Abstract
The wild relatives of rice (Oryza sativa L.) are useful sources of alleles that have evolved to adapt in diverse environments around the world. Oryza rufipogon, the known progenitor of the cultivated rice, harbors genes that have been lost in cultivated varieties through domestication or evolution. This makes O. rufipogon an ideal source of value-added traits that can be utilized to improve the existing rice cultivars. To explore the potential of the rice progenitor as a genetic resource for improving O. sativa, 33 chromosome segment substitution lines (CSSLs) of O. rufipogon (W0106) in the background of the elite japonica cultivar Koshihikari were developed and evaluated for several agronomic traits. Over 90% of the entire genome was introgressed from the donor parent into the CSSLs. A total of 99 putative QTLs were detected, of which 15 were identified as major effective QTLs that have significantly large effects on the traits examined. Among the 15 major effective QTLs, a QTL on chromosome 10 showed a remarkable positive effect on the number of grains per panicle. Comparison of the putative QTLs identified in this study and previous studies indicated a wide genetic diversity between O. rufipogon accessions.
Keywords: Oryza rufipogon, chromosome segment substitution lines (CSSLs), QTL mapping
Introduction
Food problem is a serious global issue that humans need to address. According to the World Food Program, 870 million people in the world do not have enough to eat. The Food and Agriculture Organization estimated that a 60% increase in food production would be necessary to feed the continuously growing human population, which is predicted to increase up to 9 billion in 2050 (Alexandratos and Bruinsma 2012).
Rice is one of the most important staple food, with 50% of the world population depending on rice as a main carbohydrate source. Breeding new rice varieties with high yield, resistance to pests and diseases and tolerance to abiotic stresses is a key strategy in increasing rice production and contributing in world food security.
The wild rice species serve as a treasure trove of novel QTLs and alleles underlying traits of agronomic importance. Throughout the history of rice domestication, genes controlling highly beneficial traits in rice such as the non-shattering of grains, high yield and ideal plant architecture were preferentially selected, whereas genes with negative effects on agronomic traits were selected against (Ishii et al. 2013, Konishi et al. 2006, Shomura et al. 2008, Tan et al. 2008, Wang et al. 2008, Weng et al. 2008). Early domestication by selection is biased towards traits that are easily seen hence, phenotypes that are not easily observed such as biotic and abiotic stress resistance were not as strongly targeted for selection. Because of this, wild rice species may still have genes controlling beneficial traits that were not actively selected. O. rufipogon is the known wild progenitor of O. sativa, which was originally cultivated in China (Huang et al. 2012). This species largely inhabits Asia, but is also spread throughout Africa, North America, Central America, South America and Australia. The wide genetic diversity of O. rufipogon allows it to adapt to diverse environmental conditions (Cai et al. 2004, Huang et al. 2012, Song et al. 2003, Sun et al. 2001).
To date, several quantitative trait loci (QTL) for various agronomic traits have been identified in O. rufipogon (Marri et al. 2005, McCouch et al. 2006, Onishi et al. 2007, Tan et al. 2007). In general, F2 populations, backcross inbred lines or recombinant inbred lines are used in QTL analysis. However, only QTLs with major effects were detected in these populations. Minor QTLs or those with epistatic interactions with other QTLs were not easily identified. For investigating the potential of germplasm as gene resource without concerning complex interactions among QTLs as much as possible, chromosome segment substitution lines (CSSLs) have been developed for rice (Ando et al. 2008, Doi et al. 1997, Ebitani et al. 2005, Kubo et al. 2002, Shim et al. 2010). Each line consists of one or few homozygous chromosome segments derived from a donor parent in the genetic background of a recurrent parent. Each line of CSSLs should exhibit only the effects by the introduction of each chromosome segment. Therefore, the effect of each chromosome segment on a trait can be evaluated without genetic interactions among QTLs. In addition, genetically fixed lines, such as CSSLs, can be simultaneously and repeatedly subjected to diverse analyses. These features make CSSLs a powerful genetic resource for surveying the genetic potential of donor germplasm.
Here, we report the development and evaluation of CSSLs that have chromosome segments of the wild rice O. rufipogon in the genetic background of the elite japonica cultivar Koshihikari. The CSSLs were screened for several agronomic traits to evaluate their potential as a genetic resource to improve existing cultivars. We identified 15 major effective QTLs, some of which have not been reported in previous QTL studies for O. rufipogon.
