Abstract
To compare standard PCR/cloning and single genome sequencing (SGS) in their ability to reflect actual intra-patient polymorphism of HIV-1 populations, a total of 530 HIV-1 pro-pol sequences obtained by both sequencing techniques from a set of 17 ART naïve patient specimens was analyzed. For each specimen, 12 and 15 sequences, on average, were characterized by the two techniques. Using phylogenetic analysis, tests for panmixia and entropy, and Bland-Altman plots, no difference in population structure or genetic diversity was shown in 14 of the 17 subjects. Evidence of sampling bias by the presence of subsets of identical sequences was found by either method. Overall, the study shows that neither method was more biased than the other, and providing that an adequate number of PCR templates is analyzed, and that the bulk sequencing captures the diversity of the viral population, either method is likely to provide a similar measure of population diversity.
Keywords: HIV, Single genome sequencing (SGS), pro-pol diversity, cloning and sequencing, treatment naïve
1. Introduction
Human immunodeficiency virus type 1 (HIV-1) exists as an evolving population in infected individuals (Coffin 1995). The genetic diversity of HIV-1 results from rapid, high-level virus turnover (approximately 1011 virions per day and 108 infected cells per day) and from nucleotide misincorporation during replication of the HIV-1 genome by error prone reverse transcriptase (RT), (Mansky and Temin 1995; Menendez-Arias 2002; Preston et al., 1988; Roberts et al., 1988) as well as mutagenic host factors (Smith 2005). Importantly, many mutations do not have a deleterious impact on viral fitness and thus accumulate during successive rounds of viral replication. To characterize variants making up a viral population, it has been a common practice to obtain multiple sequences by performing RT-PCR on a region of the viral genome, cloning the amplified products, and selecting at random a number of clones for sequencing. Because primer DNA sequences used in PCR are pre-defined, PCR imposes a selection which may underestimate actual intra-patient diversity (Liu et al., 1996). If the number of RT-PCR templates in the original specimen is low, (or poorly reactive with the primers), it is unlikely that all sequences subsequently obtained by cloning will be derived from different input templates resulting in the resampling of individual genomes in the population. PCR-based recombination has also been observed, generating sequences that are not present in the original virus population (Liu et al., 1996; Shao et al., 2009). Single genome sequencing (SGS, also called SGA) permits individual cDNA molecules derived from defined portions of the genome to be PCR amplified and sequenced in bulk thus eliminating the effects of PCR-based recombination and the re-sampling of multiple clones from the same initial template molecule; and greatly reducing the error rate due to PCR (Palmer et al., 2005). The SGS assay error rate has been estimated to be 0.003% and the assay recombination rate was estimated to be less than one crossover between two closely related templates in 66,000 bp analyzed (Palmer et al., 2005). Previously a comparison of genetic diversity obtained from sequences derived by SGS and PCR was published (Salazar-Gonzalez et al., 2008); however, no comparative analysis of the PCR/cloning and SGS in their ability to reflect intra-patient HIV-1 diversity has been published. The present study compares the genetic diversity among HIV-1 pro-pol sequences derived from a set of patient specimens using these two methods.
2. Materials and Methods
2.1 Patients and virological endpoints
Single plasma specimens from seventeen ART naïve individuals over the age of 18 were obtained from patients attending the Tufts Medical Center infectious disease clinic or from an established cohort of ART naive HIV-1 infected prisoners in the Commonwealth of Massachusetts (Table 1) (Stone et al., 2002). The study was approved by the Institutional Review Board at Tufts Medical Center, the Human Research Review Committee for the Massachusetts Department of Public Health, Lemuel Shattuck Hospital and the Massachusetts Department of Corrections Health Service Unit, and the Office of Human Subjects Protection at the National Institutes of Health. All subjects provided written informed consent for participation and testing of specimens. All patients were antiretroviral naïve by self-report, chart review, and/or primary physician report. The median HIV-1 RNA level was 34,000 copies/ml (490- 300,000 copies/ml); and the median CD4 count cells was 393 cells/μl. Subjects’ estimated year of HIV infection, by self-report, ranged from 1988-2003. All plasma specimens were obtained from July 2000 to July 2001 except for the specimens from patient 15 and patient 16 which were obtained in 2004. Estimated times from seroconversion to specimen collection ranged from 6 months to 12 years.
