended time intervals (at least 11 mo) after transplantation, the distribution of both old and young HSC clones across multiple skeletal sites is highly skewed. This is in line with data showing relatively slow rates of HSC equilibration in parabiotic mice (Abkowitz et al., 2003). However, our findings are in contrast with a study that suggested that 1–5% of HSC pool circulates daily (Bhattacharya et al., 2009). Simple dilution model simulation indicates that at such rates the clonal make-up of the HSC pool would equalize within 4–6 mo (unpublished data). This prediction strongly contrasts our experimental observations. Notably, previous studies were never performed on the clonal HSC level, and therefore inequality in the level of egress of distinct HSC clones within one mouse was never addressed.
Strikingly, a single G-CSF mobilization regimen resulted in uniform spreading of HSC clones, which was preserved for many weeks after treatment. Here our data are quite distinct from results obtained in parabiotic models, in which only partial equilibration of the HSC pool occurs (Abkowitz et al., 2003). Our data confirm that mobilized HSCs retain the ability to reengraft BM niches. They also strongly suggest that all young HSCs are equally prone to mobilization; we did not find any evidence of mobilization of only a selected subset of stem cell clones.
Heterogeneity of the spatial clonal distribution of stem cells, as demonstrated here, is likely the combined result of multiple factors, such as the number, size, and migratory potential of distinct HSCs. It has recently been shown that multiple (biologically and physically) separate niches may exist (Lo Celso and Scadden, 2011; Ding and Morrison, 2013; Greenbaum et al., 2013). One might speculate that certain HSCs home and expand in one but not another niche. Although we have not addressed whether specific clones were found in specific niches, differential niche preference could potentially contribute to heterogeneity in the observed localization patterns as well. This issue could potentially be resolved by further subfractionation of HSCs from increasingly more defined physical regions of the BM if it allows separation of different niches. It is recognized that G-CSF induces mobilization through affecting specific ligand-receptor interactions and by inducing proteolytic cleavage of receptors (Lapid et al., 2012). G-CSF affects not only HSCs, but also mature hematopoietic cells, as well as other cell types collectively referred to as the niche (osteoblasts, osteoclasts, and mesenchymal cells; Lapid et al., 2012). G-CSF treatment will inevitably cause modification of the niche. How this modification is contributing to the randomization of the clonal distribution is currently not known.
The fact that G-CSF efficiently equilibrates practically all clones indicates that each individually labeled HSC achieved sufficient clonal outgrowth to guarantee redistribution of its offspring across all bones tested. This observation also predicts that naturally occurring infections in mice, when endogenous G-CSF levels are elevated, will induce spatial equilibration of HSC clones. The physiological relevance of HSC redistribution remains unexplored.
Previous studies suggest differences in mobilization abilities of young and aged HSCs (Morris et al., 2003; Xing et al., 2006; Pozotrigo et al., 2013), but extrinsic and intrinsic effects could not be discriminated. Here, we observed similar migratory abilities of young and old populations in co-transplantation settings, in transplants of only young or old cells, and upon mobilization, demonstrating that putative changes in mobilization with age must be primarily stem cell extrinsic, highlighting the possible role of niche ageing in modulating HSC mobilization.
Although barcode clonal analysis is potentially a powerful tool, several putative caveats must be considered. First, HSC activation during gene transfer procedures could affect their function. Although we (inevitably) did use ex vivo transduction and transplantation to follow HSCs, we demonstrate that these cells retain the ability for multilineage differentiation in irradiated hosts. The extent of heterogeneity of repopulating ability and developmental capacity of individual HSCs observed is in agreement with experiments with transplantation of unmanipulated single cells (Ema et al., 2005; Dykstra et al., 2007). Therefore, we are confident that qualitatively our observations are not affected by the method, but rather seem to reflect the natural clonal behavior of repopulating HSCs. Second, we determined barcode identities only in single LSK48−150+ cells that were able to form a colony in vitro. Although this approach allowed us to confirm high proliferative activity of purified cells, bona fide stem cells that fail to produce a colony in our culture conditions would remain unnoticed. Alternatively, single-cell barcode analysis could be used; however, it is currently problematic because of the low success rate of PCR barcode amplification in a single cell. Third, this research was performed in irradiated recipients, and it remains a subject of investigation whether it affects HSCs migration.
