Abstract
A novel xanthomonadin-dialkylresorcinol hybrid named arcuflavin was identified in Azoarcus sp. BH72 by a combination of feeding experiments, HPLC-MS and MALDI-MS and gene clusters encoding the biosynthesis of this non-isoprenoid aryl-polyene containing pigment are reported. A chorismate-utilizing enzyme from the XanB2-type producing 3- and 4-hydroxybenzoic acid and an AMP-ligase encoded by these gene clusters were characterized, that might perform the first two steps of the polyene biosynthesis. Furthermore, a detailed analysis of the already known or novel biosynthesis gene clusters involved in the biosynthesis of polyene containing pigments like arcuflavin, flexirubin and xanthomonadin revealed the presence of similar gene clusters in a wide range of bacterial taxa, suggesting that polyene and polyene-dialkylresorcinol pigments are more widespread than previously realized.
Introduction
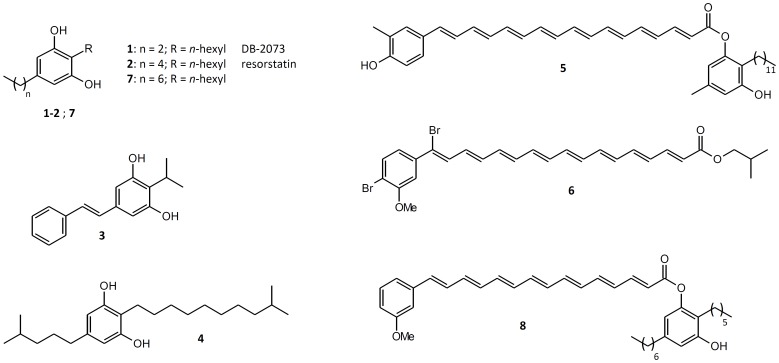
An underexplored class of natural compounds are the 2,5-dialkylresorcinols (DAR), microbial secondary metabolites, which are derived from a condensation of two fatty acid metabolism intermediates [1]. DAR examples with known bioactivities (Figure 1) are the free radical scavengers DB-2073 (1) [2]–[4], resorstatin (2) [4], a multipotent isopropylstilbene (3) [5] or the mammalian cell growth stimulating factor resorcinin (4) [6] which can also be found as part of the flexirubins from Flavobacterium johnsoniae [7]. Flexirubins (5) [8] are orange pigments made of a DAR esterified with a non-isoprenoid aryl-polyene carboxylic acid and are used as chemotaxonomic marker for bacteria from the Bacteroidetes phylum. Another class of aryl-polyene pigments, the xanthomonadins, has been found in bacteria of the genus Xanthomonas. Besides the dibrominated xanthomonadin I (6) [9], derivatives with different levels of bromination, methylation and chain length are known [10], all lacking a DAR-moiety. The gene cluster for xanthomonadin biosynthesis (Figure 2.B) is conserved in several strains of Xanthomonas [11]–[13] and was recently studied in Lysobacter enzymogenes [14], another bacterium from the family xanthomonadaceae. The gene cluster encodes an acyltransferase, a ketoreductase, a dehydratase, a predicted ketosynthase, an unusual chain-length factor like protein [14] as well as a chorismate-utilizing enzyme [15]–[17] suggesting, a type II fatty acid synthase like biosynthesis of xanthomonadin. Whereas the biological role of flexirubin is unknown, it could be shown that xanthomonadins protect Xanthomonas against photooxidative damage and lipidperoxidation [18]–[20]. Similar results were obtained for the xanthomonadin-like pigment from L. enzymogenes, which protects the bacterium from UV/visible-light and H2O2 [14].
Figure 1. Natural occurring DARs (1–4) [2]–[6], polyene containing pigments like flexirubin (5) from Chitinophaga filiformes [8] or xanthomonadin I (6) [9], DAR (7) heterologously produced with Azoarcus DAR proteins [1] and the proposed structure of the DAR-xanthomonadin hybrid pigment named arcuflavin (8) from Azoarcus sp. BH72 identified in this study.
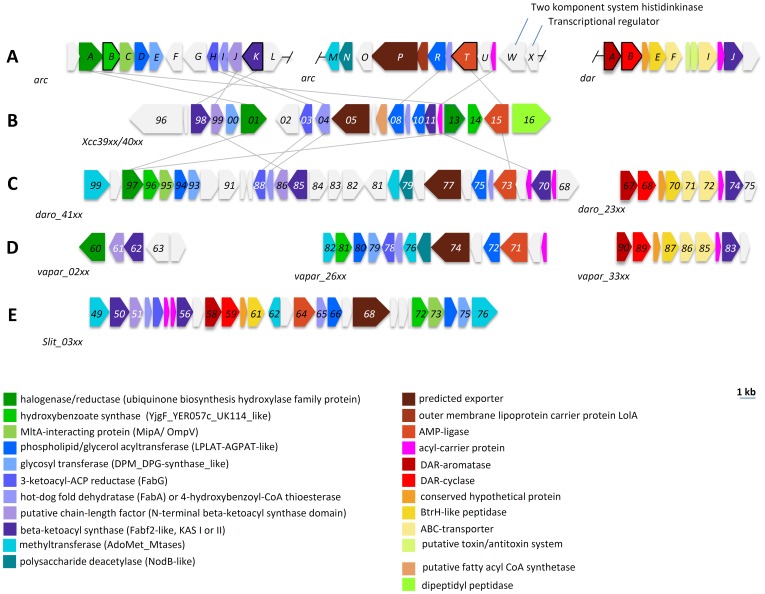
Figure 2. Known and proposed gene clusters involved in biosynthesis of DARs, xanthomonadin and xanthomonadin-like compounds in Azoarcus sp. BH72 (A), Xanthomonas campestris pv campestris ATCC 33913 (B), Dechloromonas aromatica RCB (C) Variovorax paradoxus S110 (D) and Sideroxydans lithotrophicus ES-1 (E).