Table 1.
Patient demographic data
| Patient Number |
Gender1 | Risk Factor2 | HIV-1 RNA copies/ml |
CD4 cells/μl | Estimated Year of Infection |
Estimated Time Seroconversion to Specimen Collection (years) |
|---|---|---|---|---|---|---|
| 1 | F | Unknown | 99,000 | NA | 2000 | 1 |
| 2 | F | HS | 19,467 | 393 | 1997 | 3 |
| 3 | M | HS | 45,708 | 351 | 1997 | 3 |
| 4 | M | MSM or HS | 61,022 | 1,196 | 1999 | 2 |
| 5 | M | MSM or HS | 200,000 | 89 | 1988 | 12 |
| 6 | M | HS | 17,796 | NA | 1998 | 3 |
| 7 | M | IDU | 154,000 | 421 | 1988 | 12 |
| 8 | M | IDU or HS | 34,000 | NA | 1996 | 4 |
| 9 | M | HS | 5,323 | 735 | 1996 | 4 |
| 10 | M | IDU | 3,446 | 530 | 1996 | 4 |
| 11 | M | IDU | 54,018 | 135 | 2001 | 1 |
| 12 | F | IDU | 3,401 | 384 | 1994 | 6 |
| 13 | F | IDU | 679 | 293 | 1990 | 11 |
| 14 | M | HS | 490 | 677 | 1999 | 2 |
| 15 | M | MSM | 300,000 | 194 | 2003 | 0.5 |
| 16 | M | MSM | 34,000 | NA | 2003 | 1 |
| 17 | M | IDU | 750 | 751 | 1998 | 2 |
Median HIV RNA 34,000 copies/ml and median CD4 count 393 cells/μl. Estimated year of infection: Range (1998-2003). All specimens were obtained from July 2000 to July 2001. NA=Data not available;
F=Female; M=Male;
HS=heterosexual contact; MSM=men who have sex with men; IDU=intravenous drug user
2.2 PCR/Cloning and sequencing
HIV RNA was harvested using a standard guanidinium isothyocyanate extraction method (Zhang et al., 1991). Population based sequencing was performed using a previously described protocol using MULV reverse transcriptase and platinum Taq (Invitrogen, Carlsbad, CA, USA). A 1.4 kb fragment of gag- pol was amplified by a 35-cycle RT-PCR and subsequent 25-cycle nested PCR (NPCR) using a previously described protocol and primer sets initially designed to amplify HIV-1 subtype B at low levels of viraemia (Coakley et al., 2002). PRL-f (nt. 1800 HXB2; 5′GGGACCAGCGGCTACACTAGAAGAAATGATGACAGCATGTCAGG3′), pRev (nt. 2514 HXB2; 5′AATCTGAGTCAACAGATTTCTTCC3) and Pro1.8-f (nt. 1897 HXB2; 5′GAAGCAATGAGCCAAGTAACAAAT3′), pRev (nt. 2514 HXB2; 5′AATCTGAGTCAACAGATTTCTTCC3) (Coakley et al., 2002).
NPCR products generated as described above were cloned using a TOPO TA cloning vector (Invitrogen, Carlsbad, CA, USA) following manufacturer’s instructions. Sequencing of plasmid DNA isolated from randomly chosen individual bacterial colonies (7-20 per specimen) was performed by standard dideoxy methods using conserved primers (Macrogen, Rockville, MD, USA).
2.3 Single Genome Sequencing
HIV RNA was extracted using standard guanidinium extraction methods [7]; cDNA was synthesized using random hexamers and diluted to an average of one amplifiable molecule per 3 wells of a microtiter plate and PCR amplified using a previously described methodology and primer sets (Palmer et al., 2005). A 1.4 kb fragment of gag-pol was (p6-RT region; HXB2 bases 2253-3257) was amplified and analyzed. Sequencing of DNA produced by SGS was performed by standard dideoxy methods using conserved primers (Macrogen, Rockville, MD, USA).