Our findings have important implications for the design and interpretation of experimental and clinical transplantation protocols. First, we show that the current routine of using a single femur and tibia as a source for BM cells to initiate secondary transplants in mice results in an underestimation of the clonal repertoire and will exclude (potentially dominant) clones located elsewhere. This can contribute to a lack of consistency between the clonal make-up of BM and blood, and the use of cells obtained from multiple skeletal bones is advisable. Second, the same limitations apply for using BM biopsies to monitor clonal fluctuations in mice, cats, dogs, monkeys, or indeed in human gene therapy patients. Previous research on xenotransplantation of human cells into immunodeficient mice indicated that the clonal composition of BM in individual bones varied (Mazurier et al., 2003). However, the authors interpreted local clonal dominance as a sign of functional differences between subsets of human HSCs, rather than reflecting general patterns of HSC distribution (Mazurier et al., 2003).
In conclusion, the data presented here demonstrate that upon transplant functional HSC clones preferentially expand in certain skeletal locations, exhibiting only limited migration toward other niches. High cytokine levels, which are characteristic of infections and are clinically widely used to induce stem cell mobilization, result in rapid and permanent homogenization of clonal stem cell distribution.
MATERIALS AND METHODS
Mice.
C57BL/6 mice were purchased from Harlan, and C57BL/6.SJL and C57BL/6.SJLxC57BL/6 (all referred to as B6) mice were bred in the Central Animal Facility of the University Medical Center Groningen. C57BL/6J-kitW-41J/kitW-41J (W41) mice were originally obtained from E. Dzierzak (Erasmus University Medical Center, Rotterdam, Netherlands) and were crossed with B6.SJL to obtain CD45.1 W41 as described previously (Dykstra et al., 2011). All experiments were approved by the University of Groningen Animal Experimentation Committee.
Purification and transduction of BM cells.
LSK48−150+ cell sorting and transduction were performed as described previously (Verovskaya et al., 2013). In brief, BM cells were isolated from bones of hind legs, spines, and sterna of naive B6 mice and stained with antibodies against Sca1, c-Kit, CD48, CD150, CD3ε, Gr1, CD11b, Ter119, and B220. LSK48−150+ cells were sorted using MoFlo XDP and MoFlo Astrios (Beckman Coulter) cell sorters and prestimulated in StemSpan medium (STEMCELL Technologies) supplemented with 300 ng/ml stem cell factor (SCF), 1 ng/ml Flt3 ligand (both Amgen), and 20 ng/ml IL-11 (R&D Systems) for 24 h. Cells were transduced with viral supernatant containing barcoded MIEV vectors in RetroNectin-covered plates (Takara Bio Inc.) in the presence of 2 µg/ml polybrene (Sigma-Aldrich).
BM cell transplantation.
