The development of serrated polyps in the cecum is driven by the interplay among genetic changes in the host, an inflammatory response, and a host-specific microbiota.
Abstract
The preferential localization of some neoplasms, such as serrated polyps (SPs), in specific areas of the intestine suggests that nongenetic factors may be important for their development. To test this hypothesis, we took advantage of transgenic mice that expressed HB-EGF throughout the intestine but developed SPs only in the cecum. Here we show that a host-specific microbiome was associated with SPs and that alterations of the microbiota induced by antibiotic treatment or by embryo transfer rederivation markedly inhibited the formation of SPs in the cecum. Mechanistically, development of SPs was associated with a local decrease in epithelial barrier function, bacterial invasion, production of antimicrobials, and increased expression of several inflammatory factors such as IL-17, Cxcl2, Tnf-α, and IL-1. Increased numbers of neutrophils were found within the SPs, and their depletion significantly reduced polyp growth. Together these results indicate that nongenetic factors contribute to the development of SPs and suggest that the development of these intestinal neoplasms in the cecum is driven by the interplay between genetic changes in the host, an inflammatory response, and a host-specific microbiota.
The preferential localization of some neoplasms in specific areas of the intestine suggests that nongenetic factors may be important for their development. In contrast to adenomas, which occur throughout the large intestine, serrated polyps (SPs) occur in specific areas of the gut in a subtype-specific manner (Huang et al., 2004; Noffsinger, 2009). SPs encompass a heterogeneous set of lesions and are associated with perturbations of the MAPK pathway through, e.g., activating mutations in KRAS or BRAF (Noffsinger, 2009).
We have recently shown that EGFR activation is associated with SPs in human biopsies and that expression of the EGFR ligand HB-EGF in transgenic mice (HBUS mice) promotes development of SPs that mostly resemble human hyperplastic polyps (Bongers et al., 2012). Strikingly, despite expression of the HB-EGF transgene throughout the gut, SPs were only observed in the cecum, suggesting that beside genetic alterations, environmental factors played a pivotal role in their development.
In the intestine, the microbiota is in close proximity to the intestinal epithelial cells that form a protective barrier separating commensal bacteria from the host. Changes in the microbiota have been associated with inflammatory conditions and cancer (Plottel and Blaser, 2011; Honda and Littman, 2012; Schwabe and Jobin, 2013). For example, activation of TLR4 by the intestinal microbiota is important for the promotion of hepatocellular carcinoma induced by the carcinogen diethylnitrosamine/CCl4 (Dapito et al., 2012). In the gastrointestinal (GI) tract, the presence of Helicobacter pylori is strongly associated with an increased risk for the development of peptic ulcers, gastric mucosa–associated lymphoid tissue tumors, and gastric adenocarcinomas (McColl, 2010). The colonic microbiota has also been suggested to play a role in the pathogenesis of colorectal cancer (Cho and Blaser, 2012), either by provoking an inflammatory response or alterations in metabolic processing (Plottel and Blaser, 2011). Specifically, activation of Th17 cells by the enterotoxigenic Bacteroides fragilis has been shown to promote colonic tumorigenesis in the APCmin mouse model (Wu et al., 2009). More recently, the polyketide synthase genotoxic island expressed by Escherichia coli NC101 was implicated in the development of carcinomas in Il10−/− mice treated with the colonic carcinogen azoxymethane (Arthur et al., 2012).
In this study we explored the possibility that the topological distribution of SPs was dependent on the microbiota. We show that alterations of the microbiota induced by antibiotic treatment or by embryo transfer rederivation attenuated the formation of cecal SPs in HBUS mice. SP development was associated with bacterial invasion of the lamina propria, which was associated with production of Il-17 by innate immune cells, neutrophil recruitment, and production of antimicrobials. Together these results indicate that the interplay between genetic changes in the host, an inflammatory response, and a host-specific microbiota accounts for the development of SPs in HBUS mice.
RESULTS
Antimicrobial defense response genes are induced in SPs
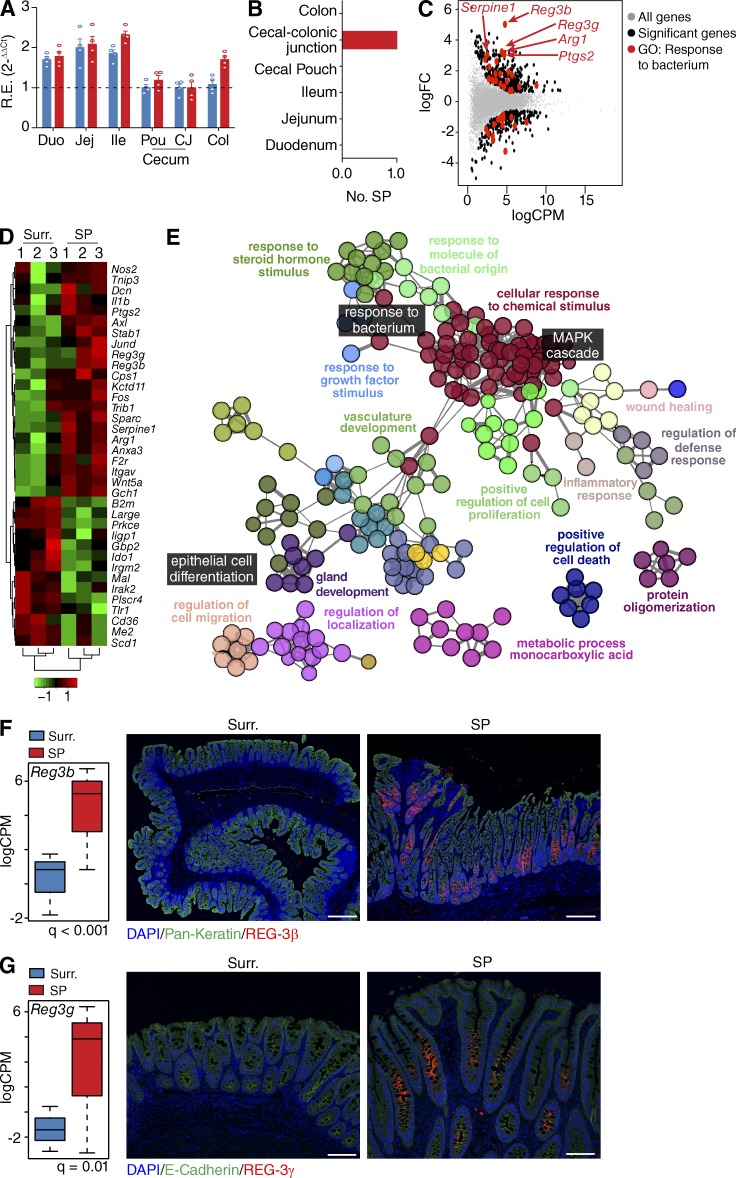
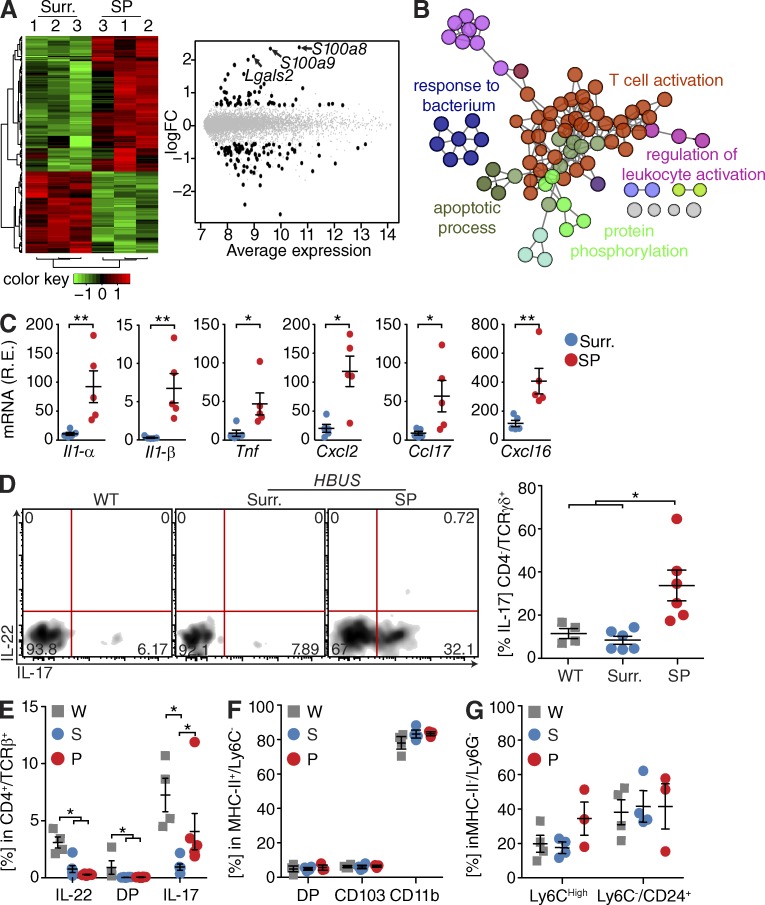
Expression of the transgenes in HBUS mice (Bongers et al., 2012) is highest in the small intestine (Fig. 1 A), but HBUS mice only develop SPs at the cecal–colonic junction (Fig. 1 B). To further elucidate potential pathways involved in the formation of SPs, we compared the transcriptome of SPs in HBUS mice with adjacent histologically normal cecal tissue (n = 3). Whole transcriptome sequencing (RNA-Seq) revealed 404 genes with increased and 272 genes with decreased expression in SPs, respectively (Fig. 1 C and Table S1; false discovery rate [FDR] Q < 0.05). Gene Ontology (GO) analysis of all 676 differentially regulated genes using ClueGO (Bindea et al., 2009) found significant enrichment of 59 overview GO terms such as “inflammatory response” and “cellular response to chemical stimulus” that encompassed the “MAPK cascade” GO term (Fig. 1 E and Table S2; Q < 0.05). Consistent with the genetic alterations in HBUS mice and our earlier observations (Bongers et al., 2012), we see significant enrichment of the overview GO term “response to growth factor stimulus” and induction of genes associated with HB-EGF and EGFR signaling, including the Egfr and its ligands Ereg, Areg, and Nrg1.
Figure 1.
Expression changes in SPs of HBUS mice. (A) Expression of HBUS transgenes (n = 4) US28 (red) and HB-EGF (blue) in duodenum (Duo), jejunum (Jej), Ileum (Ile), cecal pouch (Pou), cecal–colonic junction (CJ), and colon (Col). The experiment was performed once. R.E., relative expression. (B) Number of SPs found in various gut segments of HBUS mice (n = 210). (C) HBUS SPs and surrounding (Surr.) cecal tissue were analyzed by RNA-Seq/edgeR using a paired design (n = 3/group). Plot of logFC (log fold change) versus logCPM (log counts per million) of all detected transcripts. Points are colored according to expression status: nonsignificant genes (gray), significant genes (676 genes; Q < 0.05; black), and defense response genes (red). The experiment was performed once. (D) Z-scored heat map of genes associated with the GO term “response to bacterium”; red indicates increased and green decreased expression in SPs compared with surrounding tissue. (E) ClueGO analysis of significantly regulated genes shown in C. Shown are the GO overview terms selected by %Genes/Term in color and relevant individual GO terms in black (Q < 0.05; terms > 25 genes; kappa 0.5). (F and G) Increased transcript levels (left, n = 3/group) and immunoreactivity (right, n = 8/group) of REG-3β (F) and REG-3γ (G) in SPs compared with surrounding tissue (representative figure of three independent experiments). All histology sections were counterstained with DAPI (F and G) and pan-Keratin (F) or E-cadherin (G). Bars: (F) 250 µm; (G) 100 µm.