Annotation colors follow the Azoarcus gene clusters and genes are connected by grey lines if their identity was ≥40% in a BLAST-P analysis. All genes are scaled to the depicted size and are listed in Table S1, S2, S3, S4.
Recently we reported that DAR biosynthesis is encoded by a conserved gene cluster which was found in 89 bacterial strains with most of them being associated with other organisms [1]. Among them is Azoarcus sp. BH72, a rod shaped motile bacterium which was isolated from surface-sterilized roots of Kallar grass (Leptochloa fusca) [21] where it lives as diazotroph mutualistic endophyte. Since heterologous expressed DAR-biosynthesis proteins of Azoarcus sp. BH72 led to DAR (7) (Figure 1) production [1] but no DAR was detected in Azoarcus sp. BH72 extracts, we were interested about its fate in the bacterial secondary metabolism. As it is known that Azoarcus sp. BH72 produces a cell-bound yellow pigment [21] we speculated that its DAR might be connected to the yellow compound in a flexirubin-like manner.
Here we report gene clusters responsible for the pigment biosynthesis in Azoarcus sp. BH72 and show that gene clusters containing polyene-biosynthesis associated genes are widespread in bacteria. Furthermore, we show by a combination of labeling experiments, HPLC-MS and MALDI-MS that the pigment from Azoarcus sp. BH72 contains the already reported DAR 7, which is connected to a xanthomonadin-like polyene. Additionally, a benzoate synthase and an AMP-ligase encoded by the Azoarcus pigment biosynthesis gene clusters were characterized that might perform the first two reactions of the polyene assembly.
Materials and Methods
Cultivation of strains
E. coli and Azoarcus sp. BH72 strains were cultivated on solid LB-medium (per L 0.5% yeast extract, 1% tryptone, 0.5% NaCl, 1.5% agar) or in liquid LB-medium on a rotary shaker at 200 rpm and are listed in Table S5. For protein overproduction E. coli was grown in modified auto induction medium (per L 0.5% yeast extract, 1% tryptone, 0.5% NaCl, 50 mM Na2HPO4, 50 mM KH2PO4, 25 mM (NH4)2SO4, 0.5% glycerol, 0.05% glucose, 0.2% lactose, pH 6.75) [22]. For extraction of chromosomal DNA and labeling experiments Azoarcus sp. BH72 was cultivated at 30°C and E. coli strains at 37°C. Where appropriate, kanamycin (50 µg/mL) was added to the medium.
Genome mining
For the identification of DarAB biosynthesis gene clusters, the primary sequences of the DarAB homologues from C. pinensis DSM 2588 were used for a BLAST-P identification (protein-protein-BLAST) of homologues in Azoarcus sp. BH72, Variovorax paradoxus S110, Dechloromonas aromatica RCB and Sideroxydans lithotrophicus ES-1. The xanthomonadin gene cluster from Xanthomonas campestris pv. campestris ATCC 33913 was used in a STRING-analysis (version 9.0) [23] for the identification of the putative xanthomonadin polyene cluster in Azoarcus sp. BH72, V. paradoxus S110, D. aromatica RCB and S. lithotropicus ES-1. Gene colors in Figure 2 are based on NCBI-annotation or domain guided annotations from BLAST-P with primary sequences of X. campestris pv. campestris-genes as template (see Table S1, S2, S3, S4). Grey connections between genes highlight BLAST-P results with an identity ≥40%.
Feeding experiments
400 mL liquid LB-medium was inoculated 1∶100 from an overnight culture of Azoarcus sp. BH72. After 3 h L-[methyl-2H3]methionine or 4-fluoro-3-hydroxybenzoic acid was fed in 1 mM portions to the cultures and feeding was repeated after 12, 24 and 48 h, resulting in a final concentration of 4 mM. As control, a culture without feeding was cultivated at the same time. Due to poor growth compared to the control culture, feeding of Azoarcus sp. BH72 with 4-fluoro-3-hydroxybenzoic acid was stopped after 24 h at a final concentration of 2 mM and cultures were grown for additional 48 h before harvesting. Cultures were harvested 12 h after the last feeding by centrifugation at 10.000 rpm.
Extraction and isolation of compounds
Pelleted cells of liquid cultures were shaken for 30 min with 50 mL acetone in the dark, followed by filtration to remove cell debris. The extracts were dried under reduced pressure. For structure elucidation raw extracts were fractionated using silica gel columns with hexane/ethylacetate (4∶1; 1% acetic acid). Yellow fractions were dried under reduced pressure. Samples were stored dry at −20°C in the dark and dissolved in 10–50 µL acetonitrile (ACN) before further analysis.
HPLC-MS
ESI HPLC MS analysis was performed with a Dionex UltiMate 3000 system coupled to a Bruker AmaZon X mass spectrometer and an Acquity UPLC BEH C18 1.7 µm RP column (Waters) as described previously [24].
MALDI-MS
For structure elucidation MALDI-analysis, samples were mixed 1∶2 with 1 µl of a 20 mM 4-chloro-α-cyanocinnamic acid (ClCCA) in 70% ACN with 0.1% trifluoracetic acid (TFA) or 9-aminoacridin in acetone, spotted onto a polished stainless steel target and air-dried. MALDI-MS analysis was performed with a MALDI LTQ Orbitrap XL (Thermo Fisher Scientific, Inc., Waltham, MA) equipped with a nitrogen laser at 337 nm as described previously [25]–[26]. Qual Browser (version 2.0.7; Thermo Fisher Scientific, Inc., Waltham, MA) was used for spectra analysis and to calculate possible sum formulas.
