Abstract
In lung transplant recipients (LTRs), human cytomegalovirus (HCMV) DNAaemia could be associated with HCMV disease and reduced allograft survival. In the present study we analysed whether or not HCMV-specific granzyme B (Grz-B) responses indicating CD8+ T cell cytotoxicity exert an impact on HCMV DNAaemia and relate to specific interferon (IFN)-γ secretion. HCMV-specific Grz-B responses were quantitated by enzyme-linked immunosorbent assay (ELISA) in 70 samples from 39 HCMV seropositive LTRs who were prospectively investigated for HCMV DNA plasma levels and IFN-γ kinetics using a standardized CD8+ T cell assay (QuantiFERON®-CMV assay). In all LTRs who were protected from HCMV DNAaemia by early and persistent IFN-γ responses, Grz-B responses were also detected. In LTRs who developed episodes of HCMV DNAaemia, the Grz-B responses which were detected prior to viral DNA detection differed significantly in patients who experienced episodes with high (exceeding 1000 copies/ml) and low plasma DNA levels (P = 0·0290, Fisher's exact test). Furthermore, the extent of Grz-B release prior to viral DNAaemia correlated statistically with the detected levels of IFN-γ (P < 0·0001, Spearman's rank test). Of note, simultaneous detection of Grz-B and IFN-γ secretion was associated significantly with protection from high HCMV DNA plasma levels during the subsequent follow-up (P = 0·0057, Fisher's exact test), and this association was stronger than for IFN-γ detection alone. We conclude that, in addition to IFN-γ responses, Grz-B secretion by CD8+ T cells is essential to control HCMV replication and a simultaneous measurement of IFN-γ and Grz-B could contribute to the immune monitoring of LTRs.
Keywords: cytomegalovirus, DNAaemia, granzyme B, HCMV, lung transplant recipients
Introduction
In lung transplant recipients (LTRs) human cytomegalovirus (HCMV) replication can lead to HCMV syndrome, characterized by fever, neutropenia or thrombocytopenia, or tissue-invasive HCMV disease, which manifests frequently in LTRs as pneumonia, and might also trigger chronic inflammatory processes that reduce the survival of the allograft [1,2]. For the development of HCMV disease the plasma HCMV DNA level has been identified as a prognostic marker [3]. Although viral DNA is therefore measured routinely, which factors determine emergence and magnitude of HCMV DNAaemia remain unclear. For controlling HCMV replication, specific CD8+ T cells are essential. They produce the anti-viral cytokine interferon (IFN)-γ and secrete cytoplasmic granules which, among other cytolytic molecules, contain granzyme B (Grz-B) and induce apoptosis in infected cells [4,5].
The QuantiFERON®-CMV assay is the only standardized test for monitoring HCMV-specific IFN-γ responses by CD8+ T cells [6]. Previous studies covering solid organ recipients showed that when IFN-γ responses are measured using this assay the clinical progression of HCMV DNAaemia as well as the incidence of HCMV disease can be predicted [7–9]. We applied the assay in LTRs and demonstrated that IFN-γ responses also affect the incidence and magnitude of HCMV DNAaemia [10]. None the less, although most LTRs who displayed early and persistently detectable IFN-γ responses were protected from high-level HCMV DNAaemia, we identified single patients who displayed high HCMV DNA plasma loads despite the detection of IFN-γ. Therefore, we further analysed whether or not HCMV-specific Grz-B release, indicating cytotoxicity, is associated with HCMV DNAaemia, correlating with the standardized measurement of IFN-γ secretion by the QuantiFERON®-CMV assay and can be used to predict HCMV DNAaemia.
Materials and methods
Study population
The present study included 39 HCMV seropositive patients (21 females, 18 males, mean age: 53 years), who were part of a previously described cohort and who received a lung transplant at the Medical University of Vienna from 11 HCMV seronegative and 28 seropositive donors. The patients were administered immunosuppressive treatment as well as anti-viral prophylaxis in a regimen described previously [10]. Of 39 study patients, 36 completed a follow-up of 1 year and three were followed for at least 7 months. All patients gave written consent for participation, the protocol was approved by the Institutional Review Board and the study was conducted in accordance with the Declaration of Helsinki and the guidelines of the local ethics committee (EC protocol 165/2007).