Interestingly, we found a significant enrichment of the GO overview term associated with “response to molecule of bacterial origin” (Fig. 1 E and Table S2). This encompassed the GO term for “response to bacterium” and can be triggered in response to the presence of a bacterium and includes genes associated with bactericidal activity such as Arg1, Ptgs2 (also known as COX-2), Serpine1, Reg3b, and Reg3g (Fig. 1, D and E). REG-3β and REG-3γ have previously been shown to play an important role in bacterial defense (Dessein et al., 2009; van Ampting et al., 2012) and were among the genes with the largest increase in expression in SPs compared with surrounding tissue (12- and 32-fold, respectively; Fig. 1 C). Immunostaining of cecal sections localized REG-3β and REG-3γ expression to cells in the basal epithelium of the SPs (Fig. 1, F and G), whereas they were mostly absent in unaffected surrounding cecal tissue. This localization pattern suggested that the epithelium of the SPs was actively responding to bacteria present in the local environment.
Broad-spectrum antibiotic treatment prevents SP development in HBUS mice
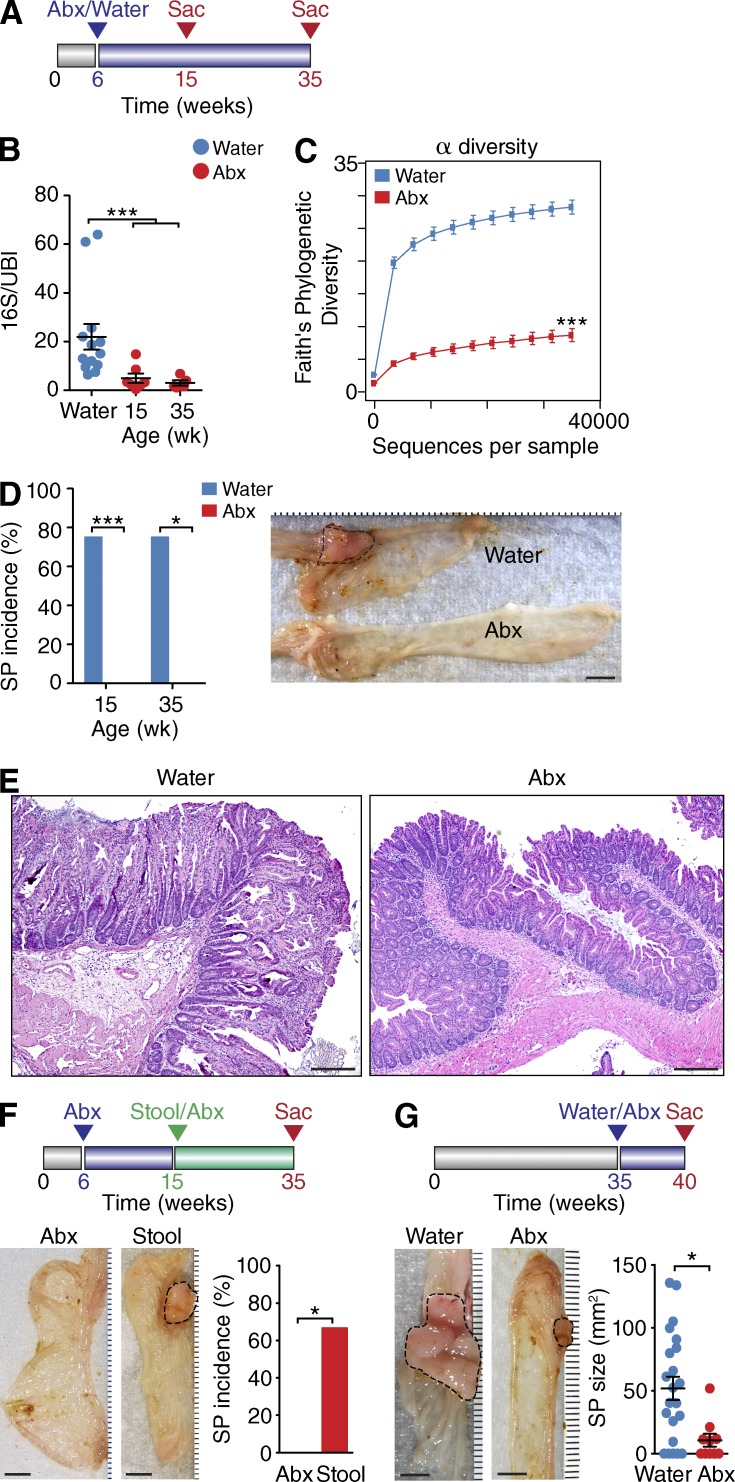
The elevated expression of antimicrobial genes in SPs prompted us to investigate whether the intestinal microbiota played a role in SP pathogenesis. To this end, we treated HBUS mice for 9 or 29 wk with a broad-spectrum antibiotic cocktail (metronidazole, ampicillin, neomycin, and vancomycin) in the drinking water and assessed the presence of SPs after 15 or 35 wk (Fig. 2 A). Treatment with antibiotics led to a significant decrease in the relative abundance of bacterial 16S ribosomal DNA (rDNA; Fig. 2 B) and α diversity (species richness, P < 0.001) within each sample of the cecal mucosa–associated microbiota of HBUS mice (Fig. 2 C), indicating that most bacteria were greatly reduced or absent altogether. Remarkably, none of the HBUS mice treated with antibiotics for 9 (n = 14) or 29 (n = 6) wk developed gross SPs (Fig. 2 D) or microscopic lesions (Fig. 2 E), suggesting that the microbiota is essential for formation of SPs.
Figure 2.
Alterations in the microbiota affect formation of SPs in HBUS mice. (A) Overview of the antibiotics (Abx) treatment plan. At 6 wk of age, HBUS mice started receiving broad-spectrum antibiotics or water and were examined for the presence of SPs at 15 and 35 wk of age. (B) Bacterial content in antibiotics-treated HBUS mice at 15 (n = 7) and 35 (n = 5) wk of age compared with water-treated HBUS mice (n = 13). Determined by pan-bacterial 16S qPCR amplification relative to host ubiquitin (UBI). ***, P < 0.001 (pairwise Wilcoxon rank sum test [FDR]; representative of two independent experiments). (C) α diversity, a measure of species richness, in antibiotics-treated HBUS mice (n = 11) and HBUS mice on regular water (n = 20). ***, P < 0.001 (Wilcoxon rank sum test; experiment was performed once). (D) SP incidence in antibiotics-treated HBUS mice after 9 (n = 14) and 29 (n = 6) wk compared with HBUS mice on regular water (75% at 15 [n = 16] and 35 wk [n = 12]). (right) Gross picture of the cecum of antibiotics and water-treated HBUS mice at 35 wk of age showing an SP (dashed lines). *, P < 0.05; ***, P < 0.001 (Fisher’s exact test; combined data of two independent experiments). (E) Representative histological section of the cecum of 35-wk-old HBUS mice treated with water or antibiotics as described in D. (F) HBUS mice were treated with antibiotics for 9 wk and subsequently gavaged with stools from unmolested HBUS mice (Abx/Stool). Control HBUS mice were maintained on antibiotics. Ceca of HBUS mice were analyzed by gross inspection and histological sectioning. SP incidence in HBUS mice treated with Abx/Stool (n = 8) and HBUS mice maintained on antibiotics (n = 9). *, P < 0.05 (Wilcoxon rank sum test; experiment was performed once). (G) At 35 wk of age, the drinking water of HBUS mice was supplemented with antibiotics, whereas control mice were maintained on regular water. After 5 wk of antibiotic treatment, HBUS mice were checked for the presence of SPs by gross and histological analysis. Graph represents SP size of HBUS mice treated with antibiotics for 5 wk starting at 35 wk of age (n = 10) compared with age-matched HBUS controls on water (n = 17). *, P < 0.05 (Wilcoxon rank sum test; experiment was performed once). Bars: (D, F, and G) 5 mm; (E) 250 µm. Error bars indicate SEM.
Next we explored whether reconstitution of the original microbiota after antibiotic treatment would restore development of SPs. After treatment of HBUS mice with antibiotics for 9 wk, we gavaged half of the mice with stool samples from untreated HBUS mice in the colony, three times per week for 6 wk, while continuing antibiotic treatment for the other HBUS mice (Fig. 2 F). At 35 wk of age, 20 wk after treatment with antibiotics was stopped, HBUS mice were examined for the presence of cecal SPs by gross inspection and histologically. As expected, none of the mice on continued antibiotic treatment developed SPs. In contrast, 67% of the mice that were first treated with antibiotics and subsequently gavaged with stools obtained from untreated HBUS mice developed SPs (n = 8; Fig. 2 F).
Finally, we investigated whether continued exposure to the gut microbiota is required for maintenance of SPs. We treated a group of animals at 35 wk of age with antibiotics for 5 wk (Fig. 2 G). At 40 wk, the size of SPs in HBUS mice subjected to antibiotic treatment (10.7 ± 4.6 mm2; n = 10) was significantly smaller compared with untreated HBUS mice (52 ± 11 mm2; n = 17). Collectively, our data suggest that the cecal microbiota is essential for both the development and maintenance of SPs in HBUS mice.
A specific cecal microbiota is necessary for development of SPs in HBUS mice
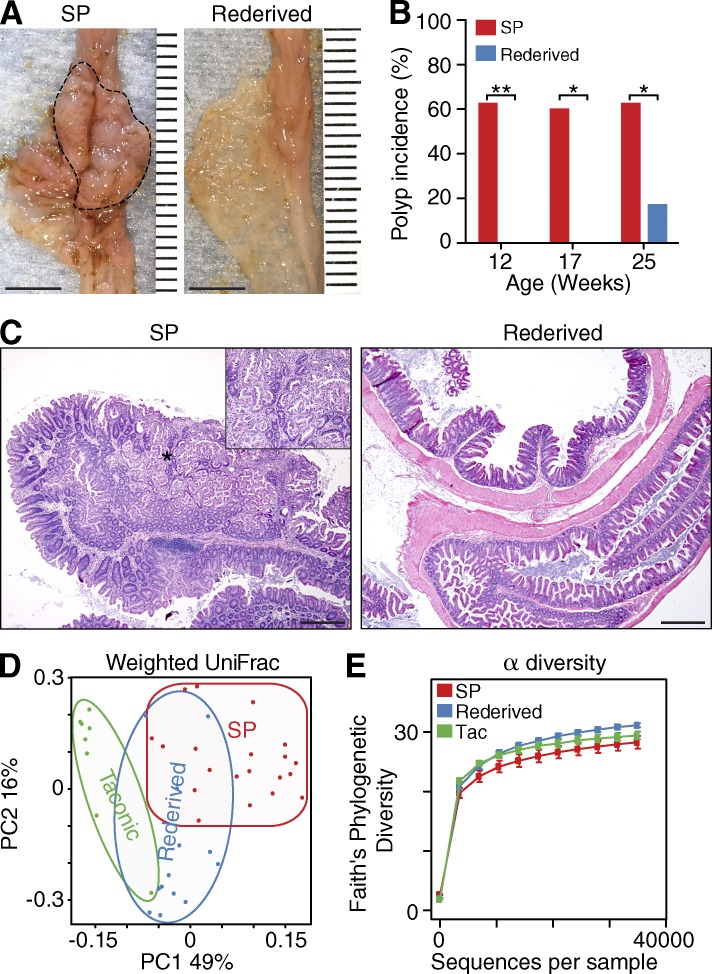
Treatment with broad-spectrum antibiotics leads to a marked reduction in microbial diversity and biomass, and long-term treatment promotes mucosal imbalance (Willing et al., 2011), which all could affect disease susceptibility. To exclude nonspecific effects of antibiotic treatment on SP development and to circumvent the massive changes associated with germ-free rederivation, we transferred one-cell embryos into pseudopregnant foster mothers of a different strain to promote adoption of a different microbiota. It is well established that the newborn gut microbiota is shaped by the maternal microbiota (Clemente et al., 2012). We reasoned that we could alter the microbiota of HBUS mice by transferring HBUS embryos (generated from and interbred with C57BL/6 mice obtained from the Jackson Laboratory) into Swiss Webster females obtained from Taconic. The results from this rederivation experiment were striking: HBUS mice rederived using Taconic mothers and reared separately for 12 (n = 7) or 17 wk (n = 8) did not develop SPs as determined by gross (Fig. 3 A) and histological analysis (Fig. 3, B and C). Interestingly, by 25 wk of age, we also observed SPs in Taconic-derived mice (n = 6), although at a significantly reduced frequency (17 vs. 63%; Fig. 3 B) and size compared with Jackson-derived HBUS mice (4.3 ± 4 vs. 57 ± 8 mm2).