2.4 Sequence alignment and distance measurements
A total of 530 sequences, 1.4 kb in length, was analyzed from the seventeen patients. For each specimen, a mean of 12 and 15 sequences was characterized by PCR/cloning and by SGS respectively. Nucleotide sequences were aligned using Clustal X (Chenna et al., 2003). All alignments were visually inspected and frameshifts were removed using BioEdit sequence editor (http://www.mbio.ncsu.edu/BioEdit/BioEdit.html). A consensus sequence for each patient sequence set was generated by the BioEdit sequence editor (http://www.mbio.ncsu.edu/BioEdit/BioEdit.html). Genetic diversity was measured by average pairwise differences (APD) within and between sequence sets derived from each specimen using MEGA 4.0 (http://www.megasoftware.net). Neighbor-joining (NJ) tree construction with 1,000 bootstrap replicates was performed using MEGA 4.0 (http://www.megasoftware.net).
2.5 Testing for Divergence
A series of tests for population subdivisions described by Hudson et al. (Hudson et al., 1992) and adapted to biological sequences by Achaz et al. (Achaz et al., 2004) was performed. This test determines the probability that HIV-1 sequences derived by SGS and by cloning are derived from the same or different populations within the viral quasispecies. The method is a non-parametric test that computes pairwise differences between all sequences from both samples and calculates the probability of panmixia (p (K*s)) (i.e. the probability that two different populations are not statistically different from each other). A p-value greater than the nominal level of significance (p (K*s) >0.003) suggests that each set of sequences is unlikely to have been derived from different populations. In this analysis, 10,000 permutations were used to obtain p-values. Testing was performed using a web-based program (http://wwwabi.snv.jussieu.fr/achaz/hudsontest.html). Bland-Altman analysis was used to determine the limits of agreement between SGS and cloning measurements. Limits of agreement between methods were defined as the mean difference ± 2 SD (Bland and Altman 1999; Dewitte et al., 2002).
2.6 Testing for Entropy
To further characterize and understand the differences observed between sequence diversity obtained by both methods a test of Shannon entropy, which applies a measure of variation in sequence alignments and compares two sets of aligned sequences to determine if there is variability in one set relative to the other, was performed; statistical confidence is achieved using a Monte Carlo randomization strategy (Efron and Tibshirani 1991; Leitner et al., 1993). One thousand randomizations were performed, comparing each set of sequences with statistical significance defined as p < 0.005. Analyses were performed on both nucleic and amino acid sequences.
3. Results
3.1 Overall sequence relationships and drug resistance
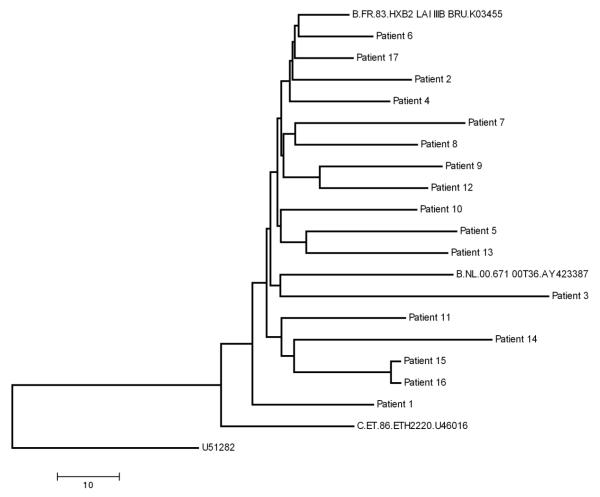
The NJ tree showed no evidence of relatedness among the virus consensus sequences from different patients with the exception of patients 15 and 16, a known transmission pair (Fig. 1). Sequences had no evidence of major HIV drug resistance mutations based on the Stanford HIV drug resistance mutation algorithm (http://hivdb.stanford.edu) except for patient 15 and 16, both of whom had K103N mutations, encoding resistance to non-nucleoside RT inhibitors. Sequences with K103N mutations were reverted to wild type when analyses were performed.