Materials and Methods
Development of the CSSLs
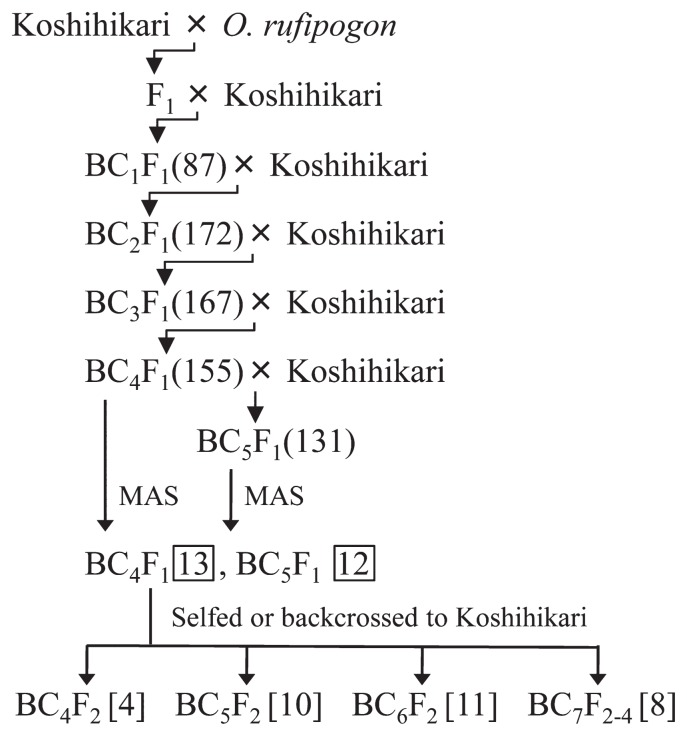
The O. rufipogon (Accession W0106) used in this study originated from India and was distributed from the National Institute of Genetics, Japan. The CSSLs, hereinafter named RSLs, were developed by crossing O. rufipogon as the donor parent with O. sativa cv. Koshihikari as the recurrent parent. The resulting F1 hybrid was backcrossed to Koshihikari to obtain 87 BC1F1 followed by successive backcrossing to the recurrent parent to produce 172 BC2F1, 167 BC3F1, 155 BC4F1 and 131 BC5F1 progenies. 155 BC4F1 and 131 BC5F1 lines were genotyped using 149 single nucleotide polymorphisms via the AcycloPrime-FP Detection System and Fluorescence Polarization Analyzer (Perkin Elmer Life Science, Boston, MA, USA) (Supplemental Table 1). The SNP markers, which were developed based on the Build 2 Pseudomolecules of cv. Nipponbare, were evenly distributed among the 12 rice chromosomes at an average marker interval of 2.63 Mb. Using marker-assisted selection (MAS), 13 BC4F1 and 12 BC5F1 were chosen for backcrossing or selfing. A total of 33 RSLs derived from BC4F2 (4 lines), BC5F2 (10 lines), BC6F2 (11 lines) and BC7F2-F4 (8 lines) were obtained through MAS. The breeding scheme used in developing the RSLs is outlined in Fig. 1.
Fig. 1.
Breeding scheme used for developing RSLs carrying O. rufipogon (W0106) chromosome segments in O. sativa L. cv. Koshihikari genetic background. MAS indicate that plants with optimal genotypes were selected by marker assisted selection. Numbers in parenthesis denote the number of lines subjected to MAS. Boxed numbers represent the numbers of candidate RSLs selected by MAS. Numbers in brackets indicate the total number of RSLs derived from the preceding backcross generation.
Measurement of agronomic traits
The RSLs were evaluated for several agronomically important traits at the experimental field of the Honda Research Institute (HRI) in Kisarazu, Chiba, Japan. Seedlings of RSLs and Koshihikari were first raised in the greenhouse and transplanted in the field 30 days after sowing. Twenty-four seedlings each of the RSLs and Koshihikari were planted in double-row plots in the field with a spacing of 20 cm between hills and 30 cm between rows. At maturity, 5 plants at the 3rd–7th hills of the 1st row were individually harvested and measured for number of grains per panicle, grain length, grain width, grain thickness, 100-grain weight and seed fertility. Data on culm length, panicle length, number of primary branches per panicle were also collected. The days-to-heading of each RSL was recorded as the number of days from sowing to the actual flowering of 50% of the plants. The 100-grain weight was determined using seeds that were air-dried in a glasshouse for at least 2 months.
Detection of QTLs
The putative QTLs in the RSLs were identified by Dunnett’s multiple comparisons test at 95% confidence interval (P < 0.05). A putative QTL was assigned in the chromosome regions carried by the RSL which showed a significantly different mean value for the trait examined compared to that of Koshihikari. Boxplots for the mean values of each RSL were generated for each trait using an implemented function in R statistics software. Outliers in the boxplot were identified as an RSL carrying a chromosome segment harboring a major effective QTL.
Results
Development of CSSLs
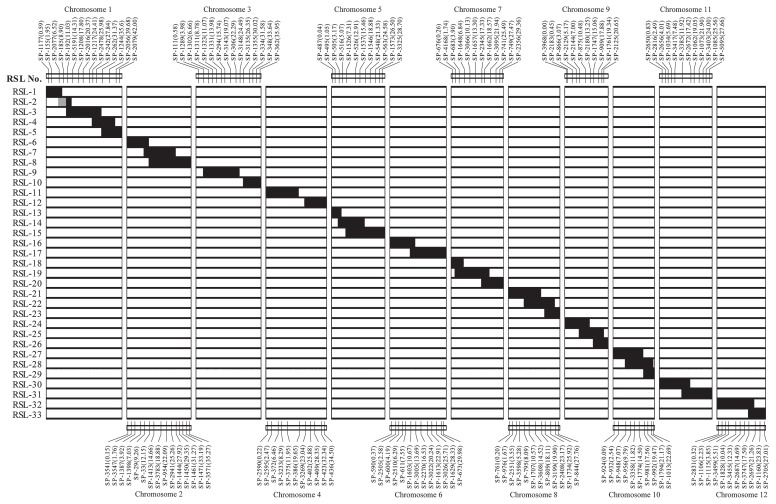
A total of 33 RSLs with homozygous chromosome segments from O. rufipogon substituted to the chromosome of Koshihikari were developed. More than 90% of the donor genome was introgressed in contiguous or overlapping chromosome segments in the RSL series. Approximately 5% of the donor genome was not successfully introgressed in the RSL set as indicated by gaps in chromosome 3 (3.98 Mb between the distal part of the short arm chromosome 3 and SP1289 and 4.06 Mb between SP306 and SP3135) and chromosome 4 (11.09 Mb between SP375 and SP3269) (Fig. 2). Chromosome segments of O. rufipogon around SP-185, an SNP marker at 8.90 Mb in chromosome 1 and SP-386, an SNP marker at 19.65 Mb in chromosome 4, could not be fixed in the Koshihikari background. These chromosome regions may contain genes involved in reproductive isolation.