Figure 3.
Biota modification through rederivation prevents SP formation in HBUS mice. Two HBUS colonies were established; one was maintained by interbreeding with C57BL/6 mice obtained from the Jackson Laboratory and developed SPs (dashed line); the second colony was generated through embryo transfer using Swiss Webster mothers freshly obtained from Taconic (rederived). (A–C) HBUS mice were examined grossly (A) and histologically (B and C) for the presence of SPs. Shown is the SP incidence of regular HBUS mice (n = 16, n = 10, and n = 12 at 12, 17, and 25 wk of age, respectively) and rederived HBUS mice (n = 7, n = 9, and n = 10 at 12, 17, and 25 wk of age, respectively) as determined by gross and histological analysis. *, P < 0.05; **, P < 0.01 (Fisher’s exact test; experiment was performed once). Inset shows a higher-magnification image of the area in C indicated by the asterisk. (D) Weighted UniFrac analysis of Taconic mothers (n = 8) compared with HBUS mice that develop SPs (n = 20) and rederived HBUS mice (n = 13; Adonis test; experiment was performed once). (E) α diversity of Taconic mothers, rederived HBUS mice, and HBUS mice with SPs (pairwise Wilcoxon rank sum test; experiment was performed once). Bars: (A) 5 mm; (C) 500 µm. Error bars indicate SEM.
Analysis of the cecal microbiota of the rederived mice by 16S sequencing and weighted UniFrac distances showed that the microbiota of rederived HBUS mice was significantly different from both the Jackson-derived HBUS mice (Adonis: P < 0.001) and their Taconic mothers (Adonis: P < 0.001; Fig. 3 D), suggesting that rederived HBUS mice had a biome that contained, as expected, elements of the Taconic maternal biota as well as the local environment. In contrast to antibiotic treatment (Fig. 2 C), rederivation did not significantly affect microbiome α diversity (P > 0.05; Fig. 3 E). Collectively, our results demonstrate that the presence of a host-specific microbial environment is key to the development of cecal SPs in HBUS mice.
Identification of a subset of bacteria enriched in cecal mucosa of SPs
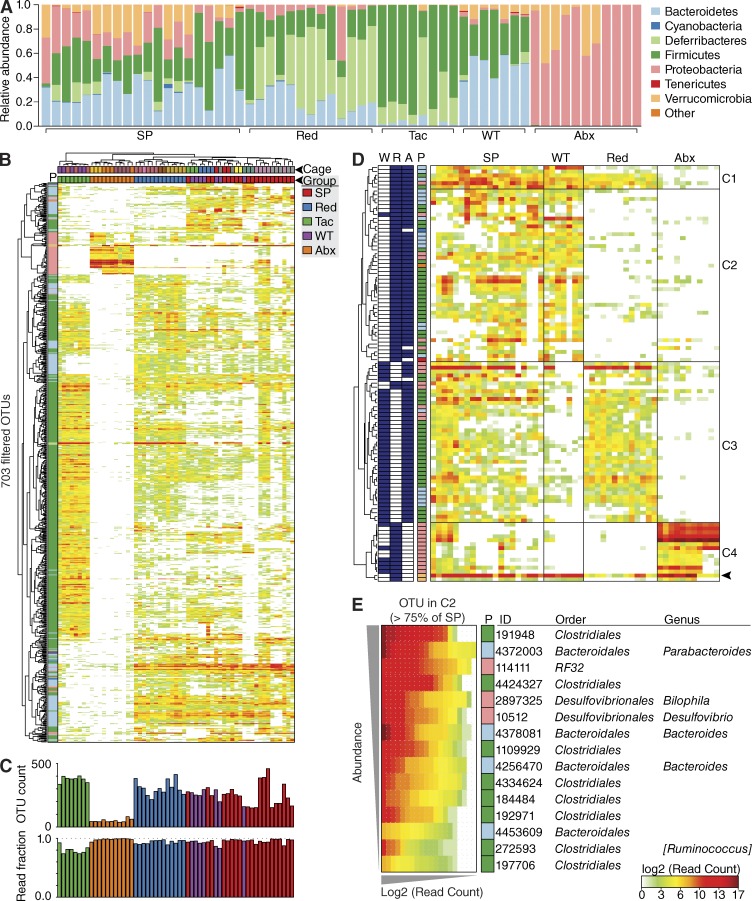
Next, we sought to more narrowly define which bacteria contributed to polyp formation. To do so, we compared the cecal mucosa–associated microbiota of affected HBUS mice with those of unaffected rederived HBUS mice, antibiotic-treated HBUS mice, and cohoused WT control littermates. We defined operational taxonomic units (OTUs) based on sequence similarity of the 16S rRNA fragments with Greengenes reference sequences (McDonald et al., 2012). This resulted in the identification of 704 unique OTUs across all samples after filtering as described in Materials and methods (Table S3). Hierarchical clustering (Fig. 4 B) and weighted UniFrac distances (Fig. 3 D) of OTU abundance profiles separated the microbiota of the individual sample groups, whereas WT mice clustered with cohoused HBUS mice that developed SPs (Fig. 4 B). At the phylum level, we found a distinct expansion of Verrucomicrobia and a decrease in Deferribacteres in SPs compared with rederived HBUS mice (Fig. 4 A). Closer inspection revealed these differences to be caused by the increased abundance of Akkermansia muciniphila (Greengenes ID: 4306262) and a decrease of Mucispirillum schaedleri (Greengenes ID: 1136443). Although enriched in SPs, A. muciniphila is one of the few species resistant to long-term treatment of HBUS mice with broad-spectrum antibiotics (Fig. 4 D, arrowhead), which makes it unlikely to be important for SP development.
Figure 4.
Biome analysis of SPs. (A) Relative abundance of phyla present in HBUS mice with SPs (n = 20), WT littermates (n = 7), rederived HBUS mice (Red; n = 13), and HBUS mice treated with antibiotics (Abx; n = 11). Data shown represent the most abundant phyla, whereas low abundant and unclassified OTUs were grouped in “Other.” (B, top) Pearson hierarchical clustering of the abundance profiles of all 703 OTUs after filtering. Phyla (P) are colored according to the legend in A. Mice were clustered (Spearman) by cage (color coded, top) and sample group (colored according to the legend). (bottom) Number of OTUs above background in each sample (OTU count) and fraction of total sample reads accounted for by the set of 703 filtered OTUs (read fraction). (D) Pearson hierarchical clustering identified four major clusters (C1–C4) in the abundance profiles of 106 OTUs that were significantly different between HBUS mice with SPs and rederived HBUS mice or WT controls (ANOVA Q < 0.05 [FDR]; Tukey P < 0.05; fold > 1.6). OTUs significantly enriched in SPs compared with rederived (R), WT (W), or antibiotics-treated (A) mice are shaded blue (left). Phyla (P) are colored according to the legend in A. OTU abundance is expressed as the log2-normalized read count in each sample. The OTU corresponding to A. muciniphila is indicated (arrowhead). (E) 15 OTUs from C2 that were present in >75% of SPs. OTUs are ranked by abundance (vertical axis) and according to abundance within each dataset (horizontal axis). OTU are annotated with Greengenes ID, color coded phyla (P) annotations (according to A), taxonomic family, and genus assignments. Each bar (A and C) or column (B, D, and E) represents a different mouse.
To determine which OTUs were specially associated with SPs, we compared the relative abundance of OTUs in SPs with cohoused WT, rederived, and antibiotic-treated HBUS mice by ANOVA (Q < 0.05) and determined differences between groups by a Tukey post-hoc test (P < 0.05, −1.6 > fold > 1.6; Table S4). Cluster analysis led to the identification of four distinct clusters (C1–C4; Fig. 4 D). OTUs present in clusters C1, C3, and C4 were also present in rederived or antibiotic-treated HBUS mice, i.e., HBUS mice that did not develop SPs. The second cluster (C2) represented 44 OTUs that were specifically enriched in SPs (Fig. 3 B). Of these, only 15 OTUs were found in >75% of all SP samples (Fig. 3 C).