Fig. 1. Relationship among virus populations in the studied patients.
All sequences obtained in this study were compiled and a single NJ tree was constructed to check for sequence overlap. The NJ tree and bootstrap resampling of 1,000 trees demonstrated separate clustering of sequences from each patient; thus excluding contamination (data not shown). An NJ tree was prepared for the consensus sequences of the virus in all patients. Patients 15 and 16 are a known transmission pair. All sequences are HIV-1 subtype B. Clustering of SGS derived sequences and sequences derived by PCR/cloning within each patient cluster was evident with no evidence of contamination (data not shown). Additional subtype B and C reference sequences obtained from the Los Alamos National HIV Database were included in the analysis for comparison.
3.2 Average pairwise distance observed within and between assays
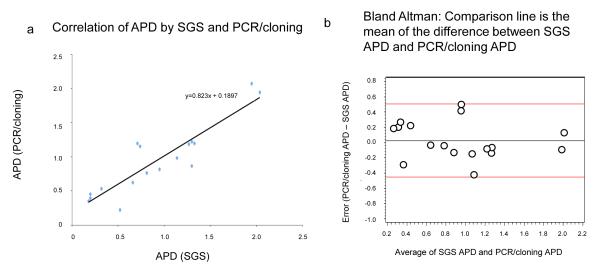
Intrapopulation APD observed by SGS ranged from 0.20% to 2.04% with a median of 0.81%. APD values obtained by PCR/cloning had a similar range, 0.23% to 2.08% with a median of 0.87% (Table 2). The mean pairwise difference between the two assays ranged from 0.03% to 1.27% with a median difference of 0.15%. The diversity values obtained by SGS and by PCR/cloning were highly correlated, r2=0.82; p=<0.000001 (Student t-test) (Fig 2a); the correlation was robust irrespective of plasma RNA level, and remained statistically significant with removal of the values with highest diversity (p<0.0003) suggesting that outliers were not driving the correlation.
Table 2.
Genetic distance of HIV-1 sequences derived by SGS and PCR/cloning
| Patient | HIV RNA copies/ml |
Average Pairwise Difference SGS, % |
Average Pairwise Difference PCR/Cloning, % |
Average Pairwise Difference Between Assays, % |
p(K*s) |
|---|---|---|---|---|---|
| 1 | 99,000 | 1.27% | 1.19% | 0.08% | 0.76 |
| 2 | 19,467 | 1.30% | 0.87% | 0.43% | 0.06 |
| 3 | 45,708 | 0.52% | 0.23% | 0.29% | 0.27 |
| 4 | 61,022 | 0.20% | 0.46% | 0.26% | 0.024 |
| 5 | 200,000 | 2.04% | 1.95% | 0.09% | 0.95 |
| 6 | 17,796 | 0.20% | 0.40% | 0.20% | 0.01 |
| 7 | 154,000 | 1.95% | 2.08% | 0.13% | 0.15 |
| 8 | 34,000 | 0.95% | 0.82% | 0.13% | 0.67 |
| 9 | 5,323 | 0.81% | 0.77% | 0.04% | 0.01 |
| 10 | 3,446 | 1.33% | 1.20% | 0.13% | 0.0001+ |
| 11 | 54,018 | 0.71% | 1.20% | 0.49% | 0.0008+ |
| 12 | 3,401 | 0.66% | 0.63% | 0.03% | 0.0001+ |
| 13 | 679 | 1.30% | 1.23% | 0.07% | 0.034 |
| 14 | 490 | 0.74% | 1.16% | 0.42% | 0.0031 |
| 15 | 300,000 | 0.18% | 0.36% | 0.18% | 0.0268 |
| 16 | 34,000 | 0.32% | 0.54% | 0.22% | 0.18 |
| 17 | 750 | 1.14% | 0.99% | 0.15% | 0.62 |
Statistically significant difference p(K*s) <0.003
Fig. 2. Relationship of sequence diversity to virus load for the two methods.
a) Correlation of APD values obtained by SGS and PCR/cloning. b) Bland-Altman plot of the difference in average pairwise difference between the two assays as a function of diversity.