Fig. 2.
Graphical representation of the genotypes of the 33 RSLs. Black bars and white bars represent homozygous segments from O. rufipogon and O. sativa, respectively. The gray bar indicates a heterozygous segment of O. rufipogon and O. sativa. The SNP markers used in marker-assisted selection are indicated as SP series. Numbers in parenthesis indicates the physical distance (Mb) of each SNP markers from the upper end of each chromosome.
Evaluation of RSLs
Data on the morphometric measurements of the agronomic traits in the 33 RSLs are presented in Table 1.
Table 1.
Morphometric measurements for different agronomic traits of 33 RSLs carrying chromosome segments from O. rufipogon
| Chr. No. | Lines | Days-to-heading (days) | Culm length (cm) | Panicle length (cm) | Number of primary branches/panicle | Number of grains/panicle | Grain length (mm) | Grain width (mm) | Grain thickness (mm) | 100-grain weight (g) | Fertility (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Koshihikari | 94.80 | 89.32 | 18.08 | 11.40 | 116.00 | 7.31 | 3.21 | 2.14 | 2.23 | 95.92 | |
| 1 | RSL1 | 92.00* | 75.56* | 19.56 | 12.40 | 158.00** | 7.34 | 3.26 | 2.17 | 2.23 | 94.28 |
| 1 | RSL2 | 91.60* | 80.76* | 20.48** | 12.40 | 138.80 | 7.41 | 3.36 | 2.25** | 2.37 | 96.04 |
| 1 | RSL3 | 94.80 | 90.08 | 18.74 | 10.60 | 112.40 | 7.32 | 3.16 | 2.14 | 2.25 | 97.20 |
| 1 | RSL4 | 98.00** | 91.72 | 20.55** | 10.00 | 108.50 | 7.42 | 3.30 | 2.20 | 2.50** | 92.13 |
| 1 | RSL5 | 96.00 | 82.00* | 19.16 | 10.60 | 100.00 | 7.30 | 3.24 | 2.17 | 2.26 | 95.24 |
| 2 | RSL6 | 95.60 | 70.44* | 18.24 | 9.80 | 123.20 | 7.29 | 3.17 | 2.05* | 2.08* | 94.96 |
| 2 | RSL7 | 93.20 | 72.24* | 18.26 | 10.80 | 135.20 | 7.34 | 2.92*† | 2.13 | 2.00* | 91.84 |
| 2 | RSL8 | 96.40 | 84.84 | 18.14 | 11.00 | 131.80 | 7.03 | 2.88*† | 2.09 | 1.91* | 90.74 |
| 3 | RSL9 | 100.00** | 82.08* | 20.30** | 11.60 | 100.00 | 8.44**† | 3.17 | 2.13 | 2.38** | 84.24* |
| 3 | RSL10 | 117.80**† | 70.68* | 19.06 | 10.20 | 88.80* | 7.35 | 3.33 | 2.10 | 2.13 | 76.32*† |
| 4 | RSL11 | 95.60 | 83.80 | 18.58 | 10.20 | 113.40 | 7.45 | 3.26 | 2.21** | 2.28 | 93.36 |
| 4 | RSL12 | 95.80 | 88.00 | 17.02 | 8.80* | 81.00* | 7.39 | 3.36 | 2.22** | 2.49** | 91.54 |
| 5 | RSL13 | 96.40 | 81.32* | 19.12 | 11.40 | 120.40 | 7.29 | 3.26 | 2.22** | 2.30 | 95.96 |
| 5 | RSL14 | 92.60 | 79.28* | 19.58 | 10.60 | 134.00 | 7.57 | 2.88*† | 2.07* | 2.00* | 95.66 |
| 5 | RSL15 | 94.00 | 94.28 | 21.08** | 11.00 | 119.20 | 7.73** | 3.30 | 2.25** | 2.55** | 87.20* |
| 6 | RSL16 | 116.00**† | 93.48 | 20.32** | 11.60 | 123.40 | 7.43 | 3.31 | 2.15 | 2.23 | 79.80*† |
| 6 | RSL17 | 97.00 | 100.56** | 21.10** | 13.20 | 157.20** | 7.38 | 3.24 | 2.15 | 2.23 | 94.18 |
| 7 | RSL18 | 95.20 | 88.52 | 19.42 | 12.00 | 122.40 | 7.16 | 3.37** | 2.22** | 2.32 | 94.54 |
| 7 | RSL19 | 105.80**† | 90.80 | 19.96** | 13.20 | 128.40 | 7.29 | 3.39** | 2.16 | 2.25 | 88.48 |
| 7 | RSL20 | 98.40** | 88.20 | 20.42** | 10.40 | 107.60 | 7.66** | 3.44** | 2.17 | 2.44** | 90.88 |
| 8 | RSL21 | 104.00** | 83.00* | 20.56** | 11.20 | 112.20 | 7.77** | 3.41** | 2.08 | 2.16 | 89.76 |
| 8 | RSL22 | 94.00 | 80.48* | 18.86 | 10.20 | 113.40 | 7.43 | 3.43** | 2.19 | 2.38** | 93.82 |
| 8 | RSL23 | 102.60** | 90.68 | 18.70 | 9.60 | 108.60 | 7.81**† | 3.40** | 2.09 | 2.28 | 92.32 |