Bacterial infiltration SPs and decreased barrier function in SPs
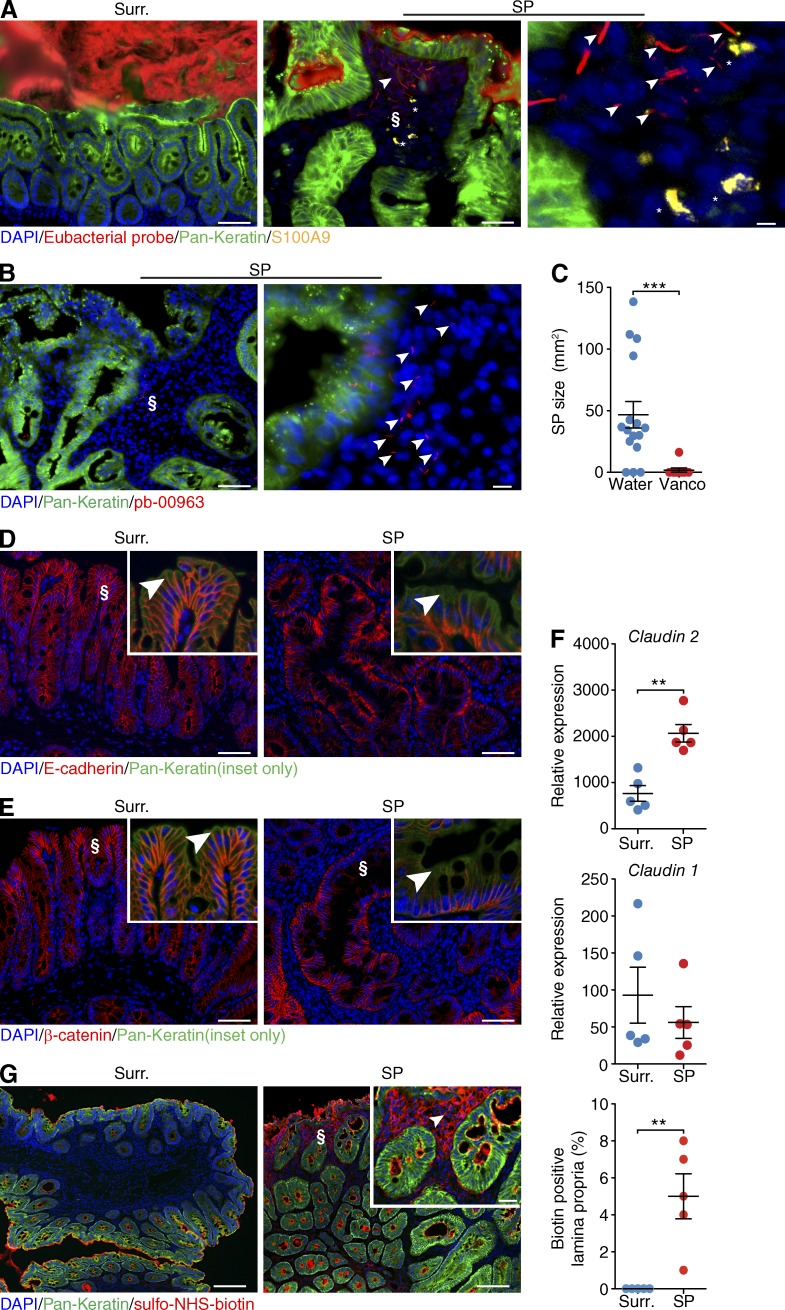
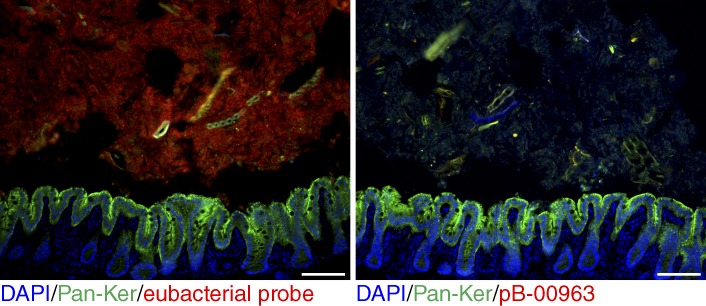
Various bacteria have tissue-invasive properties (Pizarro-Cerdá and Cossart, 2006; Sartor, 2008). To examine whether bacteria were present within SPs, we performed in situ hybridization with a eubacterial probe. As expected, probe signal was observed throughout the cecal lumen. In the lamina propria of SPs we observed the presence of bacteria, but not in unaffected surrounding cecal tissue. The invasive bacteria were found in close proximity to infiltrating neutrophils (Fig. 5 A). The majority (7/15) of OTUs enriched in HBUS SPs belonged to the order of Clostridiales. To test whether the invasive bacteria corresponded to these OTUs, we performed in situ hybridization with a Clostridium coccoides–Eubacterium rectale probe (Clostridium cluster XIVa and XIVb, pb-00963) that targets all the Clostridiales that were increased in HBUS SPs compared with the rederived HBUS mice (Fig. 5 B). Analysis of HBUS SPs and surrounding tissue revealed positive signal for this probe within the lamina propria of eight out eight tested HBUS polyps (Fig. 4 B). In antibiotic-treated HBUS mice, as expected, no signal was observed (Fig. 6). To examine whether Clostridiales would be of relevance to the formation of SPs in HBUS mice, we treated HBUS mice for 9 or 29 wk with 0.5 mg/ml vancomycin in the drinking water and assessed the presence of SPs after 15 or 35 wk. Treatment with vancomycin targets gram-positive bacteria and shifts the composition of the microbiota, particularly the family Clostridiales/Lachnospiraceae, without affecting the total number of cecal bacteria (Sekirov et al., 2008; Ubeda et al., 2010; Willing et al., 2011). Vancomycin-treated HBUS mice showed a significant decrease in incidence (11%, P = 0.002, n = 9) and size of SPs (Fig. 5 C).
Figure 5.
Bacterial infiltration SPs and decreased barrier function in SPs. (A and B) In situ hybridization with a eubacterial probe (A) or Clostridium cluster XIVa and XIVb (pb-00963; B) on frozen sections obtained from HBUS mice. Shown is a representative image obtained from two independent experiments of surrounding cecal tissue (Surr.) and an SP (middle; higher magnification of § on right; n = 6). S100A9-positive cells are indicated by asterisks. Arrowheads indicate bacteria that invaded the lamina propria (A) or bacteria recognized by pb-00963 that invaded the lamina propria (B). (C) At 35 wk of age, the drinking water of HBUS mice was supplemented with 1 mg/ml vancomycin, whereas control mice were maintained on regular water. After 4 wk of treatment, HBUS mice were checked for the presence of SPs by gross and histological analysis. Shown is SP size of HBUS mice treated with vancomycin for 4 wk starting at 35 wk of age (n = 9) compared with age-matched HBUS controls on water (n = 17). ***, P < 0.001 (Wilcoxon rank sum test; experiment was performed once). (D and E) Immunofluorescent analysis of the adherens junction protein E-cadherin (D) and its binding partner β-catenin (E) in surrounding cecal tissue (left) and an SP (right). Insets show a higher-magnification image of §. Arrowheads indicate the apical adherens junction. Representative images of three independent experiments are shown (n = 5). (F) Expression of Claudin-2 and Claudin-1 mRNA in SPs of HBUS mice compared with surrounding tissue (n = 5/group). **, P < 0.01 (Wilcoxon rank sum test, experiment was performed once). (G) Immunofluorescent analysis of proteins labeled with sulfo-NHS-biotin in the ceca of HBUS mice. Shown is a representative image of two independent experiments of biotin signal in cecal surrounding tissue and SPs of HBUS mice. The arrowhead indicates biotin signal in the lamina propria (inset shows higher-magnification image of §). Immunofluorescent sections were counterstained with DAPI and pan-Keratin (A, B, D, E, and G). Bar graphs represent combined data of two independent experiments (n = 5). Bars: (A [left and middle], B [left], D, and E) 50 µm; (A, right) 2 µm; (B, right) 10 µm; (G) 100 µm; (G, inset) 25 µm. Error bars indicate SEM.
Figure 6.
Clostridial probe signal in antibiotic-treated HBUS mice. Representative image of an experiment performed once showing in situ hybridization with a eubacterial probe (left) or Clostridium cluster XIVa and XIVb (pb-00963) on frozen sections obtained from HBUS mice treated with antibiotics (n = 3). Bars, 50 µm.
Next, we examined whether bacterial invasion was associated with a decreased barrier function. We observed an apical loss of adherens junction proteins E-cadherin and β-catenin in SPs of HBUS mice compared with unaffected surrounding cecal tissue (Fig. 4, C and D). Besides the 54 genes associated with the GO term “biological adhesion,” such as Claudin-4, Claudin-23, and Cadherin-1 (Fig. 1), we also found altered expression of tight junction protein Claudin-2, but not Claudin-1, in SPs compared with unaffected surrounding cecal tissue (Fig. 4 E). To examine whether changes in junction molecules had functional consequences, we treated ceca obtained from HBUS mice with sulfo-NHS-biotin. Sulfo-NHS-biotin can be used to examine permeability of tight junction proteins ex vivo (Tamura et al., 2008; Ding et al., 2011; Lei et al., 2012). Sulfo-NHS-biotin (molecular weight 443.43) is a membrane- and tight junction–impermeable molecule that efficiently biotinylates primary amine-containing macromolecules such as proteins. Ceca from HBUS mice were dissected and treated with sulfo-NHS-biotin solution. Sulfo-NHS-biotin signal was only observed in SPs of HBUS mice and not in adjacent unaffected cecal tissue (n = 5; Fig. 5 G). These results suggest that bacterial invasion of SPs is facilitated by a decreased intestinal epithelial barrier function.
The inflammatory response in SPs is consistent with bacterial invasion
The combination of bacterial invasion and the increased expression of inflammatory and antimicrobial defense genes in SPs prompted us to further characterize the immune response in SPs of HBUS mice. Therefore, we enriched for CD45+ cells from SPs and surrounding unaffected cecal tissue and analyzed gene expression profiles using the Illumina BeadArray system. We found significant changes in expression of 153 genes in SPs compared with unaffected cecal tissue (Q < 0.05, −1.5 > fold > 1.5; Fig. 7 A and Table S5), consistent with a signature of activated leukocytes, as well as “response to bacterium” (Fig. 7 B and Table S6). Analysis of cytokine and chemokine expression by BeadArray and quantitative PCR (qPCR) showed that the proinflammatory cytokines Il1-α, Il1-β, and Tnf and the chemokines Ccl1, Ccl2, Ccl17, Cxcl2, and Cxcl16 were significantly up-regulated in SPs compared with unaffected cecal tissue (n = 5; Fig. 5 C).
Figure 7.
Marked inflammatory changes in SPs. (A) Leukocytes (CD45+ cells) isolated from HBUS SPs and surrounding (Surr.) tissue were analyzed by Illumina BeadArray/limma. Quantile-normalized expression values were analyzed using a paired design (n = 3/group) and filtered for Q < 0.05 and −1.5 > fold change > 1.5. Z score–normalized data were subjected to hierarchical clustering (left): red indicates increased and green indicates decreased expression in SPs compared with surrounding tissue. Plot of logFC (log fold change) versus mean expression (right) of all detected transcripts (gray) and significant genes (153 genes; black). (B) ClueGO analysis of significantly regulated genes in an Illumina BeadArray analysis of SPs of HBUS mice compared with unaffected surrounding cecal tissue. Shown are GO overview terms selected by %Genes/Term in color (Q < 0.05; terms > 15 genes; kappa 0.5). (C) Cytokine and chemokine mRNA expression in tissue obtained from SPs compared with unaffected surrounding proximal cecal tissue (n = 5/group). *, P < 0.05; **, P < 0.01 (Wilcoxon rank sum test; experiment was performed twice). R.E., relative expression. (D) Il-17 production by TCR-γδ cells in SPs (n = 6) from HBUS mice compared with WT (n = 4) and unaffected surrounding HBUS cecal tissue (n = 6); cells were gated on CD45+/CD4−/TCR-γδ+. *, P < 0.05 (Wilcoxon rank sum test; experiment was performed once). Shown are representative FACS plots and a summary scatter dot plot. (E–G) FACS analysis in SPs from HBUS mice (P; n = 3) compared with unaffected surrounding HBUS cecal tissue (S; n = 4) and WT cecal tissue (W; n = 4). For production of IL-17 and IL-22 by CD4/TCR-αβ, cells were gated on CD45+/CD4+/TCR-αβ+ (E). For DC subsets, cells were gated on CD45+/CD11b+/MHC-II+/Ly6C− (F). For monocytes and eosinophils as defined by Ly6C and CD24 expression, cells were gated on CD45+/CD11b+/MHC-II−/Ly6G− (G). Shown are representative summary scatter dot plots. *, P < 0.05 (Wilcoxon rank sum test; experiment was performed once). Error bars indicate SEM.
Next we examined the cecal leukocyte composition of WT and HBUS mice by FACS analysis. No differences in the relative abundance of T cells, DCs, or macrophage subsets were observed between SPs and unaffected surrounding tissue or WT ceca. The most enriched GO overview term was associated with T cell activation (Fig. 7 B). Detailed analysis of the T cell subsets showed no change in the frequency of Th17 cells, but there was an increase in the frequency of IL-17–producing TCR-γδ cells in SPs compared with unaffected surrounding tissue (Fig. 5, D and E).