HCMV DNA quantification and HCMV-specific IFN-γ responses
Blood samples from the LTRs were collected at routine visits to the out-patient clinic during the post-transplant follow-up. The samples were investigated for HCMV DNA plasma loads (Cobas Amplicor HCMV Monitor-Test Kit; Roche Molecular Systems, Branchburg, NJ, USA) and HCMV-specific IFN-γ responses using the QuantiFERON®-CMV assay (Cellestis GmbH – a Qiagen company, Darmstadt, Germany) [10]. As described previously, one aliquot of 1 ml whole blood was stimulated with a mix of 21 HCMV peptides, derived from immunodominant human leucocyte antigen (HLA)-restricted CD8+ T cell epitopes of various HCMV proteins; one aliquot was stimulated with mitogen (positive control) and one was incubated in the presence of sterile phosphate-buffered saline (unstimulated negative control) [10]. All patients displayed HLA haplotypes which included the epitopes that were covered by the QuantiFERON®-CMV assay. After incubation for 24 h at 37°C, serum supernatants were harvested and kept frozen at −20°C. After thawing, HCMV-specific IFN-γ responses were quantified (IU/ml) using an included enzyme-linked immunosorbent assay (ELISA) and used according to the manufacturer's instructions. In total, 620 plasma samples (mean 15·9 per patient) were quantitated for HCMV DNA loads at a mean interval of 26·3 days, and 422 heparinized whole-blood samples (mean 10·8 samples per patient) were measured for specific IFN-γ responses at a mean interval of 33·2 days. According to the protocol of our centre, pre-emptive valganciclovir therapy was initiated after acquisition of samples when HCMV DNA loads exceeded 1000 copies/ml.
HCMV-specifc granzyme B responses
Seventy samples with at least one assessment per patient (mean of 1·8 per patient) were analysed for Grz-B levels using the Human Granzyme B Platinum ELISA (eBiosciences, Bender MedSystems, Vienna, Austria). In each patient who developed HCMV DNAaemia (n = 32), the sample which was obtained at the last time-point before HCMV DNA became detectable was quantitated for HCMV-specific Grz-B production (mean interval 31·2 days). In patients who did not develop HCMV DNaemia during the follow-up, Grz-B levels were determined at cessation of anti-viral prophylaxis and at least another time-point during the follow-up (mean 2·9 samples per patient). Grz-B levels were analysed in supernatants that were acquired using the QuantiFERON®-CMV assay after incubation of whole blood with HCMV peptides from various CD8+ T cell epitopes, with mitogen (positive control) and without stimulation (negative control) [6,10]. HCMV-specific Grz-B responses were evaluated by subtracting levels in unstimulated from HCMV-stimulated samples. A cut-off value (mean Grz-B levels at cessation of anti-viral prophylaxis + ×3 standard deviations) was determined in 10 donor and recipient HCMV seronegative LTRs who did not show any evidence of HCMV replication during the post-transplant follow-up and who were not part of the HCMV seropositive study cohort. Responses which exceeded this cut-off and were at least twice as high as unstimulated levels were interpreted as positive. Test results were interpreted as valid only when unstimulated levels were < 15 pg/ml and mitogen-stimulated levels exceeded this value by 10 times.
Statistical methods
Fisher's exact test was used to evaluate whether or not specific HCMV-specific Grz-B responses in preceding samples differed between patients who subsequently developed HCMV DNAaemia with high and low HCMV DNA levels. Spearman's rank test was used to estimate the correlation of Grz-B and IFN-γ levels that were detected prior to HCMV DNAaemia episodes. A P-value of < 0·05 was considered statistically significant. GraphPad Prism version 5.0 software was used for statistical analyses.