| 9 | RSL24 | 94.00 | 77.92* | 19.44 | 10.40 | 117.00 | 7.28 | 3.23 | 2.08 | 2.18 | 90.78 |
| 9 | RSL25 | 95.40 | 75.08* | 18.12 | 10.60 | 96.40 | 7.20 | 3.24 | 2.08 | 2.07* | 94.44 |
| 9 | RSL26 | 95.20 | 76.72* | 18.62 | 10.60 | 107.40 | 7.33 | 3.32 | 2.07* | 2.12 | 87.80* |
| 10 | RSL27 | 101.60** | 36.56*† | N.D. | N.D. | N.D. | 6.84*† | 2.87*† | 2.08 | 1.77* | N.D. |
| 10 | RSL28 | 94.60 | 82.08* | 21.06** | 11.80 | 170.40**† | 7.51 | 3.42** | 2.18 | 2.46** | 97.86 |
| 10 | RSL29 | 93.40 | 77.28* | 19.24 | 11.80 | 126.80 | 7.32 | 3.31 | 2.15 | 2.21 | 93.16 |
| 11 | RSL30 | 93.60 | 75.48* | 19.08 | 11.00 | 117.40 | 7.68** | 3.35 | 2.17 | 2.33 | 96.60 |
| 11 | RSL31 | 93.60 | 77.76* | 20.56** | 11.60 | 136.60 | 7.26 | 3.35 | 2.11 | 2.16 | 93.94 |
| 12 | RSL32 | 97.40** | 87.48 | 18.58 | 11.80 | 102.80 | 7.49 | 3.39** | 2.22** | 2.44** | 96.14 |
| 12 | RSL33 | 96.20 | 84.16 | 18.86 | 11.40 | 130.40 | 7.06 | 3.25 | 2.12 | 1.92* | 7.28*† |
significantly higher than Koshihikari,
significantly lower than Koshihikari,
outlier in the boxplot (Fig. 3)
siginificance calculated at P < 0.05. b values shown are means of 5 samples.
Days-to-heading
RSL1 and 2 exhibited significantly earlier flowering (92.00 and 91.60 days, respectively), whereas another 10 RSLs showed significantly later flowering (95.20–117.80 days) compared to Koshihikari. RSL10 exhibited the latest flowering which was 23 days later than Koshihikari. This line has a chromosome segment of O. rufipogon between SP-3148 (at 24.49 Mb) and the distal end of the long arm of chromosome 3. RSL2 exhibited the earliest flowering, which was 3.2 days earlier than Koshihikari.
Culm and panicle length
The culm length of 20 lines was significantly different from that of Koshihikari. The culms of RSL17 were significantly longer by 11.24 cm, whereas the culms of 15 lines were 7.1–59.1% shorter than that of Koshihikari. In particular, RSL27 showed a severe reduction in culm length which was less than half of that of Koshihikari. RSL27 has a genomic fragment of O. rufipogon between the distal end of the short arm of chromosome 10 and SP981 (at 17.56 Mb). Although RSL28 has a partially overlapped introgressed segment with RSL27, no reduction in the culm length of RSL28 was observed. This indicates that the O. rufipogon genomic segment between the end of the short arm of chromosome 10 and SP948 (at 7.07 Mb) which spans approximately 7 Mb causes the severe reduction in the culm length of RSL27. Eleven lines had 10.4–16.7% longer panicles compared to Koshihikari. RSL17, which carries an introgressed segment from O. rufipogon in the long arm of chromosome 6, recorded the longest panicle (21.1 cm).
Number of primary branches per panicle and number of grains per panicle
RSL12 had 22.8% less primary branches compared to Koshihikari which recorded an average of 11.4 branches per panicle. This line has a chromosome segment from O. rufipogon between SP386 (at 19.65 Mb) and the distal part of the long arm of chromosome 4. No other lines showed significantly different value for number of primary branches compared to Koshihikari. For number of grains per panicle, significant increases ranging from 35.5 to 46.9% were observed in RSL1, 17 and 28, whereas significant reductions ranging from 23.4 to 30.2% were observed in RSL10 and 12.