Molecular-biological methods
Molecular-biological experiments were performed according to standard procedures [27]. The phusion high-fidelity polymerase (Thermo-scientific) was used for polymerase chain reactions (PCR) according to the manufacturer's instructions and with oligonucleotides (Table S6) obtained from Sigma-Aldrich. Plasmid isolation was performed with the GeneJet™ Plasmid Extraction Kit (Fermentas) and DNA extraction from agarose gels with the GeneJet™ Gel Extraction Kit (Fermentas).
Generation of mutants of Azoarcus sp. strain BH72
Genes azo3911 (arcK) and azo0260 (arcT) were disrupted via plasmid integration with plasmid pK18GGST [28]. Briefly, internal gene fragments of the 5′ region of the respective genes were amplified and cloned into the expression vector pK18GGST through specific restriction sites, and a single recombination event with the wild type chromosome resulted in plasmid integration and thus a polar mutation. Growth conditions for mutant generation were as previously described [29]. The primers used for amplification were for azo0260 azo0260KOFor (XbaI restriction site) and azo0260KORev (HindIII restriction site), and for azo3911 azo3911KOFor (XbaI restriction site) and azo3911KORev (HindIII restriction site), respectively. The resulting fragments 0260KOfragment and 3911KOfragment cloned into the XbaI-HindIII restriction sites of pK18GGST spanned nucleotide positions of 7–639 nt of gene azo0260, or of 16–625 nt of gene azo3911, respectively. The correct plasmid sequences were verified by sequencing. Plasmids were transferred to strain BH72 by biparental mating, and the resulting plasmid integration mutants BH0260 and BH3911 were selected by their kanamycin resistance. Integration of the plasmid into the correct target site in the chromosome was verified by Southern blot hybridization.
Construction of plasmids for heterologous expression of ArcB and ArcT
An expression cassette encoding the TEV-protease cleavage site and a cherrytag (DelphiGenetics SA) with c-terminal His6-tag was digested with the restriction endonucleases NcoI and AvrII and ligated into the similar treated pCOLA-DUET1 resulting in pCATI1. The genes arcB (azo3920) and arcT (azo0260) were amplified by PCR from genomic DNA isolated from Azoarcus sp. BH72, resulting in 3920fragment and 0260fragment respectively. For construction of pCATI-arcB, pCATI1 was linearized by PCR giving pCATI1fragment which was fused with azo3920fragment using the Gibson assembly cloning kit (NEB). For construction of pCATI-arcT, 0260fragment was digested with the restriction endonucleases NcoI and XhoI and ligated with the similar treated pCATI1. The inserts of the plasmids pCATI-arcB and pCATI-arcT were sequenced by SeqIT GmbH (Kaiserslautern).
Overproduction and purification of ArcB and ArcT
E. coli BL21 (DE3) Star cells were transformed with pCATI-arcB or pCATI-arcT resulting in strain TS3920 and TS0260, respectively. 200 mL auto induction medium was inoculated 1∶100 from an overnight culture of TS3920 or TS0260 and grown to a OD600 of 0.6–0.8 at 37°C, followed by 16 h at 18°C. After centrifugation, the cell pellets were frozen at −20°C. Cells were lysed by sonification in lysis buffer (500 mM NaCl, 20 mM imidazole, 20 mM tris(hydroxymethyl)aminomethane (Tris), 0.01% Triton X-100, pH 7,5) which additionally contained lysozyme (10 kU/mL) and benzonase (2,5 U/ml). After filtration of the soluble fraction through 0.6 µM syringe filters it was applied to a ÄKTAexplorer™-System (GE-Healthcare) equipped with a HisTrap™ HP 1 mL affinity-chromatography column. Binding and elution buffer contained 500 mM NaCl, 20 mM imidazole, 20 mM HEPES pH 7,5 and 500 mM NaCl, 500 mM imidazole, 20 mM HEPES pH 7,5, respectively. Fusion tags were cleaved with TEV-Protease in elution buffer (12 h, room temperature) and tag-less proteins were purified as mentioned above. Purified proteins were analyzed by SDS-PAGE, protein concentrations were determined on a Nanovue Plus Spectrophotometer and proteins were stored at −80°C in storage buffer (100 mM NaCl, 50 mM Tris (pH 7,5), 1 mM dithiothreitol, 1 mM EDTA, 10% glycerol).
Benzoate synthase activity assays with ArcB
For in vivo activity assays with ArcB, 1 mL auto induction medium was inoculated 1∶100 from pre-cultures of E. coli BL21 (DE3) Star containing pCATI-arcB (strain TS3920) or the empty vector (pCATI1). After incubation on a rotary shaker (16 h, 37°C) cultures were extracted with 1 volume ethylacetate, dried and dissolved in 500 µL acetonitrile followed by derivatization with N-methyl-N-trimethylsilyl-trifluoroacetamid (MSTFA) (55°C, 1 h) and assay products were analyzed by GC-MS. For in vitro assays with ArcB, the tag-less purified protein was buffered in 50 mM Tris (pH 7.4). 100 µL assays (2 µg ArcB, 1 mM chorismic acid (Sigma-Aldrich), 100 mM Tris pH 7.4) were incubated at 37°C and stopped after 10 or 60 minutes by incubation at 100°C for 5 min. Stopped assays were adjusted to pH 4 and extracted 2× with 1 volume of ethylacetate. Dried organic phases were dissolved in 40 µL acetonitrile, derivatized with MSTFA (55°C, 1 h) and analyzed by GC-MS. Assay products were detected on a 7890A gas chromatograph (Agilent) equipped with a DB5ht column (30 m×250 µm×0.1 µM, Agilent) coupled with a 5975C mass selective detector (Agilent). The method parameters were the following: Carrier gas: Helium (1 mL/min); injection volume: 1 µL; inlet temperature: 250°C; injection mode splitless; oven starting temperature 5 min 70°C, then 5°C/min to 300°C. Ionization of the analyte molecules were carried out by electron impact ionization at 70 eV. Products were identified with the “Automated Mass Deconvolution and Identification Software” (AMDIS) version 2.64 in combination with the NIST-library.