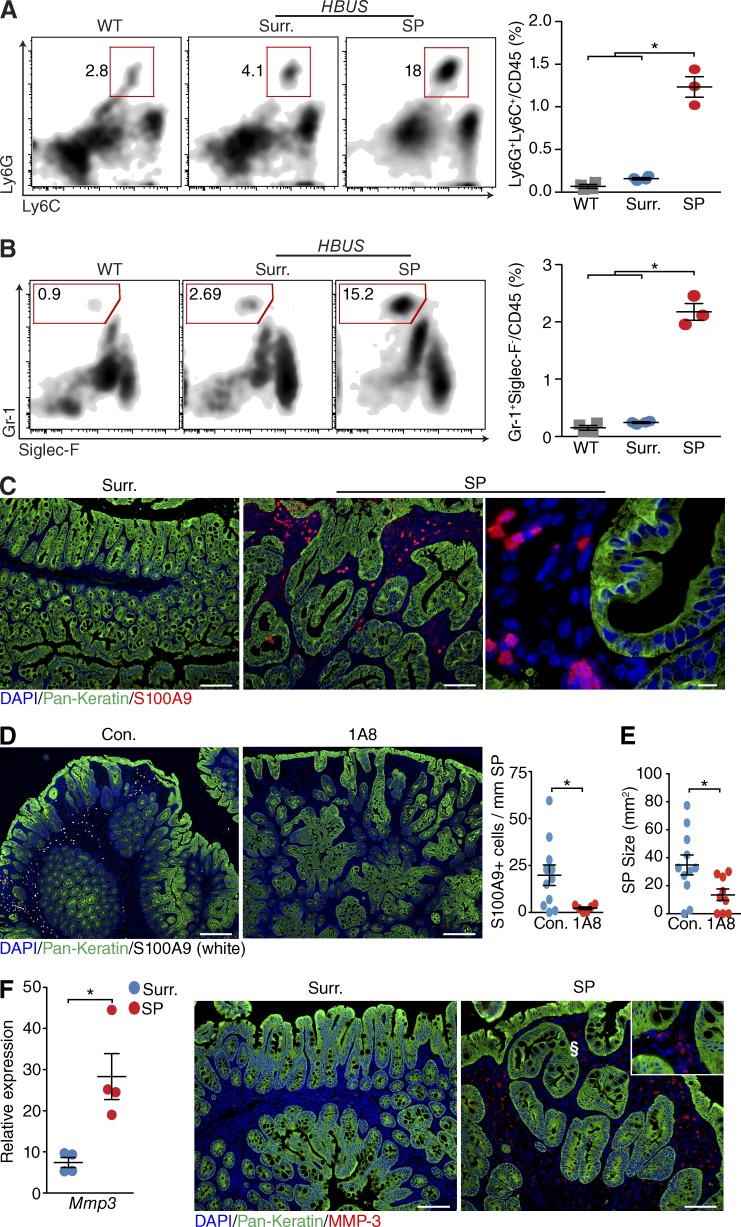
Among the most pronounced induced genes were S100a8, S100a9, and Lgals2, which are expressed by neutrophils (Kehl-Fie et al., 2011). To further evaluate this, we examined the CD11b/c+/MHCII− myeloid subsets that consist of neutrophils, eosinophils, and monocytes. No significant differences were observed in the Ly6G− subset composed of eosinophils and monocytes (Fig. 7, F and G); however, the number of Ly6G and Ly6C double-positive cells was increased in SPs (Fig. 8 A). Further analysis identified these cells as Ly6C/G+ and Gr-1High/Siglec-F− neutrophils (Fig. 8 B), consistent with the increased expression of S100a9 observed in the CD45 BeadArray analysis (Fig. 7 A). To further characterize the neutrophil infiltrate, we stained cecal sections of HBUS mice with an antibody against S100A9, which stains neutrophils. Strong staining was observed close to the lumen in SPs, but not in unaffected cecal surrounding tissue (Fig. 8 C). To determine whether neutrophils play a role in the pathology of SPs, we subjected 35-wk-old HBUS mice to anti–Ly-6G (1A8) treatment and examined the SP size after 30 d. Treatment with Ly-6G–specific (1A8) antibodies can be used to deplete neutrophils in mice (Daley et al., 2008). Treatment with 1A8 antibody led to a significant decrease in the number of S100A9+ neutrophils in SPs (10-fold) that was accompanied by a significant decrease in SP size (2.5-fold). These results suggest that neutrophils are functionally relevant for the growth of SPs in HBUS mice.
Figure 8.
Marked neutrophil infiltration in SPs. (A and B) Relative number of neutrophils determined by the number of Ly6G/Ly6C double-positive cells (A; gated on CD45+/CD11b+/MHC-II−) and Gr-1–positive, Siglec-F–negative cells (B; gated on CD45+/CD11b+/MHC-II−) in SPs (n = 3) of HBUS mice compared with unaffected surrounding cecal tissue (Surr.; n = 4) and WT littermate controls (n = 4). Shown are representative FACS plots and a summary scatter plot. *, P < 0.05 (Wilcoxon rank sum test; experiment was performed once). (C) Representative image of three independent experiments showing S100A9-positive cells in SPs and unaffected surrounding cecal tissue (n = 6). (D and E) At 35 wk of age, HBUS mice were treated with 0.4 mg anti-1A8 i.p. every other day for 30 d. Shown are representative immunofluorescent images (left) and quantification (right) of the number of S100A9-positive cells in SPs (D) and the size of SPs (E) in control (n = 11) and 1A8-treated (n = 9) HBUS mice. *, P < 0.05 (Wilcoxon rank sum test; experiment was performed once). (F) Expression of Mmp3 mRNA by qPCR analysis and immunofluorescent staining (MMP-3) in SPs and surrounding tissue (n = 4/group). Inset shows higher-magnification image of the area indicated by §. *, P < 0.05 (Wilcoxon rank sum test; experiment was performed twice). Sections were counterstained with DAPI and pan-Keratin. Bars: (C [left and middle] and F) 100 µm; (C, right) 10 µm; (D) 250 µm. Error bars indicate SEM.
Inflammation can affect the expression of MMP-3, a matrix metalloproteinase shown to cleave HB-EGF and thereby promote autocrine/paracrine EGFR signaling (Suzuki et al., 1997). We therefore examined MMP-3 mRNA expression in SPs and compared it with the nonaffected surrounding tissue. We observed an increased mRNA expression of MMP-3 in SPs. Immunostaining of SPs with an anti–MMP-3 antibody showed an increased number of MMP-3–positive cells in the lamina propria (>50 per 10× field) compared with unaffected cecal tissue where MMP-3–positive cells were only found in the submucosa (<5 per 10× field; Fig. 8 E).
In summary, the presence of bacteria in the lamina propria of SPs triggered an inflammatory response that included expression of several cytokines, chemokines, and expression of an HB-EGF–processing metalloproteinase. These findings mechanistically link microbe-induced inflammation, HB-EGF/EGFR signaling, and development of SPs in HBUS mice.
DISCUSSION
Intestinal SPs occur at specific locations in the gut in a subtype-specific manner (Huang et al., 2004; Noffsinger, 2009). In this study, we show that the development of SPs in the cecum of HBUS mice (Bongers et al., 2012) depends on the presence of a host-specific microbiota. Alteration of the cecal microbiota through antibiotic treatment or embryo transfer–mediated rederivation significantly attenuated the formation of SPs in HBUS mice. To our knowledge, this study shows for the first time that relatively minor changes in the microbiota can dramatically affect the formation of neoplasms.
The GI tract is home to a complex set of microbial communities that differ between the various regions (Sartor, 2008). Longitudinal sampling of the GI tract has shown clear differences in the bacterial composition of samples from the mouth, stomach, duodenum, colon, and stool (Stearns et al., 2011). Proximal and distal colon samples differ both in their microbiota composition as well as in abundance of OTUs (Wang et al., 2010). Overall, nutrient availability, pH, bile salts, or other host factors can regulate the composition of the microbiota in different regions of the GI tract (Merritt and Donaldson, 2009; von Rosenvinge et al., 2013). Here, we show that the exclusive penetrance of the polyp phenotype in the cecum is related to bacteria living at this particular location. Antibiotic treatment completely prevents development of SPs, and conversely, reconstitution of the flora using stools from mice with SPs promoted SP development. Furthermore, treatment of animals with well-developed SPs with antibiotics reduced polyp size, indicating that the presence of cecum bacterial communities was important for SP maintenance and growth. Importantly, our results indicate that there is a requirement for a host-specific cecum flora for development of SPs. We show here that a relatively minor change in the cecal microbiome markedly affected the incidence of SPs. Transfer of HBUS embryos into Taconic foster mothers promoted partial adoption of a Taconic cecal microbiome by the offspring, as verified by weighted UniFrac and OTU analysis. These effects were not dominant because after 25 wk of age we observed SPs in rederived HBUS mice, likely reflecting a progressive drift from the maternal microbiota to the local environment.
Detailed analysis of the SP-associated microbiome indicated that 15 OTUs, from the four orders Clostridiales, Bacteroidales, RF32, and Desulfovibrionales, were consistently different between SPs and unaffected rederived and antibiotic HBUS mice. While association does not prove that these OTUs are causative of SP formation, members of the order of Clostridiales have been known to predominantly colonize the cecum and proximal colon and promote the production of MMPs (Honda and Littman, 2012). Similarly, toxin B produced by Clostridium difficile has been show to promote EGFR-mediated activation of ERK1/2 in human colonocytes in an MMP- and TGF-α–dependent manner (Na et al., 2005). In the lamina propria of SPs in HBUS mice, we observed increased MMP-3 expression, which has been shown to cleave HB-EGF (Suzuki et al., 1997). This raises the possibility that Lachnospiraceae promotes expression of MMP-3 in the SPs that facilitates the cleavage of HB-EGF and subsequent activation of the EGFR–MAPK pathway. Previously, we have shown that this pathway plays a key role in the development of SPs in HBUS mice (Bongers et al., 2012). Alternatively, MMP-3 production and release could also be increased as part of a general inflammatory response (Louis et al., 2000).
Among the OTUs that were enriched in SPs was the genus Bilophila whose members have been associated with bile resistance and are often recovered from appendicitis specimens (Finegold and Jousimies-Somer, 1997). Bilophila wadsworthia has recently been shown to flourish on diet-induced taurine-conjugated bile acid and promote colitis in IL10−/− mice. The mechanism by which B. wadsworthia induces colitis is unknown but was proposed to involve an immune response to tissue damage induced by the bacterial byproducts, such as H2S or secondary bile acids (Devkota et al., 2012). Interestingly, most bile acids secreted by the host are absorbed in the terminal ileum, whereas secondary bile acids produced by bacteria increase in more distal sections of the intestine (Ridlon et al., 2006; Hamilton et al., 2007). Therefore, a specific niche of cecal bile acid–transforming bacteria could explain the specific cecal localization of SPs.
The bacterial invasion we observed in SPs could be caused by properties encoded by the bacteria or specific interactions with the genetic alterations in the host; e.g., EGFR signaling can be used by infectious agents to increase their survival and promote invasion (Galán et al., 1992; Fiske et al., 2009). HB-EGF signaling has also been associated with changes in expression of tight junction proteins such as Claudin-2 (Singh et al., 2007), which we also found in this study. Currently, it is unclear whether apical loss of the adherens junction–associated proteins β-catenin and E-cadherin we observed in SPs are functionally related to the changes in Claudin-2 expression, as shown for Claudin-7 (Lioni et al., 2007), or brought about by changes in inflammation-associated cytokine expression (Bruewer et al., 2003).