Grain related traits and fertility
Significant increases in grain length were observed in 6 lines, whereas increases in grain width and thickness were observed in 8 and 7 lines, respectively. On the other hand, grain length, width and thickness were reduced in 1, 4 and 3 lines, respectively. One-hundred-grain weight significantly increased in 8 lines but decreased in another 7 lines. Nine lines had either an increase or decrease in grain size, without any effect on the 100-grain weight. A 6.7–14.3% increase in 100-grain weight, along with a corresponding expansion in grain size were observed in RSL9, 12, 15, 20, 22, 28 and 32. A 6.7–20.6% reduction in 100-grain weight, along with a corresponding reduction in grain size was observed in RSLs 6, 7, 8, 14 and 27. RSL4, 25 and 33 showed an increase and a decrease in 100-grain weight, respectively, without changes in grain size. This indicates that the chromosome segments possessed by these RSLs may affect the filling rate or the density of starch in the grains. RSL9, 10, 15, 16, 26 and 33 showed significant reductions in fertility, with RSL33 exhibiting an average of 92.4% sterility.
Identifying major effective QTLs
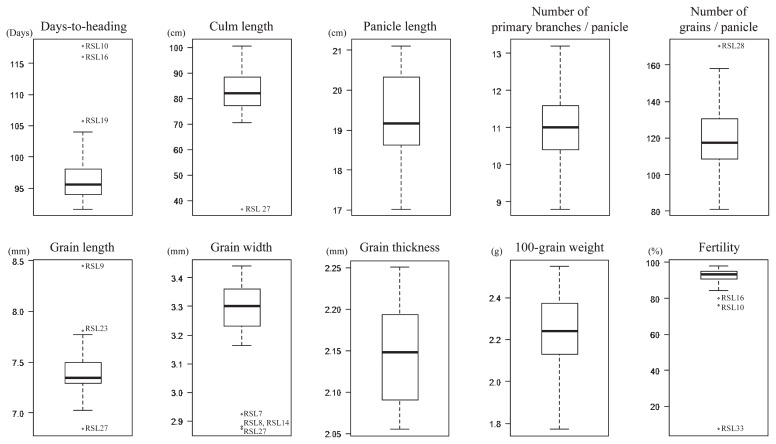
A total of 99 putative QTLs affecting several agronomic traits were detected in the RSLs, although some putative QTLs showed only small effects (Table 1). To identify the major effective QTLs, we drew boxplots using the mean values of each RSL for each trait. Outliers in the boxplots were identified as the RSLs harboring a major effective QTL (Table 1 and Fig. 3). The major effective QTLs were compared to QTLs that have been reported in previous studies using O. rufipogon (Table 2).
Fig. 3.
Boxplots of the mean values for each trait. Boxplots for all traits measured in this study were shown. Dots in boxplots indicate outliers showing mean values that are significantly different from others. These outliers were identified as RSLs that have a chromosome segment harboring a major effective QTL derived from O. rufipogon.
Table 2.
Comparison of the major effective QTLs identified in this study with previously reported QTLs from O. rufipogon
| Trait | CSSL | Chr. | Start positiion | End position | Effecta | QTLb | Referencec |
|---|---|---|---|---|---|---|---|
| Days-to-heading | RSL10 | 3 | 24,488,442 | End | 24.3% ↑ | ||
| RSL16 | 6 | End | 10,671,175 | 22.4% ↑ | dth6.1 | 1 | |
| RSL19 | 7 | 3,898,132 | 17,327,528 | 11.6% ↑ | dth7.1 | 2, 3, 4, 6 | |
|
| |||||||
| Culm length | RSL27 | 10 | End | 3,552,082 | 59.1% ↓ | ph10.1 | 3 |
|
| |||||||
| Grain number | RSL28 | 10 | 2,541,199 | 17,555,159 | 46.9% ↑ | ||
|
| |||||||
| Grain length | RSL9 | 3 | 57,726 | 24,488,442 | 15.5% ↑ | gl3 | 5 |
| RSL23 | 8 | 23,167,567 | End | 6.8% ↑ | |||
| RSL27 | 10 | End | 3,552,082 | 6.4% ↓ | |||
|
| |||||||
| Grain width | RSL7 | 2 | 9,256,584 | 27,919,903 | 9.0% ↓ | ||
| RSL8 | 2 | 9,256,584 | End | 10.3% ↓ | |||
| RSL14 | 5 | 3,173,133 | 7,305,956 | 10.3% ↓ | qSW5 | 6 | |
| RSL27 | 10 | End | 3,552,082 | 10.6% ↓ | |||
|
| |||||||
| Fertility | RSL10 | 3 | 24,488,442 | End | 20.4% ↓ | pss3.1 | 1, 4 |
| RSL16 | 6 | End | 10,671,175 | 16.8% ↓ | pss6.1 | 4 | |
| RSL33 | 12 | 17,499,935 | End | 92.4% ↓ | |||
↑ and ↓ indicate an increase or a decrease in the trait value compared to Koshihikari, respectively.
Names of QTLs reported in previous studies.
Reference numbers are as follows; 1 Xiao et al. 1998, 2 Moncada et al. 2001, 3 Septiningsih et al. 2003, 4 Thomson et al. 2003, 5 Lee et al. 2005, 6 Onishi et al. 2007.