ArcT in vitro assays
To test the adenylation activity of ArcT in vitro, purified tag-less protein was buffered in 20 mM Tris (pH 7.4) and incubated with labeled [γ-18O4]-ATP, various benzoic acid derivatives (listed in Figure S5), MgCl2 and pyrophosphate as described previously [26]. Assays were analyzed by MALDI-MS as reported above.
Results
Gene clusters for xanthomonadin and DAR biosynthesis
During our investigations of the DAR biosynthesis we could show that heterologous overexpressed DAR-biosynthesis proteins of Azoarcus sp. BH72 led to DAR 7 production in Escherichia coli [1], but the corresponding DAR was not detectable in Azoarcus extracts. However, Azoarcus sp. BH72 extracts contained a yellow hydrophobic compound that showed no color shift with alkali, indicating that the pigment was not a flexirubin [30]. The gene cluster for xanthomonadin biosynthesis (Figure 2.B, Table S4) is conserved within the gammaproteobacteria in several strains of Xanthomonas [11]–[13] and was also studied in L. enzymogenes [14]. We identified two gene clusters in the genome of the betaproteobacterium Azoarcus sp. BH72 (Rhodocyclales) encoding proteins with high similarity to those of the xanthomonadin biosynthesis (Figure 2.A, Table 1). Both putative arc loci (azo3910-azo3921 and azo0253-0264) were not physically linked. In addition to their already reported DAR biosynthesis gene clusters [1], the putative polyene biosynthesis gene clusters were also found in other betaproteobacteria like Dechloromonas aromatica RCB (Rhodocyclales) (Figure 2.C, Table S1), Variovorax paradoxus S110 (Burkholderiales) (Figure 2.D, Table S2) and Sideroxydans lithotrophicus ES-1 (Gallionellales) (Figure 2.E, Table S3). Within the identified gene clusters several genes are highly conserved (for example a gene encoding a glycosyl transferase, an outer membrane lipoprotein carrier protein (LolA), a phospholipid/glycerol acyltransferase and an adjacent exporter like protein) and we also found these genes in flexirubin biosynthesis gene clusters [31]. BLAST-P and String analysis with proteins from the gene clusters identified in Azoarcus revealed that gene clusters encoding these polyene biosynthesis associated genes can be found within several different proteobacteria, bacteria from the Bacteroidetes phylum and in one strain from the Spirochaetes (Table S7). In addition, 16 of the 39 reported bacterial genomes in Table S7 contained DAR biosynthesis gene clusters, with nine examples where the DAR biosynthesis genes are located within or adjacent to the polyene biosynthesis associated gene cluster.
Table 1. Predicted gene clusters for arcuflavin biosynthesis in Azoarcus sp. BH72. Domain guided annotation is based on conserved domains detected by BLAST-P.
| Gene | Genelocus [azo] | NCBI annotation | domain guided annotation |
| arcL | 3910 | hypothetical protein | peptidoglycan-binding protein |
| arcK | 3911 | 3-oxoacyl-ACP synthase | ketosynthase |
| arcJ | 3912 | hypothetical protein | N-terminal beta-ketoacyl synthase domain |