Previous studies have shown that bacteria can regulate cytokine production by specific leukocyte subsets. For example, segmented filamentous bacteria and Clostridium species have been shown to induce IL-17 production by Th17 and regulatory T cells, respectively (Ivanov et al., 2009; Nagano et al., 2012). In SPs of HBUS mice, we identified TCR-γδ T cells as the main source of the increased IL-17 production. IL-17 production by resident TCR-γδ cells is critical for the recruitment of neutrophils and for the clearance of E. coli (Shibata et al., 2007).
Development of SPs in HBUS mice was associated with a distinct neutrophilic infiltrate, potentially as a result of increased levels of CXCL2, e.g., induced by the TNF or IL-17 produced by TCR-γδ cells. Neutrophils are the final effectors of an inflammatory response with an important role in the clearance of pathogens. There is mounting evidence that tumor-associated neutrophils (TANs) play an important role in tumor initiation and progression (Mantovani et al., 2011). Depletion experiments indicate that neutrophils play a role in the growth of SPs in HBUS mice.
It is clear that the pathway to formation of SPs at specific locations in the gut is complex and involves both bacterial and host factors, given that prevention of EGFR signaling (Bongers et al., 2012) or alterations of the microbiota are sufficient to decrease the incidence of SPs. Based on our observations, we propose that SPs in HBUS mice are likely initiated by increased basal HB-EGF/EGFR signaling, leading to a decreased barrier function by altering tight/adherens-associated proteins. This in turn would facilitate the invasion of specific bacterial species that trigger a host antibacterial response characterized by the production of antimicrobial molecules and an innate immune response. Subsequently, bacterial or inflammatory signals promote an expression of MMP-3 that in turn further facilitates HB-EGF/EGFR signaling, resulting in a positive feedback loop that ultimately promotes growth of SPs.
In summary, we have identified a requirement for interaction between host genetic alterations and host-specific microbiota for development of SPs. The discovery of this mechanism may help to explain the preferential localization of SPs in specific locations in the human intestine (Noffsinger, 2009).
MATERIALS AND METHODS
Mice.
VS28, HBGF, and HBUS mice were described previously (Bongers et al., 2010, 2012). No statistical method was used to determine sample size, and when applicable, mice were assigned to a treatment group using a simple randomization (coin flip). Histological analysis was performed blinded to the treatment. All experiments involving mice were performed in accordance with the guidelines of the Animal Care and Use Committee of Mount Sinai School of Medicine.
Antibiotic treatment.
Mice were treated for the indicated time with 1 g/liter ampicillin, 1 g/liter neomycin, 1 g/liter metronidazole (all Sigma-Aldrich), and 0.5 g/liter vancomycin (Western Medical Supply) ad libitum in the drinking water for the indicated time.
In vivo cell depletion.
For depletion of Ly6G+-expressing neutrophils, anti-Ly6G mAb clone 1A8 (Bio X Cell) was injected at 0.4 mg i.p. every other day for 30 d, as previously described (Daley et al., 2008; Garcia et al., 2010).
Pan-bacterial PCR amplification.
Total genomic DNA was isolated from tissue using the DNeasy Blood and Tissue kit (QIAGEN). qPCR analysis and primer sequences were described previously (Hartman et al., 2009). Relative quantity was calculated by the ΔCt method and normalized by the presence of mouse ubiquitin.
RNA extraction and qPCR.
Total RNA was extracted using the RNeasy Mini kit (QIAGEN) according to the manufacturer’s protocol. RT was performed using 3 µg total RNA. RT-PCR was conducted in triplicates using SYBR green (Fermentas). Relative expression levels were calculated by the ΔCt method and normalized by the presence of mouse ubiquitin. Primers were designed using Primer Express 2.0 software (Applied Biosystems) or Primer3Plus (Untergasser et al., 2007).
Rederivation.
Specific pathogen–free mice were produced by in vitro fertilization using standard procedures. In brief, sperm from VS28 males and oocytes from HBGF females were implanted into naturally ovulating pseudopregnant Swiss Webster female mice freshly obtained from Taconic.
Isolation of leukocytes.
Intestines were cut into 2-cm pieces and washed in ice-cold PBS. To release the intestinal epithelial cells, the pieces were incubated in PBS containing 1.3 mM EDTA for 30 min at 4°C. After vigorous shaking, intestinal epithelial cells were collected in the supernatant and used to prepare RNA extracts using the RNeasy Mini kit (QIAGEN). The remaining tissue was digested in DMEM containing 1 mg/ml Dispase II (Roche) for 20 min at 37°C, incubated with CD45 microbeads (Miltenyi Biotec), and purified using MACS columns (Miltenyi Biotec) according to manufacturer’s protocol. RNA was extracted from the CD45-enriched purified cells using the RNeasy Mini kit.
Histology and immunofluorescence.
Organs were dissected, fixed in 10% formalin, and processed for paraffin sectioning. 4-µm sections were dewaxed by immersion in xylene and hydrated by serial immersion in ethanol and PBS. Antigen retrieval was performed by incubating sections in a pressure cooker for 15 min in Target Retrieval Solution (DAKO). Sections were washed with PBS (twice for 10 min), and blocking buffer (TBS containing 10% BSA and 0.3% Triton X-100) was added for 1 h. Sections were incubated with primary antibody in blocking buffer overnight at 4°C and then incubated with Alexa Fluor 488–, Alexa Fluor 594–, or Alexa Fluor 647–labeled secondary antibodies for 1 h. Sections were mounted with Fluoromount-G (Beckman Coulter). Antibodies were obtained from R&D Systems (REG-3β), Abcam (pan-keratin, S100a9, and MMP-3), Santa Cruz Biotechnology, Inc. (REG-3γ), or Cell Signaling Technology (β-catenin and E-cadherin).
Fluorescent in situ hybridization.
Whole ceca were dissected and directly incubated overnight in fixing solution (PBS containing 1.6% paraformaldehyde and 20% sucrose), embedded in O.C.T. (Tissue-Tek), and snap frozen in 2-methylbutylene on dry-ice. 10-µm sections were washed with ice-cold PBS (twice for 10 min), and blocking buffer (TBS containing 10% BSA) was added for 30 min. Sections were incubated with 0.45 pmol/µl eubacterial oligonucleotide probe ([AminoC6 + Alexa Fluor 594] 5′-GCTGCCTCCCGTAGGAGT-3′; Operon) in prechilled hybridization buffer (Sigma-Aldrich) overnight at 4°C (Bates et al., 2006). Sections were counterstained with 30 nM DAPI (Invitrogen) in PBS for 10 min, washed for 10 min in ice-cold PBS, and mounted with Fluoromount-G. Immunofluorescent imaging was performed on the same day.
Intraepithelial cellular barrier assay.
The barrier function assay based on sulfo-NHS-biotin was performed as described previously (Tamura et al., 2008). In brief, sulfo-NHS-biotin (1 mg/ml PBS; Sigma-Aldrich) was applied to dissected ceca of HBUS mice. After 30 min of incubation in a humidified chamber at room temperature, the ceca were washed three times with PBS for 5 min, fixed overnight with 4% paraformaldehyde/20% sucrose in PBS, and frozen in OCT. Sections were stained with Alexa Fluor 594–labeled streptavidin (Invitrogen) for 30 min and counterstained with 30 nM DAPI (Invitrogen) in PBS for 10 min, washed for 10 min in ice-cold PBS, and mounted with Fluoromount-G.
Fluorescent imaging.
Immunofluorescent imaging was performed using Cy3 HYQ, FITC HYQ, and a fluorescence microscope (E600; Nikon) with Plan Apochromat objective lenses. Images were acquired using a digital camera (DXM1200F; Nikon) and Act-1 software version 2.63 (Nikon). Images were composed in Photoshop CS6 (Adobe).
FACS analysis on cecal tissue.
The cecum was cut into small pieces (<1 mm), digested with 2.5 mg/ml Collagenase D and 1 mg/ml Dispase II for 1 h, and purified using a Percoll gradient (44 and 66%). For cytokine analysis, the cells were cultured for 3 h with 10 ng/ml PMA and 1 µg/ml ionomycin and for 1.5 h with 10 µg/ml Monensin. Otherwise, the cells were immediately resuspended in Fc-Block (BD) for 20-min RT and subsequently stained with surface markers for 30 min at 4°C. For intracellular staining, cells were permeabilized in permeabilization buffer (BD) and stained with intercellular antibodies diluted in permeabilization buffer for 45 min at 4°C. DC and macrophage subsets in SPs were analyzed by FACS analysis using the markers CD45, CD11c, CD11b, MHCII, Ly6C, and CD103. FACS antibodies were obtained from BD or eBioscience.
BeadArray analysis.
BeadArray analyses were performed with 500 ng total RNA using the TotalPrep RNA Amplification kit (Illumina) and MouseRef-8 BeadChip kit 430 2.0 Expression BeadChip arrays (Illumina) with n = 3 per group. All arrays in this study were subjected to variance-stabilizing transformation (Lin et al., 2008), quantile normalization, and quality control using lumi version 2.14.0 Bioconductor/R (Du et al., 2008). Fold changes and statistical significance were determined using a paired sample analysis in limma version 3.18.7 Bioconductor/R (Smyth, 2004) and corrected for multiple testing (FDR/Benjamini and Hochberg). Probe sets were selected based on −1.5 > fold change > 1.5, Q < 0.05. The expression values were plotted with heatmap.2 (gplots version 2.11.0, CRAN/R).
DNA extraction, 16S rDNA amplification, and multiplex sequencing.
All mice used for 16S rDNA sequencing were housed in specific pathogen–free conditions that were free of Helicobacter hepaticus and H. bilis. Rederived (n = 13) and antibiotic-treated HBUS mice (n = 11) were house in separate cages, whereas WT control littermates (n = 7) were cohoused with HBUS mice that developed SPs (n = 20). Each sample was obtained from a different mouse. HBUS mice never developed more than one SP and were only found at the cecal–colonic junction. Before resection, surface stool was removed and SPs from HBUS mice or 4-mm2 piece of the cecal–colonic junction was processed directly using the DNeasy Blood and Tissue kit (QIAGEN). Bacterial 16S rRNA genes were amplified using the primers as described in Caporaso et al. (2012). Each sample was amplified in quadruplicate, combined, and cleaned using the Agencourt AMPure XP beads (Beckman Coulter). PCR reactions contained 0.5 µM for each primer, 100 ng genomic DNA, and Phusion High-Fidelity PCR Master Mix (New England Biolabs, Inc.). Reactions were held at 98°C for 30 s, proceeding to 35 cycles at 98°C for 10 s, 50°C for 30 s, and 72°C for 30 s and a final extension of 10 min at 72°C. Cleaned amplicons were quantified using Picogreen dsDNA, and a composite sample for sequencing was created by combining equimolar ratios of amplicons from the individual samples, agarose gel purifying (QIAquick Gel Extraction; QIAGEN), and mixing with 50% PhiX (Illumina). The sample concentration was verified by qPCR and loaded on Illumina MiSeq or HiSeq sequencer (members of all groups were run on both machines) for sequencing as previously described (Caporaso et al., 2012).
Analysis of 16S rDNA sequences.