3.3 Assessment of sampling bias using an automated test of panmixia
Sampling bias between SGS and PCR/cloning was further assessed using a web-based test of panmixia (http://www.abi.snv.jussieu.fr/achaz/hudsontest.html). The algorithm assumes that if a bias had been introduced by the method of analysis, the groups of sequences obtained by SGS and cloning would be significantly different with probabilities of panmixia (K*s) less than 0.003. Fourteen of the seventeen sets of sequences demonstrated no such sampling bias. For three patients, namely 10, 11, and 12, sequences had probabilities of panmixia less than this value suggesting the possibility of bias in one method relative to the other (Table 2). Bland-Altman analysis showed no evidence of bias (i.e. the difference in APD between the two assays) as a function of degree of diversity (Fig. 2b). The mean bias was 0.0271 and 95% limits of agreement ranging from −0.45 to 0.51. All values were within the 95% limits of agreement.
3.4 Assessment of genetic distances using Neighbor-joining trees
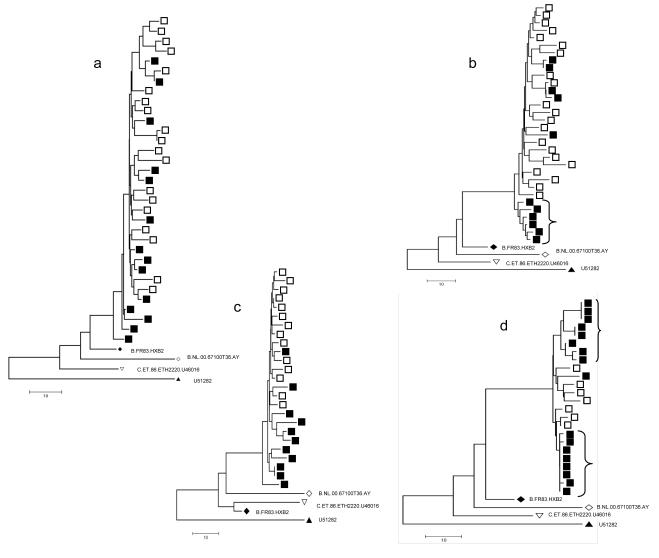
Neighbor-joining trees with all the sequences from each patient were generated to describe the genetic distance among the sequences obtained by the two techniques. Sequences from four patients, 2, 10, 11, and 12, are used to illustrate the different tree configurations observed. The NJ tree derived from the virus in patient 2 was typical of the NJ trees observed for 14 of 17 patient specimens and showed intermingling sequences with no overall difference in diversity (Fig. 3a). By contrast, for patient 10, PCR/cloning identified a relatively distinct clade of six genetically closely related sequences, none of which was similar to the genomes derived by SGS (Fig. 3b). The tree structure shows that a subpopulation of the sequences in this patient was preferentially amplified using cloning, although the bootstrap support was low and no difference in APD was observed. The APDs were similar at 1.33% and 1.20% for SGS and by PCR/cloning respectively (Student t-test, p=0.07, Table 2) with a p(K*S) of 0.001. Several assay artifacts could explain the preferential amplification including PCR amplification error, primer selectivity of both assays and/or PCR-based recombination.
Fig. 3. Neighbor-joining trees of HIV-1 populations from selected patients.