Results
Grz-B responses in relation to HCMV DNA and IFN-γ kinetics
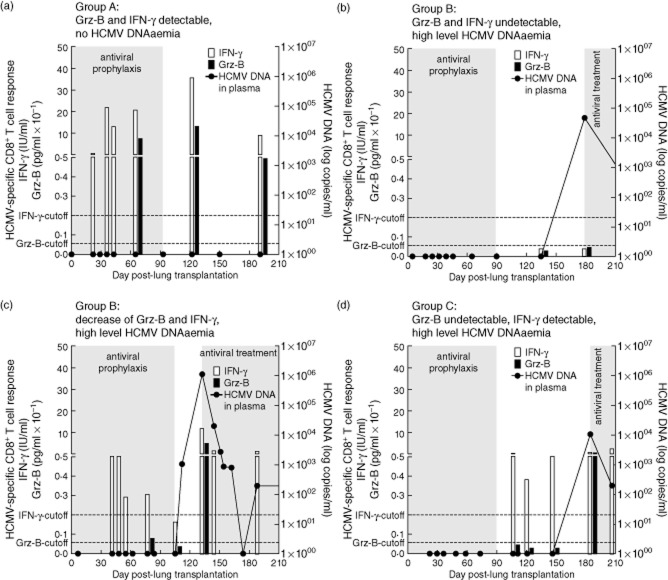
We investigated 39 R+ LTRs from a previously enrolled cohort, whose individual kinetics of plasma HCMV DNA loads and of specific IFN-γ responses had been assessed previously [10]. As shown in Table 1, Grz-B responses were analysed in six patient groups: in group A (n = 6), LTRs showed no HCMV DNAaemia during the follow-up and IFN-γ responses were detected early post-transplant and remained persistently detectable. Grz-B responses were detected in all these patients (a representative LTR is shown in Fig. 1a). Group B (n = 9) was formed by LTRs who developed high-level HCMV DNAaemia (> 1000 copies/ml) in the absence of a preceding IFN-γ response. In eight of these nine patients (88·9%), Grz-B was also undetectable prior to HCMV DNA detection (Fig. 1b). Two individuals from group B showed a decline of a previously detectable IFN-γ response prior to HCMV DNAaemia and this was also accompanied by a simultaneous Grz-B decrease below the cut-off value (Fig. 1c). Group C (n = 4) included LTRs in whom high-level HCMV DNAaemia occurred, although IFN-γ was detected simultaneously. Of note, in two of these patients (50%) there was no Grz-B response (Fig. 1d). Groups D and E consisted of 19 LTRs in whom only low HCMV DNA plasma levels (< 1000 copies/ml) were detected, and this was associated with detectable IFN-γ responses in 12 patients (group D) and absence of responses in seven LTRs (group E). Of the 12 LTRs from group D, 11 patients (91·7%) showed a detectable Grz-B response. Group F finally included a patient who displayed neither HCMV DNAaemia nor IFN-γ or Grz-B responses.
Table 1.
Human cytomegalovirus (HCMV)-specific granzyme B responses in relation to interferon-γ and HCMV DNA kinetics
| Patient group | Number of patients | HCMV DNAaemia | HCMV-specific interferon-γ response | HCMV-specific granzyme B response | Representative patient shown in | |
|---|---|---|---|---|---|---|
| A | 6 | Undetectable during follow-up | Early and persistently positive | Detectable in 100% | Fig. 1a | |
| B | 9 | > 1000 copies/ml | Prior to HCMV DNAemia | Negative | Undetectable in 88·9% | Fig. 1b,c |
| C | 4 | > 1000 copies/ml | Positive | Undetectable in 50% | Fig. 1d | |
| D | 12 | < 1000 copies/ml | Positive | Detectable in 91·7% | Not shown | |
| E | 7 | < 1000 copies/ml | Negative | Undetectable in 57·1% | Not shown | |
| F | 1 | Undetectable during follow-up | Negative during follow-up | Undetectable | Not shown | |
Figure 1.