20–22 h after transduction, transduced cells were transplanted into lethally irradiated recipients (9–9.5 Gy) in the presence of 1–2 million radioprotective cells from W41 or B6 origin. Efficiency of gene transfer, measured by flow cytometry (LSR-II; BD), was similar in all experiments and ranged from 19 to 34% GFP+ cells. CD45 congenic B6 and W41 strains were used to discriminate between donor and recipient cells and to distinguish young and aged cells in co-transplant setting. In the first young/aged cohort, aged cells were CD45.2+, young cells were CD45.1+/CD45.2+, and recipients and donors of radioprotector cells were CD45.1+. In the second cohort, aged cells were CD45.1+, young cells were CD45.2+, and recipients/radioprotector cells were CD45.1+/CD45.2+. Young donors were 4 mo old, aged donors were 24 mo old, and recipient animals were 2–6 mo old. For co-transplantations, young and aged cells were mixed in a 1:2 ratio before transduction and 20,500 cells were injected into the recipients. For young only cells, 10,000 cells were transplanted. For recipients of old HSCs, injected doses were 12,000 and 30,000 cells, respectively, in two cohorts. Transplanted transduced cells supported robust multilineage blood production. Donor and GFP chimerism were determined in blood 6 mo after transplantation as described in Clonal analysis of blood samples. In old/young co-transplants (Figs. 1 and 2 J), old cells contributed 8.0 ± 7.3% of B cells (17.2 ± 13.2% GFP), 6.3 ± 5.1% of T cells (17.1 ± 14.1% GFP), and 15.7 ± 6.8% of granulocytes (22.7 ± 16.0% GFP), whereas young HSCs generated 81.2 ± 8.5% of granulocytes (14.3 ± 10.9% GFP), 75.4 ± 12.6% of T cells (9.7 ± 6.6% GFP), and 89.5 ± 8.8% of B cells (9.7 ± 4.2% GFP). In recipients of young cells (Fig. 2, B–F; and Fig. 3), donor HSCs produced 81.0 ± 8.2% of granulocytes (15.5 ± 8.2% GFP), 65.1 ± 11.8% of B cells (9.9 ± 4.8% GFP), and 61.2 ± 8.0% of T cells (10.4 ± 4.7% GFP). In recipients of old HSCs (Fig. 2, G–I), these values were as follows: 87.8 ± 4.3% of granulocytes (23.1 ± 5.5% GFP), 75.9 ± 10.4% of B cells (16.8 ± 10.7% GFP), and 60.9 ± 16.8% of T cells (14.1 ± 15.6% GFP). For secondary transplantations, 5–8 million whole BM cells were transplanted into lethally irradiated recipients.
G-CSF administration.
To induce mobilization, we performed two injections of PEGylated G-CSF intraperitoneally (25 µg/mouse) with a 3-d interval between treatments. In two mice sacrificed 1 wk after G-CSF administration, the efficiency of mobilization was confirmed by the presence of CFUs in the blood samples.
Isolation of LSK48−150+ from distinct skeletal locations after transplantation.
Transplant recipients were sacrificed by cervical dislocation under isoflurane anesthesia. Bones were isolated separately into four groups: (1) bones of fore limbs and sternum (front); (2) left femur, tibia, and left half of pelvic bone (left hind bones); (3) right femur, tibia, and right half of pelvic bone (right hind bones); and (4) bones of spine. After removal of muscle, cells were harvested by crushing bones in lysis solution (NH4Cl) and filtered through a 100-µm filter to remove debris. Nucleated cells were washed and stained with an antibody cocktail. Cells were washed and resuspended in a 1-µg/ml solution of propidium iodide (PI). Antibodies were directed against Sca1, c-Kit, a panel of lineage markers (Ter119, CD11b, CD3ε, B220, and Gr1), CD48, and CD150 to discriminate different cell populations. CD45.1 and/or CD45.2 staining was used to further distinguish donor and recipient cells. LSK CD48− cells from different locations were isolated in “enrich mode,” and donor-derived (young and/or old) LSK48−150+ GFP+ cells were further single-cell sorted in round-bottom 96-well plates in cytokine-supplemented medium to generate monoclonal cultures. Only 60 inner wells were used to minimize medium evaporation. Approximately 120 cells were sorted per bone group per mouse.
Monoclonal expansion of LSK48−150+ cells.
Colony growth was assessed by light microscopy at low (5×) magnification at several time points 10–35 d after sorting. Most large colonies formed within 2 wk of culture. Colonies reaching ∼30,000 cells or more (colony diameter reaching ∼60% of the diameter of the viewing field) were used for barcode analysis. ∼45% of the sorted LSK48−150+ cells formed large colonies and were harvested (this efficiency varied from 38 to 49% over four cohorts of mice used for analysis). The efficiency of LSK48−150+ cells to generate a large colony was not affected by G-CSF, 10–13 wk after its administration. It constituted 50% and 52% for untreated and G-CSF groups, respectively, in the cohort transplanted with young LSK48−150+ cells and 52% and 44% correspondingly for untreated and treated recipients of old HSCs. In two mice sacrificed at the peak of mobilization, 14% of sorted cells were capable of forming a large colony. In some cases, young/old origin of the clones was established after expansion by PCR and restriction analysis for CD45 polymorphisms (Ramos et al., 2003).