| arcI | 3913 | hypothetical protein | dehydratase |
| arcH | 3914 | 3-ketoacyl-ACP reductase | reductase |
| arcG | 3915 | putative Na/H(+) antiporter | Na+/H+-Antiporter |
| arcF | 3916 | N-acetyl-gamma-glutamyl-phosphate reductase | N-Acetyl-γ-Glutamyl-Ph-Reduktase |
| arcE | 3917 | glycosyl transferase | glycosyl transferase |
| arcD | 3918 | putative phospholipid biosynthesis acyltransferase | acyltransferase |
| arcC | 3919 | MltA-interacting protein | outer membrane protein |
| arcB | 3920 | hypothetical protein | chorismatase/XanB2 |
| arcA | 3921 | hypothetical protein | halogenase |
| arcM | 0253 | hypothetical protein | methyltransferase |
| arcN | 0254 | polysaccharide deacetylase | PS-deacetylase |
| arcO | 0255 | hypothetical protein | |
| arcP | 0256 | hypothetical protein | exporter |
| arcQ | 0257 | hypothetical protein | lolA |
| arcR | 0258 | acetyltransferase | acyltransferase |
| arcS | 0259 | hypothetical protein | dehydratase |
| arcT | 0260 | hypothetical protein | Acyl-CoA synthetase/AMP- ligases |
| arcU | 0261 | hypothetical protein | |
| arcV | 0262 | hypothetical protein | ACP |
| arcW | 0263 | two-component system histidine kinase | histidinkinase |
| arcX | 0264 | transcriptional regulator | response regulator |
| darK | 0283 | hypothetical protein | |
| darJ | 0284 | putative Beta-ketoacyl synthase family protein | ketosynthase |
| darC | 0285 | acyl carrier protein | ACP |
| darI | 0286 | putative ABC transporter permease | ABC-Transporter; Permease |
| darH | 0287 | hypothetical protein | PilT-Domain |
| darG | 0288 | hypothetical protein | Prevent host death-protein |
| darF | 0289 | ABC transporter ATP-binding protein | ABC-Transporter; ATP-Binding. |
| darE | 0290 | hypothetical protein | BtrH-like peptidase |
| darD | 0291 | hypothetical protein | conserved hypothetical protein |
| darB | 0292 | 3-oxoacyl-ACP synthase | DAR-cyclase |
| darA | 0293 | dialkylrecorsinol condensing protein | DAR-aromatase |
Verification of assigned pigment biosynthesis gene clusters
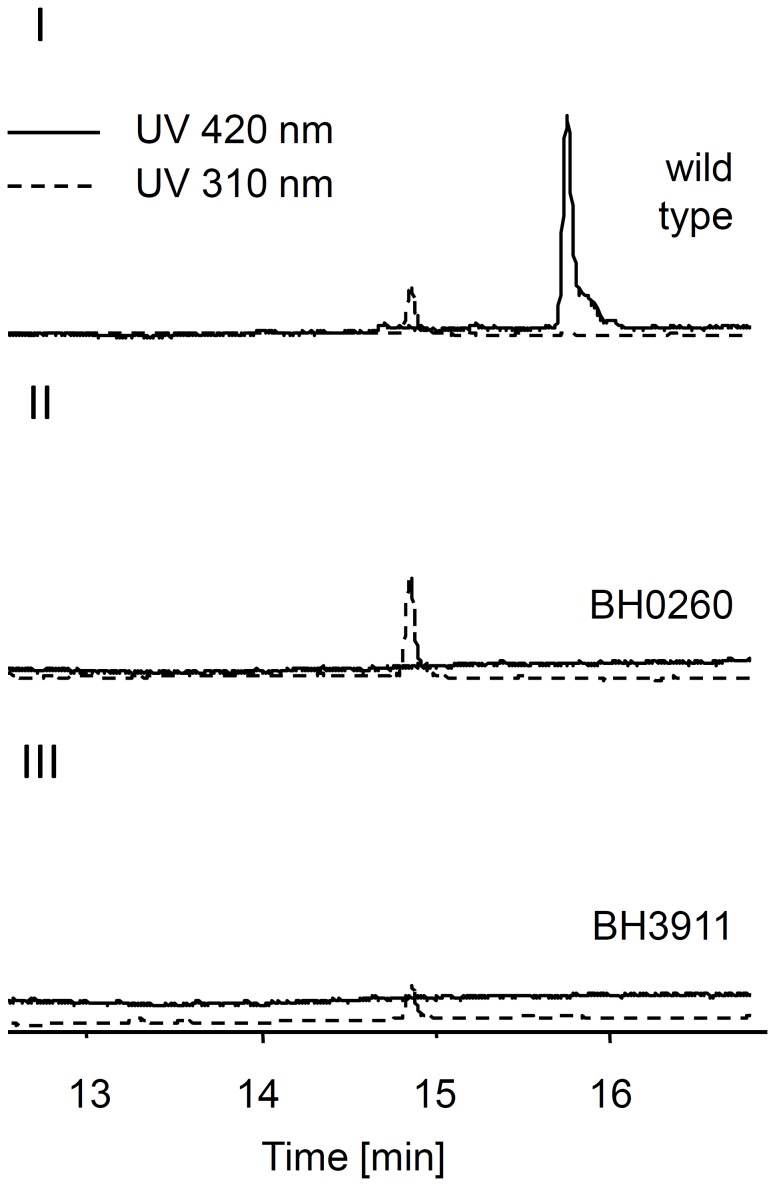
Detailed studies with Xanthomonas strains showed that the biosynthesis of xanthomonadin depends on the reported gene cluster [11]–[13] (Figure 2.B), which was also shown for the biosynthesis of the xanthomonadin-like pigment from L. enzymogenes [14]. Thus we tested whether these gene clusters encoding such homologues are essential for the pigment biosynthesis by insertion of plasmids into gene arcK (azo3911) or arcT (azo0260), respectively. The resulting mutant strains BH3911 and BH0260 appeared colorless on agar plate (Figure S1), and subsequent acetone extraction and HLPC-UV analysis showed the loss of a UV-signal at 15.7 min in the extracts from the mutants (Figure 3).
Figure 3. HPLC analysis of Azoarcus sp. BH72 wild type and mutant strains.
HPLC-UV analysis of wildtype (I), BH0260 (II) and BH3911 extracts with traces showing absorption at 420 nm (solid line) or 310 nm (dotted line). All chromatograms were drawn to the same scale.