The raw reads obtained from 150-bp paired-end MiSeq or HiSeq runs were preprocessed through the QIIME version 1.7.0 pipeline (Caporaso et al., 2010). After demultiplexing and base quality trimming (q20), a reference-based OTU picking protocol was applied and 97% OTUs were picked against the May 2013 Greengenes database (McDonald et al., 2012; prefiltered at 97% identity) using UCLUST (Edgar, 2010). Reads were assigned to OTUs based on their best match to a Greengenes sequence (89% of the reads), and reads that did not match a Greengenes sequence at 97% or greater sequence identity were discarded. The Greengenes taxonomy associated with the best match in Greengenes was assigned to each OTU, and the Greengenes tree was used for phylogenetic diversity calculations. The resulting OTU table was normalized by down-sampling to the sample with the lowest counts (35,000) and then quality filtered, discarding OTUs with less than five counts across all samples or present in less than nine samples (Bokulich et al., 2013). Filtering did not significantly affect the read fraction for each group (Fig. 4 C). Finally, filtered OTU counts were used to calculate Jackknifed Weighted UniFrac β-diversity indices (Lozupone et al., 2011) and α diversity (Faith and Baker, 2006) at an even depth of 35,000 sequences/sample and generate taxonomic plots. To evaluate differences between groups, one-way ANOVA (CRAN/R version 3.0.2) was performed on log2-transformed count data (all zeroes were set to 0.5 of the smallest nonzero entry) assuming equal variance, OTUs with Q < 0.05 (FDR/Benjamini and Hochberg) were subjected to Tukey HSD post hoc test (CRAN/R version 3.0.2), and significant differences among groups were defined as P < 0.05 and −1.6 > fold change > 1.6. Abundance profiles were hierarchically clustered using Spearman correlation as the distance metric, and heat maps were generated using R.
RNA extraction from tissues.
Approximately 200 mg of tissue sample was placed in 1.5 ml RNAlater buffer (Ambion) and snap-frozen in liquid nitrogen. RNA extraction was performed as described previously (Giannoukos et al., 2012). In brief, at time of extraction, samples were centrifuged for 10 min at 16,000 g at room temperature. Pellets were resuspended in 150 µl lysis buffer (Tris/HCl, pH 8, 1 mM EDTA, 15 mg/ml Lysozyme [Sigma-Aldrich], and 15 µl of 20 mg/ml proteinase K) and incubated at room temperature for 10 min with brief mixing every 2 min. After addition of 1.2 ml QIAGEN RLT buffer containing 1% vol/vol β-mercaptoethanol, 1 ml of 0.1-mm glass beads (BioSpec) was added, and samples were homogenized in a FastPrep at setting 5 (four pulses of 20 s). Samples were kept on ice for 1 min between pulses. Lysates were then homogenized with a QIAshredder spin column, and RNA was isolated using the AllPrep mini kit (QIAGEN), according to the manufacturer’s protocols, which included an on-column digestion with DNase I. 25 µg RNA from each tissue sample was processed with the MICROBEnrich kit (Ambion), and 5 µg of processed RNA was further depleted of rRNAs using the Meta-Bacteria RiboZero rRNA removal kit (Epicentre). The final samples consisted of a mix of host and microbial mRNAs in a 2:1 ratio.
cDNA library construction and sequencing.
rRNA-depleted RNA was prepared for Illumina paired-end sequencing using the Next mRNA Library Prep Master Mix Set for Illumina (New England Biolabs, Inc.); manufacturers’ protocols were followed with the following modifications. RNA was fragmented for 10 min and then purified with RNeasy MinElute spin columns (QIAGEN). RT was performed with SuperScript III (Invitrogen). Library preparation reactions were cleaned up using Agencourt AMPure XP beads. Size selection was performed before ligation-mediated PCR using Invitrogen E-Gel 2% with SYBR Safe staining. Excised gel fragments were purified with the QIAQuick Gel Extraction kit (QIAGEN). Adapters and primers were synthesized by IDT according to published Illumina sequences. Enrichment PCR was performed with Kapa HiFi HotStart ReadMix. Primers were used at a final concentration of 500 nM; cycling parameters were as follows: 94°C for 5 min, 15 cycles of 94°C for 1 min, 62°C for 30 s, 72°C for 45 s, and then 72°C for 10 min. Libraries were quantified using the BioAnalyzer DNA 1000 chips (Agilent Technologies), diluted to 12 pM, and sequenced for 100 cycles (paired-end) on the HiSeq 2000 (Illumina) using standard methods.
Mouse transcriptome analysis.
RNA-Seq data from HBUS tissue samples were mapped to the mouse reference genome and transcriptome (GRCm38 and the Jul 2012 ENSEMBL gene build, respectively) using Tophat (Trapnell et al., 2009). Gene-level sequence counts were extracted for all annotated protein-coding genes using htseq-count by taking the strict intersection between reads and the transcript models associated with each gene. Raw count data were filtered to remove low expressed genes with less than five counts in any sample. Remaining data (12,697 genes) were normalized with trimmed mean of M (TMM) normalization and analyzed for differentially expressed genes using the Bioconductor EdgeR package version 2.11 Bioconductor/R (Robinson et al., 2010). To take into account the experimental design where paired unaffected cecal tissue and polyp tissue samples were isolated for each mouse, we fitted an additive generalized linear model that incorporated mouse + tissue effects to adjust for any baseline differences between the mice. Statistically significant differentially expressed genes between polyp and surrounding cecal (normal) tissues (Q < 0.05) were selected in gene-wise log-likelihood ratio tests that were corrected for multiple testing by Benjamini and Hochberg FDR. GO analysis on significantly regulated genes was performed using ClueGO (Bindea et al., 2009) for GO terms containing at least 25 genes, redundancy was reduced using GO term fusion, connections were based on kappa 0.5, the leading overview GO term was selected based on %Genes/Term, and GO terms with Q < 0.05 (FDR) were considered significantly enriched.
Accession numbers.
Accession numbers for all primary array and sequencing data are available from the NCBI under BioProject accession no. PRJNA207540 and GEO accession no. GSE47736.
Statistical analysis.
Statistical analysis for BeadArray, Microbiome, and RNA-Seq was performed as described above. For all other experiments, differences among means were evaluated by a 2 × 2 contingency table using Fisher’s exact test (Prism version 5; GraphPad Software), a two-tailed Wilcoxon rank sum test, or pairwise Wilcoxon rank sum test (CRAN/R version 3.0.2); P < 0.05 was considered significant. For multiple comparisons, p-values were adjusted using Benjamini and Hochberg (FDR). No samples were excluded from analysis. All results shown represent mean ± SEM.
Online supplemental material.
Table S1, included as a separate PDF file, shows differentially regulated genes by RNA-Seq analysis in tissue isolated from HBUS polyps compared with unaffected proximal cecum, as show in the heat map. Table S2, included as a separate PDF file, shows ClueGO analysis of the RNA-Seq analysis in tissue isolated from HBUS polyps compared with unaffected proximal cecum. Table S3, included as a separate PDF file, shows a subsampled and filtered OTU table of Taconic moms, rederived HBUS mice, and HBUS and WT mice obtained through interbreeding with mice obtained from the Jackson Laboratory. Table S4, included as a separate PDF file, shows statistical analysis of OTUs in Table S3. Table S5, included as a separate PDF file, shows differentially regulated genes by BeadArray in CD45+ cells isolated from HBUS polyps compared with unaffected proximal cecum, as show in the heat map. Table S6, included as a separate PDF file, shows ClueGO analysis of the BeadArray analysis in CD45+ cells isolated from HBUS polyps compared with unaffected proximal cecum. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20131587/DC1.
Supplementary Material
Acknowledgments
We would like to thank the Genomics Core Facility of the Icahn Institute for Genomics and Multiscale Biology for help with RNA-Seq, Jeremiah J. Faith for helpful discussions, and Taciana Salviano and Alan Soto for experimental help.
We thank Jenny and Jon Steingart and Jenna and Paul Segal for a grant supporting G. Bongers and the CAPES Foundation (Brazil) for a grant supporting T.H. Geraldino. This work was supported by National Institutes of Health grants 1R01CA161373-01 and P01 DK072201 to S.A. Lira and in part by the Icahn School of Medicine at Mount Sinai for providing scientific computing resources.
The authors declare no competing financial interests.
Author contributions: G. Bongers, H. van Bakel, and S.A. Lira conceived and designed the experiments; G. Bongers, M.E. Pacer, L. Chen, Z. He, D. Hashimoto, T.H. Geraldino, K.A. Kelley, and G.C. Furtado performed the experiments; G. Bongers, L. Chen, Z. He, D. Hashimoto, J.C. Clemente, and H. van Bakel analyzed the data; J. Ochando, J.C. Clemente, M. Merad, and H. van Bakel contributed reagents/materials/analysis tools; and G. Bongers, J.C. Clemente, H. van Bakel, and S.A. Lira wrote the manuscript.