Clonal analysis of LSK48−150+ colonies.
Genomic DNA was extracted and barcoded DNA was amplified with primers against internal vector sequence (Gerrits et al., 2010). Sanger sequencing was used to detect barcodes present in the samples. When less than five colonies could be retrieved from a BM sample, data were excluded from analysis.
Clonal analysis of spleen samples.
Spleens were excised, weighed, and pressed through a 100-µm filter to generate single-cell suspensions. Red blood cells were lysed in NH4Cl solution, and cell suspensions were stained with an antibody cocktail as described in Isolation of LSK48−150+ from distinct skeletal locations after transplantation. Splenic LSK48−150+ GFP+ cells were individually sorted to generate monoclonal cultures as described in Monoclonal expansion of LSK48−150+ cells.
Clonal analysis of blood samples.
Clonal analysis in blood was performed by next-generation deep sequencing, as previously described (Verovskaya et al., 2013). In brief, blood samples were collected every 6 wk for at least 6 mo and at the moment of sacrifice. Erythrocytes were lysed in NH4Cl buffer, and cells were stained with fluorophore-conjugated antibodies against Gr1, B220, CD3ε, CD45.1, and CD45.2, washed, and resuspended in PI solution. Mature blood populations were purified by FACS. Viable (PI−) granulocytes (Gr1+ side scatter high), T cells (CD3+), and B cells (B220+) of donor origin (CD45.1+) were sorted. Barcode sequence was amplified with tagged primers to allow multiplex analysis, pooled, and analyzed on an Illumina HiSeq 2000 platform. Data extraction was performed using in-house developed scripts in Python, Perl, Bioconductor, R, and VBA, as previously described (Verovskaya et al., 2013). In short, high-quality reads were extracted and compressed into sets of unique sequences. Samples were retrieved on the basis of exactly matching the primer tag. Barcode sequences were extracted using Motif Occurrence Detection Suite (MOODS; Korhonen et al., 2009). Barcodes were sorted by read frequencies in descending order, and barcode pairs varying by a single nucleotide were compared and lower frequency barcodes removed (removing barcodes with single-nucleotide substitution). Unique barcodes with frequencies under 0.5% of total within the sample were removed (technical threshold). Samples with <1,000 reads were excluded from further analysis.
Statistical analysis.
For testing the statistical significance of variations of clone size among different bone groups, experimental data series were compared with expected random fluctuation around the mean. As the number of barcoded clones retrieved from different bones varied, we first normalized all clone frequencies to the bone location with the smallest number of successfully retrieved clones. A custom script in Python was used to test the probability of random deviation from mean (10,000 trials) for a given set of a data points (number of bone compartments), as well as total sequence reads (number of trials). The script allows us to simulate distribution of a certain number of reads (normalized number of reads related to a single clone) over several bone groups. Based on 10,000 trials, the script calculates probability of observing a deviation from the mean in a single bone. When the probability to detect the observed deviation was less than P < 0.05, this deviation was considered significant. This protocol allows us to identify all significantly deviating barcodes individually.
For correlation and linear regression analyses, coefficients of correlation (in the text r for Pearson, and ρ for Spearman) and p-values were derived using Prism (GraphPad Software). For linear regression analysis, when the slope deviated from zero, this was used as an indicator of linear relationship between the populations. For comparing RSD values in G-CSF–treated and nontreated animals, we used the nonparametric Mann–Whitney test, which allows comparison of not-normally distributed data. P-values were calculated using Prism.
Throughout this paper we mostly used correlation coefficients (Pearson and Spearman) as a measure of similarity between clonal composition of different samples. These functions are quantitative and therefore have particular limitations, especially in the case of poorly overlapping clone sets and sets with small numbers of barcodes. Nevertheless, they are well known and widely used, and when used properly provide sufficient statistical information.