Structure elucidation of the pigment
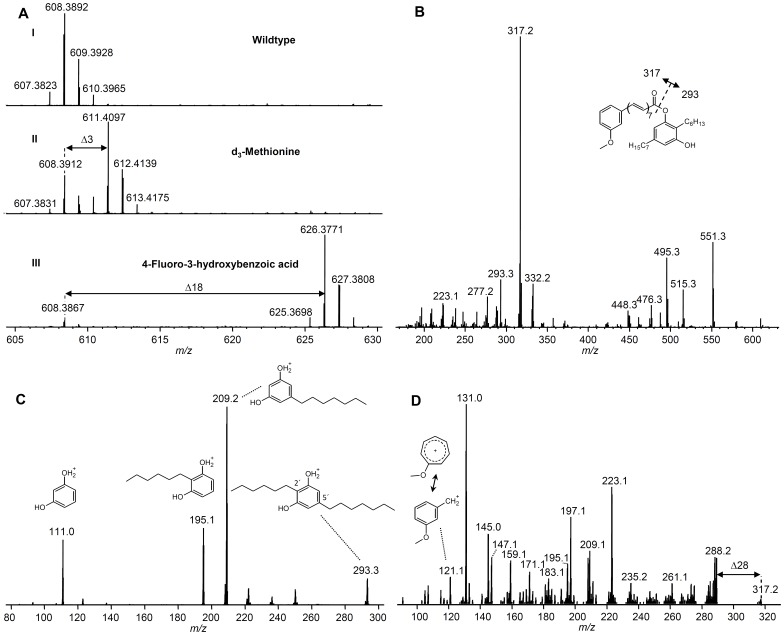
Further analysis of acetone extracts of Azoarcus sp. BH72 led to the identification of pigment 8 with a mass of m/z 608.38633 [M]+• (Figure 4) leading to a sum formula of C41H52O4 (calc. m/z 608.38601 [M]+•; Δppm 0.5) and UV absorption at 420 nm (Figure S2.A). MALDI-MS2 of 8 led to a base peak of m/z 317.2 and m/z 293.3 (Figure 4.B) and MALDI-MS3 of m/z 293.3 (Figure 4.C) revealed a fragmentation pattern identical to what has been observed for the heterologous produced DAR 7 [1]. MALDI-MS3 of m/z 317.2 (Figure 4.D) led to a complex fragmentation pattern with a m/z 121 key-fragment, a mass shift of Δ28 Da (-CO) from m/z 317.2 and Δ78 Da (-C6H6) or Δ26 Da (-C2H2) mass shifts. The conserved structural feature of xanthomonadin is the ω-(3-hydroxyphenyl)-polyene carboxylic acid that is usually methylated at the hydroxyl group. When Azoarcus was grown in the presence of L-[methyl-2H3]methionine or 4-fluoro-3-hydroxybenzoic acid (4F-3HBA) (a fluorinated derivative of the putative xanthomonadin precursor 3-hydroxybenzoic acid (3HBA)), an additional M+3 (Figure 4.AII) or M+18 signal (Figure 4.AIII; Figure S2.AII) was detectable, respectively. In contrast, if 3-fluoro-4-hydroxybenzoic acid (3F-4HBA) was fed to the culture no M+18 signal was detected (data not shown). MALDI-MS2 analysis of both labeled pigments led to the corresponding Δ3 Da and Δ18 Da mass shifts at the previously m/z 317.2 fragments (Figure S2.B) but not at the DAR fragment m/z 293.3. Subsequent MS3-analysis of the labeled fragments revealed the mass-shifts of the m/z 121 key-fragment, which is originating from the polyene moiety (Figure S2.C).
Figure 4. Mass spectra of 8 from Azoarcus sp. BH72.
A: MALDI-orbitrap mass spectra of wildtype 8 (I) and from labeling experiments with d3-methionine (II) and 4-Fluoro-3-hydroxybenzoic acid (4F-3HBA) (III). B: MALDI-iontrap-MS2 of 8. MALDI-iontrap-MS3 of m/z 293.3 (C) and m/z 317.2 (D) with m/z 608.4 [M]+• as precursor.
Initiation of the arcuflavin polyene biosynthesis
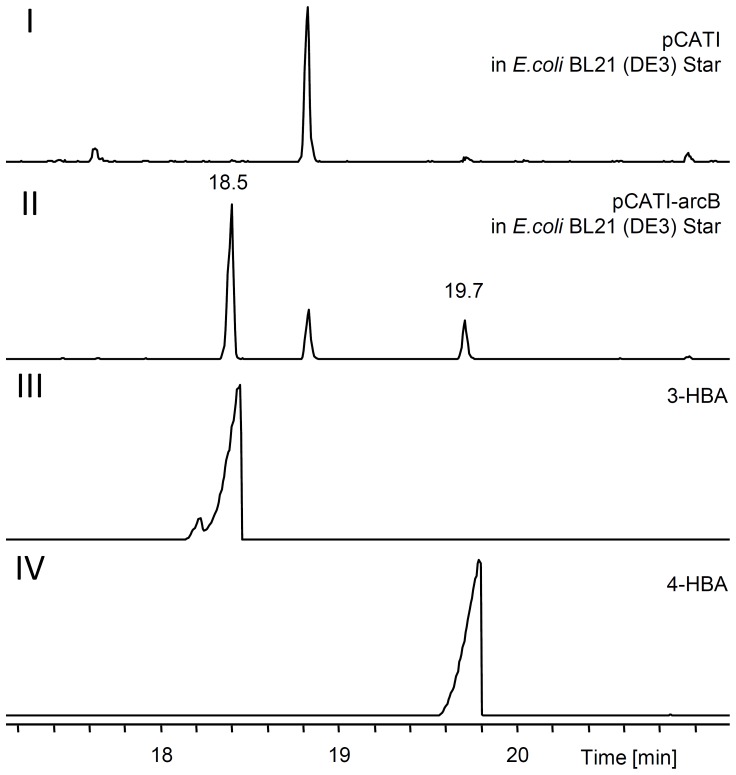
Previously it was shown that xanB2 (Xcc4014) from X. campestris is responsible for production of the diffusible factor (DF) 3-hydroxybenzoic acid (3-HBA), which is essential for xanthomonadin biosynthesis [12], [15]. Recent research revealed that xanB2 encodes a structurally novel and bifunctional chorismatase converting chorismic acid into 3-HBA, the probable biosynthetic precursor of the xanthomonadin ring moiety, and 4-hydroxybenzoic acid (4-HBA) which is needed for coenzyme Q8 biosynthesis [16], [17]. A homologue of XanB2 in Azoarcus is encoded by arcB within the postulated pigment biosynthesis gene cluster (Figure 2.A). Like XanB2, ArcB contains a YjgF-like C-terminal domain, which is also present in the chorismate utilizing enzymes Hyg5, FkbO and RapK from Streptomyces [32]. Based on their reaction products, these enzymes were sorted in three distinct types of chorismatases [33]. The FkbO type produces 3,4-dihydroxycyclohexa-1,5-dienoic acid, the Hyg5-type forms 3-HBA whereas the XanB2-type converts chorismate to 3-HBA and 4-HBA [33]. Since from sequence alignment alone it is actually not possible to predict accurately the reaction catalyzed by these chorismate-converting enzymes and to get experimental evidence for the catalyzed reaction, arcB was overexpressed in E. coli and the culture supernatants were extracted. GC-MS analysis of these extracts (Figure 5) revealed the formation of compounds with similar retention times and mass spectra (Figure S3.A) as 3-HBA and 4-HBA standards. Subsequent purification of ArcB (Figure S4) allowed its incubation with chorismic acid. In all assays 4-HBA was formed non-enzymatically (Figure S3.BIV) as reported before [16], [34], whereas 3-HBA was only detectable in assays containing ArcB (Figure S3.III). Since it was postulated that xanthomonadin biosynthesis is performed by a type II fatty acid synthase like biosynthesis using 3-HBA as polyene precursor [15], 3-HBA should be activated for the polyketide synthase (PKS) machinery. Such a reaction might be performed by the putative AMP-ligase encoded by Xcc4015 in X. campestris pv. campestris, which is essential for the pigment biosynthesis in this organism [15]. The homologue ArcT, encoded in the pigment gene cluster from Azoarcus sp. BH72, was overexpressed in E. coli and purified (Figure S4). The adenylation activity of ArcT was tested with a method measuring the isotopic back exchange of unlabelled pyrophosphate into [γ-18O4]-ATP if the enzyme adenylates a substrate [26]. MALDI-MS analysis showed ATP-label exchange in assays with 3-HBA and various other benzoic acids with modifications in 4-position which was not detected if no substrate, 3-methoxybenzoic acid or 2-substituted benzoic acids were added (Figure S5).