Solid squares represent sequences derived by PCR/cloning and open squares represent sequences derived by single genome sequencing. a) NJ tree of patient 2, representative of 14/17 trees obtained in the analyses showing intermingling of sequences obtained by PCR/cloning and sequences obtained by single genome sequencing with no overall difference in diversity by topology. Trees of sequences from the patients showing distinct populations by the two methods are shown in b-d. b) Patient 10; A cluster of 6 sequences amplified selectively by cloning and sequencing is denoted on the tree by a bracket. c) Patient 11. d) Patient 12; two distinct sub-populations of virus found by in PCR/cloning and not observed in SGS derived sequences are denoted by brackets. Additional reference sequences obtained from the Los Alamos National HIV database were included in the analysis..
In patient 11 (Fig 3c), the APD for SGS and PCR/cloning were 0.71 % and 1.20% respectively, with p(K*S) of 0.008. With a similar tree configuration, the higher APD may be a result of the small number of sequences (11) derived by PCR/cloning. This finding highlights the importance of characterizing a large number of genomes when analyzing differences in population diversity.
In patient 12, the APD was 0.66% for sequences obtained by SGS and 0.63% for PCR/ cloning, with p(K*S) of 0.001. As in patient 10, the overall diversity measured by SGS and cloning was comparable; however, the NJ tree (Fig. 3d) demonstrated two sub-populations of virus present in cloned sequences. The first sub-population detected by PCR/cloning is a cluster of identical or highly similar sequences which likely reflects re-sampling of a single template during PCR. A second cluster was detected by PCR/cloning and not by SGS. The two PCR/cloning sequence clusters and the SGS sequence cluster may reflect preferential amplification by either method.
3.5 Assessment of position-specific differences between methods using Shannon Entropy
To investigate whether there were any position-specific differences in SGS or cloning-derived measures of diversity, each nucleotide position was analyzed using an automated test for Shannon entropy (http://www.hiv.lanl.gov/content/sequence/ENTROPY/entropy.html) (Table 3). At the nucleic acid level, differences in entropy using a Monte Carlo randomization strategy were found to be statistically significant (p<0.005) in patients 9, 11, 12, 13, indicating that SGS and cloning differed in detecting genetic diversity at individual positions in pro-pol. Importantly, there was no evidence of systematic position specific bias by either method in determining genetic diversity. At the amino acid level, no differences in entropy were detected, with the exception of patient 12 at amino acid position 64 of reverse transcriptase, where all 17 PCR/clonal sequences were lysine, while arginine was present in three of the seven SGS derived sequences and lysine in the remaining four.
Table 3.
Calculation of sequence entropy
| Nucleic Acid | Amino Acid | |||||
|---|---|---|---|---|---|---|
| Patient | Position | Query consensus |
P-value at this position |
Position | Query consensus |
P-value at this position |
| 9 | 6 (RT) | C | 0.002 | |||
| 11 | 489 (RT) | A | 0.001 | |||
| 389 (RT) | A | 0.004 | ||||
| 12 | 64 (RT) | K | 0.004 | |||
| 543 (RT) | T | 0.004 | ||||
| 69 (PR) | A | 0.005 | ||||
| 13 | ||||||
| 336 (RT) | A | 0.005 | ||||
Sequences derived by cloning and sequencing were classified as background sequences and sequences derived by SGS as query sequences. One thousand randomizations were performed comparing each set of sequences with statistical significance defined as p < 0.005
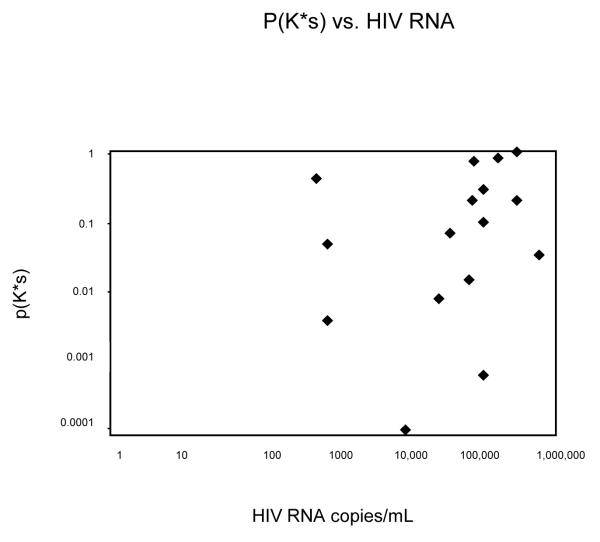
Of interest, no evidence of correlation was observed between the APD and the plasma RNA level over the range of viral loads studied 490-300,000 copies/ml (Fig. 4).