Acknowledgments
We thank H. Moes, G. Mesander, and R.-J. van der Lei for expert cell sorting assistance; E. Wojtowicz for laboratory assistance; B. Dykstra, H. Schepers, and R. van Os for valuable discussions and suggestions; and P. van der Vlies and J. Bergsma for assistance with high-throughput sequencing.
This study was supported by the Netherlands Organization for Scientific Research (VICI grant to G. de Haan and TopTalent grant to E. Verovskaya), the National Roadmap for Large Scale Infrastructure (Mouse Clinic for Cancer and Aging), and the Netherlands Institute for Regenerative Medicine (NIRM).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- HSC
- hematopoietic stem cell
- PI
- propidium iodide
- RSD
- relative SD
References
- Abkowitz J.L., Robinson A.E., Kale S., Long M.W., Chen J. 2003. Mobilization of hematopoietic stem cells during homeostasis and after cytokine exposure. Blood. 102:1249–1253 10.1182/blood-2003-01-0318 [DOI] [PubMed] [Google Scholar]
- Bhattacharya D., Czechowicz A., Ooi A.G., Rossi D.J., Bryder D., Weissman I.L. 2009. Niche recycling through division-independent egress of hematopoietic stem cells. J. Exp. Med. 206:2837–2850 10.1084/jem.20090778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Morrison S.J. 2013. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 495:231–235 10.1038/nature11885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorie M.J., Maloney M.A., Patt H.M. 1979. Turnover of circulating hematopoietic stem cells. Exp. Hematol. 7:483–489 [PubMed] [Google Scholar]
- Dykstra B., Kent D., Bowie M., McCaffrey L., Hamilton M., Lyons K., Lee S.J., Brinkman R., Eaves C. 2007. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 1:218–229 10.1016/j.stem.2007.05.015 [DOI] [PubMed] [Google Scholar]
- Dykstra B., Olthof S., Schreuder J., Ritsema M., de Haan G. 2011. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J. Exp. Med. 208:2691–2703 10.1084/jem.20111490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema H., Sudo K., Seita J., Matsubara A., Morita Y., Osawa M., Takatsu K., Takaki S., Nakauchi H. 2005. Quantification of self-renewal capacity in single hematopoietic stem cells from normal and Lnk-deficient mice. Dev. Cell. 8:907–914 10.1016/j.devcel.2005.03.019 [DOI] [PubMed] [Google Scholar]
- Gerrits A., Dykstra B., Kalmykowa O.J., Klauke K., Verovskaya E., Broekhuis M.J., de Haan G., Bystrykh L.V. 2010. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood. 115:2610–2618 10.1182/blood-2009-06-229757 [DOI] [PubMed] [Google Scholar]
- Goodman J.W., Hodgson G.S. 1962. Evidence for stem cells in the peripheral blood of mice. Blood. 19:702–714 [PubMed] [Google Scholar]
- Greenbaum A., Hsu Y.M., Day R.B., Schuettpelz L.G., Christopher M.J., Borgerding J.N., Nagasawa T., Link D.C. 2013. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 495:227–230 10.1038/nature11926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel M.J., Yilmaz O.H., Iwashita T., Yilmaz O.H., Terhorst C., Morrison S.J. 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 121:1109–1121 10.1016/j.cell.2005.05.026 [DOI] [PubMed] [Google Scholar]
- Körbling M., Burke P., Braine H., Elfenbein G., Santos G., Kaizer H. 1981. Successful engraftment of blood derived normal hemopoietic stem cells in chronic myelogenous leukemia. Exp. Hematol. 9:684–690 [PubMed] [Google Scholar]