For days-to-heading, RSL10, 16 and 19 were identified as the lines harboring major effective QTLs. These lines have an introgressed O. rufipogon chromosome segment in chromosomes 3, 6 and 7, respectively. The QTLs controlling days-to-heading from chromosomes 6 and 7 of O. rufipogon have already been reported in QTL studies using O. rufipogon (Moncada et al. 2001, Onishi et al. 2007, Septiningsih et al. 2003, Thomson et al. 2003, Xiao et al. 1998). However, this is the first report of a putative QTL in chromosome 3 of O. rufipogon controlling days-to-heading. RSL27 was the only line identified as an outlier for culm length in the boxplot, while no outlier was detected for panicle length. Despite many RSLs showing significant differences in culm and panicle length, only a few major effective QTLs were identified for these 2 traits. This result suggests the possibility that the genome of O. rufipogon possesses many QTLs which only has small effects on culm and panicle length. The QTL in the short arm of chromosome 10 of O. rufipogon affecting plant height has been previously reported as ph10.1 in a study by Septiningsih et al. (2003).
For grain number per panicle, RSL 28 was an outlier in the boxplot, producing an average of 54.4 more grains compared to Koshihikari. This line has an introgressed segment between SP932 and SP1013 in chromosome 10 (2.54–22.69 Mb). However, considering the overlaps in the substituted segments in RSL27, 28 and 29, the putative major effective QTL is located within a 3.06 Mb region between SP1774 (at 14.50 Mb) and SP981 (at 17.56 Mb). This locus only affected the number of grains per panicle without changing the number of primary branches in RSL28, which recorded a 46.9% increase in the number of grains per panicle compared to Koshihikari. No reports have been made yet regarding a QTL controlling the number of grains per panicle in the region corresponding to the genomic segment of O. rufipogon possessed by RSL28. No major effective QTLs for number of primary branches per panicle were identified.
RSL9, 23 and 27 were outliers for grain length. A putative QTL in the segment of chromosome 3 that is carried by RSL9 had already been reported (Lee et al. 2005). RSL7, 8, 14 and 27 possessed major effective QTLs for grain width. The chromosome segment derived from O. rufipogon in the short arm of chromosome 10 of RSL27 reduced the length and the width of grains, as well as the length of the culms, suggesting that this QTL globally changes plant architecture by affecting cell elongation or proliferation.
Three major QTLs controlling fertility were identified in chromosomes 3, 6 and 12 of O. rufipogon, coinciding with previous reports (Onishi et al. 2007, Xiao et al. 1998). The QTL in the long arm of chromosome 12 however, has not yet been reported before despite its major effects on fertility.
Discussion
The use of CSSLs can simplify the study of complex genetic traits and facilitate the identification of genes underlying agronomic traits. Although the effects of genetic interactions cannot be evaluated using CSSLs (Ebitani et al. 2005), these lines are very useful in the genetic study of agronomic traits which often have quantitative properties. In the past decades, several CSSLs have been constructed using elite cultivars (Ando et al. 2008, Ebitani et al. 2005, Kubo et al. 2002, Zhang et al. 2011). These CSSLs were utilized in identifying new QTLs which have the potential to improve existing rice cultivars (Adachi et al. 2010, Marzougui et al. 2012). Identification of genes controlling high-yield, good grain quality and biotic and abiotic stress resistance in elite cultivars is important in understanding the molecular basis of such traits and their use in the strategic breeding of rice.
The wild rice relatives serve as a rich reservoir of novel genes or alleles that can be used for the improvement of existing rice cultivars. One good example is the identification of Xa21 from O. longstaminata, and its successful use in breeding for bacterial blight resistance in rice (Ronald et al. 1992). To date, CSSLs derived from distant relatives of rice including O. meridionalis, O. glumepatula, O. rufipogon, O. glaberrima have been constructed (Hirabayashi et al. 2010, Shim et al. 2010, Yoshimura et al. 2010).
A previous study on the origin of current cultivars showed that O. rufipogon from southern China was the progenitor of the current rice cultivars. Domestication of O. rufipogon using only a limited number of individuals resulted in approximately 18% loss in the genetic diversity of this species (Huang et al. 2012). However, O. rufipogon that were geographically distributed outside of China may still have alleles that were lost in current cultivars. In this study, we reported the development of 33 RSLs harboring chromosome segments of O. rufipogon originating from India in the genomic background of O. sativa L. cv. Koshihikari. Field evaluation of the RSLs allowed for the identification of a total of 99 putative QTLs including QTLs with small effects. Of the 99 putative QTLs detected, 15 were identified to have major effects. Only 3 major effective QTLs showed agronomically positive effects on the traits, One QTL is for number of grains per panicle and the other two QTLs are for grain length. In particular, a putative QTL harbored by RSL28 exhibited a highly positive effect on number of grains per panicle, causing a 46.9% increase in the value of the trait examined. This putative QTL is located between 14.50 and 17.5 Mb of chromosome 10 and has the potential to improve the yield of current cultivars. Despite the large effects of the putative QTL for number of grains per panicle that was identified in this study, previous reports on such QTL from O. rufipogon have not been made (Table 2). O. rufipogon is more genetically diverse than O. sativa. Our results suggest that the use of a different accession of O. rufipogon would lead to the discovery of novel QTLs. Indeed, two sets of CSSLs for two accessions of O. rufipogon showed different QTL positions for rice blast resistance (Hirabayashi et al. 2010). Although some papers have already reported the development of CSSLs for O. rufipogon (Hirabayashi et al. 2010, Tan et al. 2007), a variety of accessions should be subjected to such genetic survey to retrieve genes which were lost through the history of domestication or evolution.