Human cytomegalovirus (HCMV)-specific granzyme B (Grz-B) responses in representative patients. Representative patient who showed early and persistent detection of specific granzyme B (Grz-B) and interferon (IFN)-γ responses in absence of HCMV DNAaemia (a). Representative patient who displayed high-level HCMV DNAaemia (plasma HCMV DNA loads exceeding 1000 copies/ml) in the absence of specific Grz-B and IFN-γ responses during the entire follow-up (b). Representative patient who developed high-level HCMV DNAaemia after fluctuating kinetics of IFN-γ and Grz-B responses which decreased prior to HCMV DNAaemia (c). Representative patient who showed occurrence of high level HCMV DNAaemia in the presence of IFN-γ, but in the absence of Grz-B responses (d).
Association of HCMV-specific Grz-B responses and HCMV DNAaemia
We then analysed whether specific Grz-B responses which were detected prior to HCMV DNAaemia differed in LTRs who subsequently developed episodes with high (> 1000 copies/ml) and low plasma DNA levels. Of 32 episodes of HCMV DNAaemia which occurred in the LTRs, 16 were preceded by a detectable Grz-B response and 16 were not. HCMV DNA loads exceeding 1000 copies/ml were detected in 62·5% (10 of 16) of the episodes which were preceded by a negative Grz-B response, but in only 18·8% (three of 16) of the episodes that were preceded by a positive response (Fisher's exact test, P = 0·0290; Fig. 2a). The extent of specific Grz-B release which was detected prior to these episodes correlated statistically with the detected levels of IFN-γ (Spearman's rank test, P < 0·0001; Fig. 2b). Finally, we analysed whether or not simultaneous detection of Grz-B and IFN-γ secretion was associated with the subsequent development of high-level HCMV DNAaemia (> 1000 copies/ml) during further follow-up. HCMV DNAaemia with viral DNA loads exceeding 1000 copies/ml occurred in 11 of 20 (55%) LTRs who previously showed either a negative Grz-B or IFN-γ response, but developed only in two of 19 (10·5%) LTRs who displayed a Grz-B and IFN-γ double-positive response (P = 0·0057, Fisher's exact test, Fig. 2c). Of note, the association of double-positive responses with protection from high-level HCMV DNAaemia showed a higher statistical significance than when IFN-γ was measured alone (high level HCMV DNAaemia developed in 52·9% of LTRs with a negative IFN-γ response and in 18·2% of patients with a positive IFN-γ response, P = 0·0392, Fisher's exact test).
Figure 2.
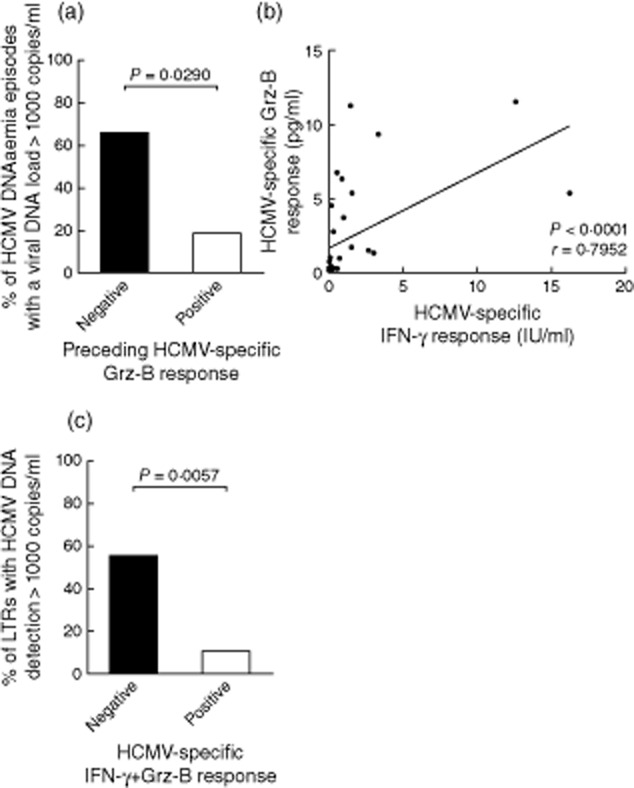
Statistical analysis of human cytomegalovirus (HCMV)-specific granzyme B (Grz-B) responses and HCMV DNAaemia. HCMV-specific Grz-B responses which were detected prior to the onset of HCMV DNAaemia differed in lung transplant recipients (LTRs) who subsequently developed DNAaemia episodes with high (> 1000 copies/ml) and low viral DNA loads. In 10 of 16 (62·5%) episodes that were preceded by a negative Grz-B response, viral DNA loads exceeded 1000 copies/ml, versus three of 16 (18·8%) episodes which were preceded by a positive Grz-B response (Fisher's exact test, P = 0·0290) (a). The levels of HCMV-specific Grz-B responses that were detected prior to the 32 HCMV DNAaemia episodes correlated with the detected levels of specific interferon (IFN)-γ (P < 0·0001, Spearman's rank test) (b). Simultaneous detection of specific Grz-B and IFN-γ responses affected the occurrence of high HCMV DNA loads (> 1000 copies/ml) during the subsequent follow-up. HCMV DNAaemia with viral DNA loads exceeding 1000 copies/ml developed in 11 of 20 (55%) LTRs who previously either showed a negative Grz-B or negative IFN-γ response, but occurred in only two of 19 (10·5%) LTRs who displayed a double-positive Grz-B and IFN-γ response (P = 0·0057, Fisher's exact test).