Figure 5. GC-MS analysis of E. coli cultures overexpressing arcB.
Total ion chromatograms of extracted supernatants of E. coli BL21 (DE3) Star harboring the vector without insert (I), the arcB overexpression plasmid (II) and of 3-hydroxybenzoic acid (3-HBA) (III) and 4-hydroxybenzoic acid (4-HBA) standards (IV).
Discussion
In previous studies the xanthomonadin biosynthesis gene cluster was found in genomes of other gamma- and betaproteobacteria and homologues of the chorismate-utilizing enzyme XanB2 (Xcc4014) were also found in genomes outside of the proteobacteria [16]. However, only the gene cluster of the closely related bacterium L. enzymogenes (family xanthomonadaceae) was studied and a non-brominated pigment was found in this bacterium, which is most likely xanthomonadin-like [14]. Here we identified for the first time similar polyene biosynthesis gene clusters in the genome of Azoarcus sp. BH72 which additionally contains a DAR-biosynthesis gene cluster. Our results with plasmid insertion mutants show that both xanthomonadin-like biosynthesis gene clusters, although they are not physically linked, are essential for pigment 8 biosynthesis in Azoarcus sp. BH72, indicating that its product is indeed a xanthomonadin-like compound.
Results from MS2 experiments with pigment 8 show that its fragmentation is similar to those of flexirubin where the basepeak was shown to result from the cleavage of the polyene-DAR ester bond [31]. Therefore we predict that DAR 7 and the xanthomonadin-like polyene are connected in a flexirubin-like manner by an ester bond between the polyene carboxylic acid and a phenolic hydroxyl group. Similar to flexirubin, the formation of a radical cation was observed that might be explained by direct absorption of the laser light by the polyene of pigment 8 instead of matrix mediated ionization mechanisms. Since the MS3 of the assigned DAR-fragment m/z 293.3 is identical to those from heterologous produced DAR 7 [1], we conclude that the Azoarcus pigment 8 has a DAR moiety identical to 7. It was postulated that 3-HBA is a biosynthetic intermediate of the xanthomonadin biosynthesis [15]. Our labeling result with 4F-3HBA now confirms that this is indeed the case in the biosynthesis of pigment 8 from Azoarcus which additionally contains a methionine derived methylgroup. Furthermore the MS3 of the assigned polyene fragment m/z 317.2 shows mass shifts of Δ28 Da (CO), Δ78 Da (C6H6) or Δ26 Da (C2H2) (Figure S2.C), already known from mass spectrometric fragmentation of flexirubin-like polyenes [31], [35]. These results suggest the structure of a nonbrominated aryl-heptaene for the polyene moiety of 8, reflecting xanthomonadin group-11 previously found in Xanthomonas [36], [37]. Based on these findings we conclude that a new xanthomonadin-DAR hybrid has been characterized, which we named arcuflavin (from Azoarcus and latin for yellow) and propose it to have the structure of 8. Since arcuflavin is only produced in low amounts and becomes insoluble in various organic solvents after enrichment, similar to the L. enzymogenes xanthomonadin-like pigment [14], further structure elucidation by means of NMR was not possible.
Combinations of xanthomonadin-like polyene gene cluster and DAR-gene cluster were also found in D. aromatica RCB, three strains of V. paradoxus and S. lithotrophicus ES-1. Whereas in previous genome screenings only the xanthomonadin-like gene clusters from D. aromatica RCB and V. paradoxus EPS and S110 were found [14], [16], we could now identify their DAR gene clusters and it will be interesting to see in future studies if the combination of both gene cluster types leads to the identification of polyene-DAR hybrids also in these organisms. In addition we found that genes associated with known polyene biosynthesis gene clusters with and without DAR biosynthesis encoding genes can be found within a wide range of bacterial genomes including medically important taxa like Burkholderia, Vibrio, Escherichia and Pseudomonas (Table S7). We therefore predict that more pigments like flexirubin, xanthomonadin or arcuflavin will be found in these organisms and that these compounds might also be involved in the pathophysiology of these organisms. Moreover, our findings suggest that these pigments cannot be used as chemotaxonomic markers, especially in the case of flexirubin for the Bacteroidetes phylum, as they seem to be much more widespread than originally thought.