Fig. 4.
Probability of p(K*S), between SGS and clonal sequences are plotted as a function of viral load. No correlation between viremia and pK is observed.
4. Discussion
Describing HIV population diversity is increasingly important for the assessment of intrahost virus evolution and its relationship to disease progression, including the existence and development of low frequency drug resistance mutations and their impact on treatment outcomes. Additionally, the accurate assessment of population diversity is essential in understanding the effects of micro-environmental pressure on population genetic variation over time and in estimating dates of HIV seroconversion based on estimates of viral diversity. The PCR/cloning technique is widely used to describe HIV-1 population diversity and detect low frequency mutations. SGS is a newer technique which is gaining popularity and this study assesses the genetic diversity obtained from techniques on plasma specimens from 17 patients.
An adequate sample size is required to estimate genetic diversity. Adequate sampling to detect minor species has been estimated using probability considerations (Salazar-Gonzalez et al., 2008); with 14 sequences there is a 10 % probability of not sampling sequences present at a frequency of <15 % (Salazar-Gonzalez et al., 2008). Carrying this model forward, binomial probability suggests that when a population is composed of two variants A and B present in equal amounts, the probability of detecting each in exactly a 50:50 mixture is 0.2. Likewise, the probability of detecting 3 of variant A and 11 of B is 0.001. Overall, both PCR/cloning and SGS detected similar levels of genetic diversity in the patients sampled, even in circumstances where sensitive statistical analysis revealed sampling differences.
Of importance is the occurrence of viral subpopulations detected by one technique and not the other. A subpopulation of homogeneous viruses may be the actual result of the selection of a fit virus variant, or on contrary it may be an artifact of the amplification step of either technique. Each technique was found to miss a viral sub-population reported by the other. Each assay employed different sets of HIV-1 subtype B specific primers and the number of subjects in the study was too small to determine if either sequencing technique preferentially selected specific subpopulations because of the primer sequences. Due to low genetic diversity among viral populations within any one individual patient and the likelihood of recombination during virus replication in vivo, it was not possible to assess PCR-based recombination. If significant recombination events had occurred during PCR in PCR/coning derived sequences, the overall measure of diversity would not be affected, but the tree topology would be severely compromised. Similarly, PCR/cloning does not permit the assessment of mutation linkage, which is feasible with SGS. Finally, while only clearly observed in one patient 10 (Fig. 3d), PCR resampling could lead to an underestimation of genetic diversity. Overall, the study demonstrates that neither method was more biased than the other, and that providing an adequate number of genomes are analyzed, either method is likely to provide similar measures of population diversity.
Acknowledgements
MRJ was supported by the National Institute for Allergy and Infectious Disease: T32 AI07389; CFAAR 1P30A142853-10; K24 A1055293-06A1; and K23 AI074423-03; and the Center for Drug Abuse and AIDS Research: P30 DA013868. JMC was a Research Professor of the American Cancer Society, with support from the George Kirby Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achaz G, Palmer S, Kearney M, Maldarelli F, Mellors JW, Coffin JM, Wakeley J. A robust measure of HIV-1 population turnover within chronically infected individuals. Mol. Biol. Evol. 2004;21(10):1902–12. doi: 10.1093/molbev/msh196. [DOI] [PubMed] [Google Scholar]
- Bland JM, Altman DG. Measuring agreement in method comparison studies. Stat Methods Med Res. 1999;8(2):135–60. doi: 10.1177/096228029900800204. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs Nucleic. Acids. Res. 2003;31(13):3497–5000. doi: 10.1093/nar/gkg500. 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coakley EP, Doweiko JP, Bellossilo NA, D’Agata EM, Albrecht MA. HIV Drug Resistance Profiles and Clinical and Virologic Outcomes among HIV-Infected Subjects with Stable Detectable Plasma Viral Loads < 1000 Copies/mL for at least 12 Months 9th Conference on Retroviruses and Opportunistic Infections.2002. p. 556-T. [Google Scholar]
- Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267(5197):483–9. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Dewitte K, Fierens C, Stöckl D, Thienpont LM. Application of the Bland-Altman plot for interpretation of method-comparison studies: a critical investigation of its practice. Clin. Chem. 2002;48(5):799–801. author reply 801-2. [PubMed] [Google Scholar]
- Efron B, Tibshirani R. Statistical Data Analysis in the Computer Age. Science. 1991;253(5018):390–395. doi: 10.1126/science.253.5018.390. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Boos DD, Kaplan NL. A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 1992;9(1):138–51. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- [Last Accessed November 3, 2009]. http://www.abi.snv.jussieu.fr/achaz/hudsontest.html.