Discussion
In the present study we analysed whether HCMV-specific production of Grz-B, which indicates CD8+ T cell cytotoxicity, controls HCMV DNAaemia in LTRs. We found that Grz-B responses which were detected prior to onset of HCMV DNAaemia differed in patients who subsequently experienced DNAaemia episodes with high and low plasma DNA levels and demonstrated that simultaneous detection of Grz-B and IFN-γ was associated clearly with the absence of high-level HCMV DNAaemia.
In LTRs high-level HCMV DNAaemia has been associated with HCMV disease development and HCMV-specific IFN-γ responses by CD8+ T cells have been shown to affect the incidence, magnitude and progression of HCMV DNAaemia [1,3,8,10]. None the less, a previous study from our group indicated that high-level HCMV DNAaemia occurs occasionally despite stable IFN-γ responses, proposing that other CD8+ T cell functions, besides IFN-γ secretion, could be critical for an efficient containment of HCMV replication [10].
The present data now indicate that HCMV-specific Grz-B responses by CD8+ T cells might exert a critical impact on limiting HCMV replication, as high-level HCMV DNAaemia occurred more frequently in the absence of Grz-B responses, while the majority of LTRs, in whom no or only low-level DNAaemia occurred, displayed significant Grz-B responses. Furthermore, simultaneous Grz-B and IFN-γ secretion showed a stronger association with protection from high level HCMV DNAaemia than IFN-γ responses alone, indicating that a measurement of Grz-B during the post-transplant surveillance of LTRs might improve the identification of functionally efficient effector cells that control HCMV replication in vivo.
Grz-B production has been associated with distinct grades of activation and differentiation in CD8+ T cell subsets and is found predominantly in recently activated effector and memory effector cells [4,5]. In HCMV-specific CD8+ T cells, Grz-B expression is linked to the production of other cytolytic enzymes, such as granzyme A and perforin, and correlates with the lytic function [4,11]. HCMV-specific Grz-B responses have been identified during HCMV primary infection of renal allograft recipients, and even during latent HCMV infection high frequencies of effector CD8+ T cells contain Grz-B [12–14]. Therefore, our finding that HCMV-specific Grz-B responses can be detected in LTRs is consistent with previous data and indicates that, even when high-dose immunsuppressive treatment is administered after lung transplantation, CD8+ T cells are capable of secreting Grz-B in response to HCMV peptides, which is a measure for HCMV-specific cytotoxicity, although cytokine-mediated bystander activation and Grz-B release by natural killer T cells cannot be ruled out completely [15].