- Korhonen J., Martinmäki P., Pizzi C., Rastas P., Ukkonen E. 2009. MOODS: Fast search for position weight matrix matches in DNA sequences. Bioinformatics. 25:3181–3182 10.1093/bioinformatics/btp554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapid K., Glait-Santar C., Gur-Cohen S., Canaani J., Kollet O., Lapidot T. 2012. Egress and mobilization of hematopoietic stem and progenitor cells: A dynamic multi-facet process. StemBook, editor. The Stem Cell Research Community, StemBook, Cambridge, MA: http://www.stembook.org/node/762 (accessed July 15, 2013) [PubMed] [Google Scholar]
- Liang Y., Van Zant G., Szilvassy S.J. 2005. Effects of aging on the homing and engraftment of murine hematopoietic stem and progenitor cells. Blood. 106:1479–1487 10.1182/blood-2004-11-4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Celso C., Scadden D.T. 2011. The haematopoietic stem cell niche at a glance. J. Cell Sci. 124:3529–3535 10.1242/jcs.074112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurier F., Doedens M., Gan O.I., Dick J.E. 2003. Rapid myeloerythroid repopulation after intrafemoral transplantation of NOD-SCID mice reveals a new class of human stem cells. Nat. Med. 9:959–963 10.1038/nm886 [DOI] [PubMed] [Google Scholar]
- Morris C.L., Siegel E., Barlogie B., Cottler-Fox M., Lin P., Fassas A., Zangari M., Anaissie E., Tricot G. 2003. Mobilization of CD34+ cells in elderly patients (≥70 years) with multiple myeloma: Influence of age, prior therapy, platelet count and mobilization regimen. Br. J. Haematol. 120:413–423 10.1046/j.1365-2141.2003.04107.x [DOI] [PubMed] [Google Scholar]
- Orkin S.H., Zon L.I. 2008. Hematopoiesis: An evolving paradigm for stem cell biology. Cell. 132:631–644 10.1016/j.cell.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozotrigo M., Adel N., Landau H., Lesokhin A., Lendvai N., Chung D.J., Chimento D., Riedel E., Chen X., Reich L., et al. 2013. Factors impacting stem cell mobilization failure rate and efficiency in multiple myeloma in the era of novel therapies: Experience at Memorial Sloan Kettering Cancer Center. Bone Marrow Transplant. 48:1033–1039 10.1038/bmt.2012.281 [DOI] [PubMed] [Google Scholar]
- Ramos C.A., Zheng Y., Colombowala I., Goodell M.A. 2003. Tracing the origin of non-hematopoietic cells using CD45 PCR restriction fragment length polymorphisms. Biotechniques. 34:160–162 [DOI] [PubMed] [Google Scholar]
- Richman C.M., Weiner R.S., Yankee R.A. 1976. Increase in circulating stem cells following chemotherapy in man. Blood. 47:1031–1039 [PubMed] [Google Scholar]
- To L.B., Haylock D.N., Simmons P.J., Juttner C.A. 1997. The biology and clinical uses of blood stem cells. Blood. 89:2233–2258 [PubMed] [Google Scholar]
- Verovskaya E., Broekhuis M.J., Zwart E., Ritsema M., van Os R., de Haan G., Bystrykh L.V. 2013. Heterogeneity of young and aged murine hematopoietic stem cells revealed by quantitative clonal analysis using cellular barcoding. Blood. 122:523–532 10.1182/blood-2013-01-481135 [DOI] [PubMed] [Google Scholar]
- Warren S., Chute R.N., Farrington E.M. 1960. Protection of the hematopoietic system by parabiosis. Lab. Invest. 9:191–198 [PubMed] [Google Scholar]
- Wright D.E., Wagers A.J., Gulati A.P., Johnson F.L., Weissman I.L. 2001. Physiological migration of hematopoietic stem and progenitor cells. Science. 294:1933–1936 10.1126/science.1064081 [DOI] [PubMed] [Google Scholar]
- Xing Z., Ryan M.A., Daria D., Nattamai K.J., Van Zant G., Wang L., Zheng Y., Geiger H. 2006. Increased hematopoietic stem cell mobilization in aged mice. Blood. 108:2190–2197 10.1182/blood-2005-12-010272 [DOI] [PMC free article] [PubMed] [Google Scholar]