- [Last Accessed November 3, 2009]. http://hivdb.stanford.edu.
- [Last Accessed November 3, 2009]. http://www.hiv.lanl.gov/content/sequence/ENTROPY/entropy.html.
- [Last Accessed April 22, 2010]. http://www.megasoftware.net.
- [Last Accessed April 22, 2010]. http://www.mbio.ncsu.edu/BioEdit/BioEdit.html.
- Leitner T, Halapi E, Scarlatti G, Rossi P, Albert J, Fenyö EM, Uhlén M. Analysis of heterogeneous viral populations by direct DNA sequencing. Biotechniques. 1993;15(1):120–7. [PubMed] [Google Scholar]
- Liu SL, Rodrigo AG, Shankarappa R, Learn GH, Hsu L, Davidov O, Zhao LP, Mullins JI. HIV quasispecies and resampling. Science. 1996;273(5274):415–6. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- Mansky LM, Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 1995;69(8):5087–94. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez-Arias L. Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases. Prog Nucleic. Acid. Res. Mol. Biol. 2002;71:91–147. doi: 10.1016/s0079-6603(02)71042-8. [DOI] [PubMed] [Google Scholar]
- Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey RT, Jr, Dewar RL, Metcalf JA, Hammer S, Mellors JW, Coffin JM. Multiple, linked human immunodeficiency virus type 1 drug Resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 2005;43(1):406–13. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston BD, Poiesz BD, Loeb LA. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242(4882):1168–71. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- Roberts JD, Bebenek K, Kunkel TA. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242(4882):1171–3. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, Manigart O, Mulenga J, Anderson JA, Swanstrom R, Haynes BF, Athreya GS, Korber BT, Sharp PM, Shaw GM, Hahn BH. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 2008;82(8):3952–70. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Boltz VF, Kearney M, Maldarelli F, Mellors JW, Stewart C, Levitsky A, Volfovsky N, Stephens RM, Coffin JM. Characterization of HIV-1 sequence artifacts introduced by bulk PCR and detected by 454 sequencing. XVIII international HIV drug resistance workshop; Fort Myers, Fl, USA. 2009. Abstract 104. [Google Scholar]
- Smith RA, Loeb LA, Preston BD. Lethal HIV mutagenesis. Virus. Res. 2005;107(2):215–228. doi: 10.1016/j.virusres.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Stone DR, Corcoran C, Wurcel A, McGovern B, Quirk J, Brewer A, Sutton L, D’Aquila RT. Antiretroviral drug resistance mutations in antiretroviral-naive prisoners. Clin. Infect. Dis. 2002;35(7):883–6. doi: 10.1086/342697. [DOI] [PubMed] [Google Scholar]
- Zhang LQ, Simmonds P, Ludlham CA, Brown AJ. Detection, quantification and sequencing of HIV-1 from the plasma of seropositive individuals and from factor VIII concentrates. AIDS. 1991;5:675–681. doi: 10.1097/00002030-199106000-00006. [DOI] [PubMed] [Google Scholar]