In the majority of the cases IFN-γ and Grz-B were secreted simultaneously when CD8+ T cells responded upon HCMV-specific stimulation. Furthermore, there was a statistically significant correlation between Grz-B and IFN-γ levels that were detected by the QuantiFERON®-CMV assay. These findings indicate that a standardized measurement of HCMV-specific IFN-γ responses, serving as an indicator for antigen recognition and cytokine production, correlates with cytotoxic T cell function, proposing that the total number of circulating HCMV-specific CD8+ T cells that exert these effector functions could also be a critical factor [16]. However, we also identified single patients in whom these effector functions were not linked. Of note, in two patients specific Grz-B production was absent while IFN-γ responses were present, and this was associated with the development of high-level HCMV DNAaemia. This could be due to a selective effect of the immunosuppressive treatment on distinct CD8+ T cell subsets or to a selective impairment of cytotoxicity, as has been demonstrated for infections with human immunodeficiency virus and hepatitis C virus [13,17–19]. Thus, polyfunctional CD8+ T cells that produce other cytokines simultaneously in addition to IFN-γ could be essential to control HCMV replication [20,21].
However, although simultaneous IFN-γ and Grz-B secretion by CD8+ T cells was associated clearly with protection from high HCMV DNAaemia, protection from high HCMV DNA plasma loads could also be dependent upon CD4+ T cell help, as it has been shown that in the absence of stimulatory signals from CD4+ T helper cells, CD8+ T cells are not capable of a sufficient containment of viral replication [22]. Thus, for an exact prediction of HCMV DNAaemia in LTRs, comprehensive immune monitoring also covering CD4+ T cell responses could prove beneficial.
None the less, we conclude that in LTRs HCMV-specific CD8+ T cell responses, comprised not only of IFN-γ secretion but also of Grz-B release, are essential for sufficient control of HCMV replication, and monitoring of Grz-B responses might improve the prediction of HCMV DNAemia during the post-transplant surveillance.
Disclosure
All authors declare that there are no conflicts of interest.
References
- 1.Humar A, Snydman D. Cytomegalovirus in solid organ transplant recipients. Am J Transplant. 2009;9(Suppl. 4):S78–86. doi: 10.1111/j.1600-6143.2009.02897.x. [DOI] [PubMed] [Google Scholar]
- 2.Westall GP, Michaelides A, Williams TJ, Snell GI, Kotsimbos TC. Bronchiolitis obliterans syndrome and early human cytomegalovirus DNAaemia dynamics after lung transplantation. Transplantation. 2003;75:2064–2068. doi: 10.1097/01.TP.0000069234.04901.A3. [DOI] [PubMed] [Google Scholar]
- 3.Emery VC, Sabin CA, Cope AV, Gor D, Hassan-Walker AF, Griffiths PD. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet. 2000;355:2032–2036. doi: 10.1016/S0140-6736(00)02350-3. [DOI] [PubMed] [Google Scholar]
- 4.Harari A, Enders FB, Cellerai C, Bart PA, Pantaleo G. Distinct profiles of cytotoxic granules in memory CD8 T cells correlate with function, differentiation stage, and antigen exposure. J Virol. 2009;83:2862–2871. doi: 10.1128/JVI.02528-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bannard O, Kraman M, Fearon D. Pathways of memory CD8+ T-cell development. Eur J Immunol. 2009;39:2083–2087. doi: 10.1002/eji.200939555. [DOI] [PubMed] [Google Scholar]
- 6.Giulieri S, Manuel O. QuantiFERON(R)-CMV assay for the assessment of cytomegalovirus cell-mediated immunity. Exp Rev Mol Diagn. 2011;11:17–25. doi: 10.1586/erm.10.109. [DOI] [PubMed] [Google Scholar]
- 7.Kumar D, Chernenko S, Moussa G, et al. Cell-mediated immunity to predict cytomegalovirus disease in high-risk solid organ transplant recipients. Am J Transplant. 2009;9:1214–1222. doi: 10.1111/j.1600-6143.2009.02618.x. [DOI] [PubMed] [Google Scholar]