Detailed studies on QTLs including those with minor effects using CSSLs may give us insights into the molecular basis of agronomic traits and also into the evolution of the genus Oryza. Minor QTLs may also be required in fine-tuning the adaptability of cultivars in diverse environments. Unlike genetically non-fixed inbred populations such as an F2 population, CSSLs can be easily evaluated for traits of interest under diverse environmental conditions. Construction of a variety of CSSLs containing genomes from wild relatives would accelerate breeding of ideal cultivars.
Acknowledgments
We thank the National Institute of Genetics for the seeds of O. rufipogon Accession W0106 in part of National Bioresource Project of MEXT, Japan and Honda Research Institute Japan Co., Ltd. for their support for developing and measuring RSLs.
This work was supported by Science and technology research promotion program for agriculture, forestry, fisheries and food industry and by the Japan Science and Technology Agency-Japan International Cooperation Agency within the framework of the SATREPS to M. A.. This work was also supported by Grants in Aid for Scientific Research (22119007 to M.A.) from the Ministry of Education, Culture, Sports and Science.
Literature Cited
- Adachi, S., Tsuru, Y., Kondo, M., Yamamoto, T., Arai-Sanoh, Y., Ando, T., Ookawa, T., Yano, M. and Hirasawa, T. (2010) Characterization of a rice variety with high hydraulic conductance and identification of the chromosome region responsible using chromosome segment substitution lines. Ann. Bot. 106: 803–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandratos, N. and Bruinsma, J. (2012) World agriculture towards 2030/2050: the 2012 revision, FAO, Rome, ESA Working Paper No. 12-03. [Google Scholar]
- Ando, T., Yamamoto, T., Shimizu, T., Ma, X.F., Shomura, A., Takeuchi, Y., Lin, S.Y. and Yano, M. (2008) Genetic dissection and pyramiding of quantitative traits for panicle architecture by using chromosomal segment substitution lines in rice. Theor. Appl. Genet. 116: 881–890 [DOI] [PubMed] [Google Scholar]
- Cai, H.-W., Wang, X.-K. and Morishima, H. (2004) Comparison of population genetic structures of common wild rice (Oryza rufipogon Griff.), as revealed by analyses of quantitative traits, allozymes, and RFLPs. Heredity 92: 409–417 [DOI] [PubMed] [Google Scholar]
- Doi, K., Iwata, N. and Yoshimura, A. (1997) The construction of chromosome substitution lines of African rice (Oryza glaberrima Steud.) in the background of Japonica rice (O. sativa L.). Rice Genet. Newsl. 14: 39–41 [Google Scholar]
- Ebitani, T., Takeuchi, Y., Nonoue, Y., Yamamoto, T., Takeuchi, K. and Yano, M. (2005) Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of indica rice cultivar “Kasalath” in a genetic background of japonica elite cultivar “Koshihikari.” Breed. Sci. 55: 65–73 [Google Scholar]
- Hirabayashi, H., Sato, H., Nonoue, Y., Kuno-Takemoto, Y., Takeuchi, Y., Kato, H., Nemoto, H., Ogawa, T., Yano, M., Imbe, T.et al. (2010) Development of introgression lines derived from Oryza rufipogon and O. glumaepatula in the genetic background of japonica cultivated rice (O. sativa L.) and evaluation of resistance to rice blast. Breed. Sci. 60: 604–612 [Google Scholar]
- Huang, X., Kurata, N., Wei, X., Wang, Z.-X., Wang, A., Zhao, Q., Zhao, Y., Liu, K., Lu, H., Li, W.et al. (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, T., Numaguchi, K., Miura, K., Yoshida, K., Thanh, P.T., Htun, T.M., Yamasaki, M., Komeda, N., Matsumoto, T., Terauchi, R.et al. (2013) OsLG1 regulates a closed panicle trait in domesticated rice. Nat. Genet. 45: 462–5, 465e1–2 [DOI] [PubMed] [Google Scholar]
- Konishi, S., Izawa, T., Lin, S.Y., Ebana, K., Fukuta, Y., Sasaki, T. and Yano, M. (2006) An SNP caused loss of seed shattering during rice domestication. Science 312: 1392–1396 [DOI] [PubMed] [Google Scholar]
- Kubo, T., Aida, Y., Nakamura, K., Tsunematsu, H., Doi, K. and Yoshimura, A. (2002) Reciprocal chromosome segment substitution series derived from japonica and indica cross of rice (Oryza sativa L.). Breed. Sci. 52: 319–325 [Google Scholar]
- Lee, S.-J., Oh, C.-S., Suh, J.-P., McCouch, S.R. and Ahn, S.-N. (2005) Identification of QTLs for domestication-related and agronomic traits in an Oryza sativa × O. rufipogon BC1F7 population. Plant Breed. 124: 209–219 [Google Scholar]