Footnotes
Abbreviations used:
- FDR
- false discovery rate
- GI
- gastrointestinal
- OTU
- operational taxonomic unit
- qPCR
- quantitative PCR
- rDNA
- ribosomal DNA
- SP
- serrated polyp
References
- Arthur J.C., Perez-Chanona E., Mühlbauer M., Tomkovich S., Uronis J.M., Fan T.J., Campbell B.J., Abujamel T., Dogan B., Rogers A.B., et al. 2012. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 338:120–123 10.1126/science.1224820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates J.M., Mittge E., Kuhlman J., Baden K.N., Cheesman S.E., Guillemin K. 2006. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol. 297:374–386 10.1016/j.ydbio.2006.05.006 [DOI] [PubMed] [Google Scholar]
- Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. 2009. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 25:1091–1093 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N.A., Subramanian S., Faith J.J., Gevers D., Gordon J.I., Knight R., Mills D.A., Caporaso J.G. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods. 10:57–59 10.1038/nmeth.2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongers G., Maussang D., Muniz L.R., Noriega V.M., Fraile-Ramos A., Barker N., Marchesi F., Thirunarayanan N., Vischer H.F., Qin L., et al. 2010. The cytomegalovirus-encoded chemokine receptor US28 promotes intestinal neoplasia in transgenic mice. J. Clin. Invest. 120:3969–3978 10.1172/JCI42563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongers G., Muniz L.R., Pacer M.E., Iuga A.C., Thirunarayanan N., Slinger E., Smit M.J., Reddy E.P., Mayer L., Furtado G.C., et al. 2012. A role for the epidermal growth factor receptor signaling in development of intestinal serrated polyps in mice and humans. Gastroenterology. 143:730–740 10.1053/j.gastro.2012.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruewer M., Luegering A., Kucharzik T., Parkos C.A., Madara J.L., Hopkins A.M., Nusrat A. 2003. Proinflammatory cytokines disrupt epithelial barrier function by apoptosis-independent mechanisms. J. Immunol. 171:6164–6172 [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7:335–336 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M., et al. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6:1621–1624 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I., Blaser M.J. 2012. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 13:260–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente J.C., Ursell L.K., Parfrey L.W., Knight R. 2012. The impact of the gut microbiota on human health: an integrative view. Cell. 148:1258–1270 10.1016/j.cell.2012.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley J.M., Thomay A.A., Connolly M.D., Reichner J.S., Albina J.E. 2008. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J. Leukoc. Biol. 83:64–70 10.1189/jlb.0407247 [DOI] [PubMed] [Google Scholar]
- Dapito D.H., Mencin A., Gwak G.-Y., Pradere J.-P., Jang M.-K., Mederacke I., Caviglia J.M., Khiabanian H., Adeyemi A., Bataller R., et al. 2012. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 21:504–516 10.1016/j.ccr.2012.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessein R., Gironella M., Vignal C., Peyrin-Biroulet L., Sokol H., Secher T., Lacas-Gervais S., Gratadoux J.-J., Lafont F., Dagorn J.-C., et al. 2009. Toll-like receptor 2 is critical for induction of Reg3 beta expression and intestinal clearance of Yersinia pseudotuberculosis. Gut. 58:771–776 10.1136/gut.2008.168443 [DOI] [PubMed] [Google Scholar]
- Devkota S., Wang Y., Musch M.W., Leone V., Fehlner-Peach H., Nadimpalli A., Antonopoulos D.A., Jabri B., Chang E.B. 2012. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 487:104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Zhang Y., Tatum R., Chen Y.H. 2011. Detection of tight junction barrier function in vivo by biotin. Methods Mol. Biol. 762:91–100 10.1007/978-1-61779-185-7_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P., Kibbe W.A., Lin S.M. 2008. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 24:1547–1548 10.1093/bioinformatics/btn224 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 26:2460–2461 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Faith D.P., Baker A.M. 2006. Phylogenetic diversity (PD) and biodiversity conservation: Some bioinformatics challenges. Evol. Bioinform. Online. 2:121–128 [PMC free article] [PubMed] [Google Scholar]
- Finegold S.M., Jousimies-Somer H. 1997. Recently described clinically important anaerobic bacteria: Medical aspects. Clin. Infect. Dis. 25(s2, Suppl 2):S88–S93 10.1086/516237 [DOI] [PubMed] [Google Scholar]
- Fiske W.H., Threadgill D., Coffey R.J. 2009. ERBBs in the gastrointestinal tract: Recent progress and new perspectives. Exp. Cell Res. 315:583–601 10.1016/j.yexcr.2008.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán J.E., Pace J., Hayman M.J. 1992. Involvement of the epidermal growth factor receptor in the invasion of cultured mammalian cells by Salmonella typhimurium. Nature. 357:588–589 10.1038/357588a0 [DOI] [PubMed] [Google Scholar]
- Garcia M.R., Ledgerwood L., Yang Y., Xu J., Lal G., Burrell B., Ma G., Hashimoto D., Li Y., Boros P., et al. 2010. Monocytic suppressive cells mediate cardiovascular transplantation tolerance in mice. J. Clin. Invest. 120:2486–2496 10.1172/JCI41628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoukos G., Ciulla D.M., Huang K., Haas B.J., Izard J., Levin J.Z., Livny J., Earl A.M., Gevers D., Ward D.V., et al. 2012. Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol. 13:r23 10.1186/gb-2012-13-3-r23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton J.P., Xie G., Raufman J.-P., Hogan S., Griffin T.L., Packard C.A., Chatfield D.A., Hagey L.R., Steinbach J.H., Hofmann A.F. 2007. Human cecal bile acids: Concentration and spectrum. Am. J. Physiol. Gastrointest. Liver Physiol. 293:G256–G263 10.1152/ajpgi.00027.2007 [DOI] [PubMed] [Google Scholar]
- Hartman A.L., Lough D.M., Barupal D.K., Fiehn O., Fishbein T., Zasloff M., Eisen J.A. 2009. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc. Natl. Acad. Sci. USA. 106:17187–17192 10.1073/pnas.0904847106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Littman D.R. 2012. The microbiome in infectious disease and inflammation. Annu. Rev. Immunol. 30:759–795 10.1146/annurev-immunol-020711-074937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.S., O’brien M.J., Yang S., Farraye F.A. 2004. Hyperplastic polyps, serrated adenomas, and the serrated polyp neoplasia pathway. Am. J. Gastroenterol. 99:2242–2255 10.1111/j.1572-0241.2004.40131.x [DOI] [PubMed] [Google Scholar]
- Ivanov I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139:485–498 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl-Fie T.E., Chitayat S., Hood M.I., Damo S., Restrepo N., Garcia C., Munro K.A., Chazin W.J., Skaar E.P. 2011. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe. 10:158–164 10.1016/j.chom.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z., Maeda T., Tamura A., Nakamura T., Yamazaki Y., Shiratori H., Yashiro K., Tsukita S., Hamada H. 2012. EpCAM contributes to formation of functional tight junction in the intestinal epithelium by recruiting claudin proteins. Dev. Biol. 371:136–145 10.1016/j.ydbio.2012.07.005 [DOI] [PubMed] [Google Scholar]
- Lin S.M., Du P., Huber W., Kibbe W.A. 2008. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 36:e11 10.1093/nar/gkm1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioni M., Brafford P., Andl C., Rustgi A., El-Deiry W., Herlyn M., Smalley K.S.M. 2007. Dysregulation of claudin-7 leads to loss of E-cadherin expression and the increased invasion of esophageal squamous cell carcinoma cells. Am. J. Pathol. 170:709–721 10.2353/ajpath.2007.060343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis E., Ribbens C., Godon A., Franchimont D., De Groote D., Hardy N., Boniver J., Belaiche J., Malaise M. 2000. Increased production of matrix metalloproteinase-3 and tissue inhibitor of metalloproteinase-1 by inflamed mucosa in inflammatory bowel disease. Clin. Exp. Immunol. 120:241–246 10.1046/j.1365-2249.2000.01227.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Lladser M.E., Knights D., Stombaugh J., Knight R. 2011. UniFrac: An effective distance metric for microbial community comparison. ISME J. 5:169–172 10.1038/ismej.2010.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A., Cassatella M.A., Costantini C., Jaillon S. 2011. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 11:519–531 10.1038/nri3024 [DOI] [PubMed] [Google Scholar]
- McColl K.E.L. 2010. Clinical practice. Helicobacter pylori infection. N. Engl. J. Med. 362:1597–1604 10.1056/NEJMcp1001110 [DOI] [PubMed] [Google Scholar]
- McDonald D., Price M.N., Goodrich J., Nawrocki E.P., DeSantis T.Z., Probst A., Andersen G.L., Knight R., Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6:610–618 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt M.E., Donaldson J.R. 2009. Effect of bile salts on the DNA and membrane integrity of enteric bacteria. J. Med. Microbiol. 58:1533–1541 10.1099/jmm.0.014092-0 [DOI] [PubMed] [Google Scholar]
- Na X., Zhao D., Koon H.W., Kim H., Husmark J., Moyer M.P., Pothoulakis C., LaMont J.T. 2005. Clostridium difficile toxin B activates the EGF receptor and the ERK/MAP kinase pathway in human colonocytes. Gastroenterology. 128:1002–1011 10.1053/j.gastro.2005.01.053 [DOI] [PubMed] [Google Scholar]
- Nagano Y., Itoh K., Honda K. 2012. The induction of Treg cells by gut-indigenous Clostridium. Curr. Opin. Immunol. 24:392–397 10.1016/j.coi.2012.05.007 [DOI] [PubMed] [Google Scholar]
- Noffsinger A.E. 2009. Serrated polyps and colorectal cancer: new pathway to malignancy. Annu. Rev. Pathol. 4:343–364 10.1146/annurev.pathol.4.110807.092317 [DOI] [PubMed] [Google Scholar]
- Pizarro-Cerdá J., Cossart P. 2006. Bacterial adhesion and entry into host cells. Cell. 124:715–727 10.1016/j.cell.2006.02.012 [DOI] [PubMed] [Google Scholar]
- Plottel C.S., Blaser M.J. 2011. Microbiome and malignancy. Cell Host Microbe. 10:324–335 10.1016/j.chom.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon J.M., Kang D.-J., Hylemon P.B. 2006. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47:241–259 10.1194/jlr.R500013-JLR200 [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. 2010. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor R.B. 2008. Microbial influences in inflammatory bowel diseases. Gastroenterology. 134:577–594 10.1053/j.gastro.2007.11.059 [DOI] [PubMed] [Google Scholar]
- Schwabe R.F., Jobin C. 2013. The microbiome and cancer. Nat. Rev. Cancer. 13:800–812 10.1038/nrc3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I., Tam N.M., Jogova M., Robertson M.L., Li Y., Lupp C., Finlay B.B. 2008. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76:4726–4736 10.1128/IAI.00319-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata K., Yamada H., Hara H., Kishihara K., Yoshikai Y. 2007. Resident Vδ1+ γδ T cells control early infiltration of neutrophils after Escherichia coli infection via IL-17 production. J. Immunol. 178:4466–4472 [DOI] [PubMed] [Google Scholar]
- Singh A.B., Sugimoto K., Dhawan P., Harris R.C. 2007. Juxtacrine activation of EGFR regulates claudin expression and increases transepithelial resistance. Am. J. Physiol. Cell Physiol. 293:C1660–C1668 10.1152/ajpcell.00274.2007 [DOI] [PubMed] [Google Scholar]
- Smyth G.K. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:e3. [DOI] [PubMed] [Google Scholar]
- Stearns J.C., Lynch M.D.J., Senadheera D.B., Tenenbaum H.C., Goldberg M.B., Cvitkovitch D.G., Croitoru K., Moreno-Hagelsieb G., Neufeld J.D. 2011. Bacterial biogeography of the human digestive tract. Sci Rep. 1:170 10.1038/srep00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Raab G., Moses M.A., Fernandez C.A., Klagsbrun M. 1997. Matrix metalloproteinase-3 releases active heparin-binding EGF-like growth factor by cleavage at a specific juxtamembrane site. J. Biol. Chem. 272:31730–31737 10.1074/jbc.272.50.31730 [DOI] [PubMed] [Google Scholar]
- Tamura A., Kitano Y., Hata M., Katsuno T., Moriwaki K., Sasaki H., Hayashi H., Suzuki Y., Noda T., Furuse M., et al. 2008. Megaintestine in claudin-15-deficient mice. Gastroenterology. 134:523–534: e3 10.1053/j.gastro.2007.11.040 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 25:1105–1111 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C., Taur Y., Jenq R.R., Equinda M.J., Son T., Samstein M., Viale A., Socci N.D., van den Brink M.R., Kamboj M., Pamer E.G. 2010. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 120:4332–4341 10.1172/JCI43918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A.M. 2007. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35(Web Server):W71–W74 10.1093/nar/gkm306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ampting M.T.J., Loonen L.M.P., Schonewille A.J., Konings I., Vink C., Iovanna J., Chamaillard M., Dekker J., van der Meer R., Wells J.M., Bovee-Oudenhoven I.M.J. 2012. Intestinally secreted C-type lectin Reg3b attenuates salmonellosis but not listeriosis in mice. Infect. Immun. 80:1115–1120 10.1128/IAI.06165-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Rosenvinge E.C., Song Y., White J.R., Maddox C., Blanchard T., Fricke W.F. 2013. Immune status, antibiotic medication and pH are associated with changes in the stomach fluid microbiota. ISME J. 7:1354–1366 10.1038/ismej.2013.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Devkota S., Musch M.W., Jabri B., Nagler C., Antonopoulos D.A., Chervonsky A., Chang E.B. 2010. Regional mucosa-associated microbiota determine physiological expression of TLR2 and TLR4 in murine colon. PLoS ONE. 5:e13607 10.1371/journal.pone.0013607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing B.P., Russell S.L., Finlay B.B. 2011. Shifting the balance: Antibiotic effects on host-microbiota mutualism. Nat. Rev. Microbiol. 9:233–243 10.1038/nrmicro2536 [DOI] [PubMed] [Google Scholar]
- Wu S., Rhee K.J., Albesiano E., Rabizadeh S., Wu X., Yen H.R., Huso D.L., Brancati F.L., Wick E., McAllister F., et al. 2009. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15:1016–1022 10.1038/nm.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.