- 8.Lisboa LF, Kumar D, Wilson LE, Humar A. Clinical utility of cytomegalovirus cell-mediated immunity in transplant recipients with cytomegalovirus viremia. Transplantation. 2012;93:195–200. doi: 10.1097/TP.0b013e31823c1cd4. [DOI] [PubMed] [Google Scholar]
- 9.Manuel O, Husain S, Kumar D, et al. Assessment of cytomegalovirus specific cell-mediated immunity for the prediction of cytomegalovirus disease in high-risk solid-organ transplant recipients: a multicenter cohort study. Clin Infect Dis. 2013;56:817–824. doi: 10.1093/cid/cis993. [DOI] [PubMed] [Google Scholar]
- 10.Weseslindtner L, Kerschner H, Steinacher D, et al. Prospective analysis of human cytomegalovirus DNAemia and specific CD8+ T-cell responses in lung transplant recipients. Am J Transplant. 2012;12:2172–2180. doi: 10.1111/j.1600-6143.2012.04076.x. [DOI] [PubMed] [Google Scholar]
- 11.Chattopadhyay PK, Betts MR, Price DA, et al. The cytolytic enzymes granyzme A, granzyme B, and perforin: expression patterns, cell distribution, and their relationship to cell maturity and bright CD57 expression. J Leukoc Biol. 2009;85:88–97. doi: 10.1189/jlb.0208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wever PC, Spaeny LH, van der Vliet HJ, et al. Expression of granzyme B during primary cytomegalovirus infection after renal transplantation. J Infect Dis. 1999;179:693–696. doi: 10.1086/314629. [DOI] [PubMed] [Google Scholar]
- 13.Gamadia LE, Rentenaar RJ, Baars PA, et al. Differentiation of cytomegalovirus-specific CD8(+) T cells in healthy and immunosuppressed virus carriers. Blood. 2001;98:754–761. doi: 10.1182/blood.v98.3.754. [DOI] [PubMed] [Google Scholar]
- 14.Hertoghs KM, Moerland PD, van Stijn A, et al. Molecular profiling of cytomegalovirus-induced human CD8+ T cell differentiation. J Clin Invest. 2010;120:4077–4090. doi: 10.1172/JCI42758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jukes JP, Wood KJ, Jones ND. Bystander activation of iNKT cells occurs during conventional T-cell alloresponses. Am J Transplant. 2012;12:590–599. doi: 10.1111/j.1600-6143.2011.03847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Remmerswaal EB, Havenith SH, Idu MM, et al. Human virus-specific effector-type T cells accumulate in blood but not in lymph nodes. Blood. 2012;119:1702–1712. doi: 10.1182/blood-2011-09-381574. [DOI] [PubMed] [Google Scholar]
- 17.Northfield JW, Kasprowicz V, Lucas M, et al. CD161 expression on hepatitis C virus-specific CD8+ T cells suggests a distinct pathway of T cell differentiation. Hepatology. 2008;47:396–406. doi: 10.1002/hep.22040. [DOI] [PubMed] [Google Scholar]
- 18.Hersperger AR, Martin JN, Shin LY, et al. Increased HIV-specific CD8+ T-cell cytotoxic potential in HIV elite controllers is associated with T-bet expression. Blood. 2011;117:3799–3808. doi: 10.1182/blood-2010-12-322727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ribeiro-Dos-Santos P, Turnbull EL, Monteiro M, et al. Chronic HIV infection affects the expression of the 2 transcription factors required for CD8 T-cell differentiation into cytolytic effectors. Blood. 2012;119:4928–4938. doi: 10.1182/blood-2011-12-395186. [DOI] [PubMed] [Google Scholar]
- 20.Clari MA, Munoz-Cobo B, Solano C, et al. Performance of the QuantiFERON-cytomegalovirus (CMV) assay for detection and estimation of the magnitude and functionality of the CMV-specific gamma interferon-producing CD8(+) T-cell response in allogeneic stem cell transplant recipients. Clin Vaccine Immunol. 2012;19:791–796. doi: 10.1128/CVI.05633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betts MR, Nason MC, West SM, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerna G, Lilleri D, Chiesa A, et al. Virologic and immunologic monitoring of cytomegalovirus to guide preemptive therapy in solid-organ transplantation. Am J Transplant. 2011;11:2463–2471. doi: 10.1111/j.1600-6143.2011.03636.x. [DOI] [PubMed] [Google Scholar]