- Marri, P.R., Sarla, N., Reddy, L.V. and Siddiq, E.A. (2005) Identification and mapping of yield and yield related QTLs from an Indian accession of Oryza rufipogon. BMC Genet. 6: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzougui, S., Sugimoto, K., Yamanouchi, U., Shimono, M., Hoshino, T., Hori, K., Kobayashi, M., Ishiyama, K. and Yano, M. (2012) Mapping and characterization of seed dormancy QTLs using chromosome segment substitution lines in rice. Theor. Appl. Genet. 124: 893–902 [DOI] [PubMed] [Google Scholar]
- McCouch, S.R., Sweeney, M., Li, J., Jiang, H., Thomson, M., Septiningsih, E., Edwards, J., Moncada, P., Xiao, J., Garris, A.et al. (2006) Through the genetic bottleneck: O. rufipogon as a source of trait-enhancing alleles for O. sativa. Euphytica 154: 317–339 [Google Scholar]
- Moncada, P., Martínez, C.P., Borrero, J., Chatel, M., Gauch, H.Jr, Guimaraes, E., Tohme, J. and McCouch, S.R. (2001) Quantitative trait loci for yield and yield components in an Oryza sativa × Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor. Appl. Genet. 102: 41–52 [Google Scholar]
- Onishi, K., Horiuchi, Y., Ishigoh-Oka, N., Takagi, K., Ichikawa, N., Maruoka, M. and Sano, Y. (2007) A QTL cluster for plant architecture and its ecological significance in Asian wild rice. Breed. Sci. 57: 7–16 [Google Scholar]
- Ronald, P.C., Albano, B., Tabien, R., Abenes, L., Wu, K.S., McCouch, S. and Tanksley, S.D. (1992) Genetic and physical analysis of the rice bacterial blight disease resistance locus, Xa21. Mol. Gen. Genet. 236: 113–120 [DOI] [PubMed] [Google Scholar]
- Septiningsih, E.M., Prasetiyono, J., Lubis, E., Tai, T.H., Tjubaryat, T., Moeljopawiro, S. and McCouch, S.R. (2003) Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor. Appl. Genet. 107: 1419–1432 [DOI] [PubMed] [Google Scholar]
- Shim, R.A., Angeles, E.R., Ashikari, M. and Takashi, T. (2010) Development and evaluation of Oryza glaberrima Steud. chromosome segment substitution lines (CSSLs) in the background of O. sativa L. cv. Koshihikari. Breed. Sci. 60: 613–619 [Google Scholar]
- Shomura, A., Izawa, T., Ebana, K., Ebitani, T., Kanegae, H., Konishi, S. and Yano, M. (2008) Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40: 1023–1028 [DOI] [PubMed] [Google Scholar]
- Song, Z.P., Xu, X., Wang, B., Chen, J.K. and Lu, B.-R. (2003) Genetic diversity in the northernmost Oryza rufipogon populations estimated by SSR markers. Theor. Appl. Genet. 107: 1492–1499 [DOI] [PubMed] [Google Scholar]
- Sun, C.Q., Wang, X.K., Li, Z.C., Yoshimura, A. and Iwata, N. (2001) Comparison of the genetic diversity of common wild rice (Oryza rufipogon Griff.) and cultivated rice (O. sativa L.) using RFLP markers. Theor. Appl. Genet. 102: 157–162 [Google Scholar]
- Tan, L., Liu, F., Xue, W., Wang, G., Ye, S., Zhu, Z., Fu, Y., Wang, X. and Sun, C. (2007) Development of Oryza rufipogon and O. sativa introgression lines and assessment for yield-related quantitative trait loci. J. Int. Plant Biol. 49: 871–884 [Google Scholar]
- Tan, L., Li, X., Liu, F., Sun, X., Li, C., Zhu, Z., Fu, Y., Cai, H., Wang, X., Xie, D.et al. (2008) Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40: 1360–1364 [DOI] [PubMed] [Google Scholar]
- Thomson, M.J., Tai, T.H., McClung, A.M., Lai, X.-H., Hinga, M.E., Lobos, K.B., Xu, Y., Martinez, C.P. and McCouch, S.R. (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 107: 479–493 [DOI] [PubMed] [Google Scholar]
- Wang, E., Wang, J., Zhu, X., Hao, W., Wang, L., Li, Q., Zhang, L., He, W., Lu, B., Lin, H.et al. (2008) Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 40: 1370–1374 [DOI] [PubMed] [Google Scholar]
- Weng, J., Gu, S., Wan, X., Gao, H., Guo, T., Su, N., Lei, C., Zhang, X., Cheng, Z., Guo, X.et al. (2008) Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18: 1199–1209 [DOI] [PubMed] [Google Scholar]
- Xiao, J., Li, J., Grandillo, S., Ahn, S.N., Yuan, L., Tanksley, S.D. and McCouch, S.R. (1998) Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics 150: 899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, A., Nagayama, H., Kurakazu, T., Sanchez, P.L., Doi, K., Yamagata, Y. and Yasui, H. (2010) Introgression lines of rice (Oryza sativa L.) carrying a donor genome from the wild species, O. glumaepatula Steud. and O. meridionalis Ng. Breed. Sci. 60: 597–603 [Google Scholar]
- Zhang, H., Zhao, Q., Sun, Z.-Z., Zhang, C.-Q., Feng, Q., Tang, S.-Z., Liang, G.-H., Gu, M.-H., Han, B. and -Q, Q. (2011) Development and high-throughput genotyping of substitution lines carring the chromosome segments of indica 9311 in the background of japonica Nipponbare. J. Genet. Genomics 38: 603–611 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.