We investigated the first steps of the arcuflavin-polyene biosynthesis and could show that ArcB has in vitro the assumed catalytic activity of a chorismate-utilizing enzyme that produces 3-HBA and 4-HBA. This is similar to the reaction performed by its homologue in the xanthomonadin biosynthesis pathway [16], [17] and makes it the third biochemically characterized example of the XanB2-type of chorismate-utilizing enzymes. Furthermore the activity of the AMP-ligase ArcT with 3-HBA could be shown in vitro. The result that 3-methoxybenzoic acid was not a good substrate for ArcT suggests that the methylation of pigment 8 takes place after the activation of the 3-HBA starter. Although ArcT showed in vitro a low substrate specificity, no other derivatives of arcuflavin 8 were found in the extracts of Azoarcus, showing that in vivo the arcuflavin-biosynthesis machinery is more selective in the precursor or intermediate selection.
Conclusions
Due to their structural similarity one might conclude a functional similarity for arcuflavin with xanthomonadins and DARs. Xanthomonadins from Xanthomonas oryzae pv. oryzae and X. campestris pv. campestris are located in the bacterial outer membrane [13], [38]and have been shown to protect their producers from photooxidative damage and lipid peroxidation [18]–[20]. On the other hand, the DARs DB-2073 (1) and resorstatin (2) were reported to act as free radical scavengers to protect against lipid peroxidation [4] and show striking structural similarity to the DAR from arcuflavin. Therefore DAR or polyene-DAR hybrids like arcuflavin might protect the cell against oxidative damage, suggesting the fusion of two protective moieties in polyene-DAR hybrids. Bacteria from the genus Xanthomonas are well known plant pathogens, Azoarcus sp. BH72 is a diazotroph endophyte of rice and other grasses [39], whereas V. paradoxus S110 is a growth-promoting endophyte of various plants [40]. As transient production of reactive oxygen species is found in most plant-microorganism interactions [41], arcuflavin and similar polyene pigments might play a role in establishing or maintaining the symbiotic life of the bacterium with its host.
Supporting Information
Predicted gene clusters for arcuflavine-like biosynthesis in Dechloromonas aromatica RCB.
(DOCX)
Predicted gene clusters for arcuflavin-like biosynthesis in Variovorax paradoxus S110.
(DOCX)
Predicted gene clusters for arcuflavin-like biosynthesis in Sideroxydans lithotrophicus ES-1.
(DOCX)
Gene cluster for xanthomonadin biosynthesis in Xanthomonas campestris pv. campestris ATCC33913.
(DOCX)
Strains and plasmids used in this work.
(DOCX)
Primers and PCR products used in this study.
(DOCX)
Predicted gene clusters encoding polyene-associated biosynthesis and DAR-biosynthesis proteins.
(DOCX)
Phenotype of Azoarcus sp. BH72 wildtype and mutant strains. Growth and pigmentation of wildtype Azoarcus sp. BH72 and insertional mutants on VM-agar (with ethanol) plate (left) or a kanamycin containing VM-agar (with ethanol) plate (right).
(TIF)
HPLC-MS-analysis and mass spectra of 8 from Azoarcus sp. BH72. A: HPLC-MS-analysis of wildtype 8 (I) and after feeding with 4-fluoro-3-hydroxybenzoic acid (4F-3HBA) (II) in raw extracts. Traces show EIC of m/z 607.5 [M-H+] (orange), m/z 625.5 [M-H+] (green) and UV at 420 nm (blue). Chromatograms are drawn to the same scale. B: MALDI-iontrap-MS2 of wildtype 8 (I), m/z 611.4 [M]+• (II) and m/z 626.4 [M]+• (III) from feeding experiments with d3-methionine or 4F-3HBA, respectively. C: MALDI-iontrap-MS3 mass spectra of the polyene-fragments m/z 317.3 (I), m/z 320.2 (II) and m/z 335.3 (III), that were obtained from the precursors m/z 608.4 [M]+•(I), m/z 611.4 [M]+• (II) and m/z 626.4 [M]+•(III). Mass shifts mentioned in the text are indicated.
(TIF)
GC-MS analysis of E. coli cultures overexpressing arcB and in vitro assays with purified ArcB. A: Mass spectra of 3-hydroxybenzoic acid (3-HBA) standard (I) and the compounds identified at min 18.5 (II) and min 19.7 (III) in E. coli cultures overexpressing arcB. B: Total ion chromatograms of 4-hydroxybenzoic acid (4-HBA) standard (I) and 3-HBA standard (II) and chorismic acid containing in vitro assays incubated with (III) or without (IV) ArcB and stopped after 10 (blue) or 60 min (red) incubation, respectively.
(TIF)
SDS-PAGE analysis of purified Azoarcus proteins. M: PageRuler™ Unstained protein ladder (Fermentas). Lane 1–8: Fractions of ArcB purification with the purified protein in Lane 8 (expected size 35,24 kDA) Lane 10: Purified ArcT (expected size: 47,1 kDa). Stars indicate protein bands with the expected size.
(TIF)
MALDI-MS analysis of [γ-18O4]-ATP containing assays with ArcT and various substrates. A mass shift from m/z 514 to m/z 506 indicates the exchange of the ATP-label and therefore activity of ArcT with the substrate. Substrates: A: none B: 3-hydroxybenzoic acid C: 4-fluoro-3-hydroxybenzoic acid D: 4-fluorobenzoic acid E: 4-hydroxybenzoic acid F: 4-methoxybenzoic acid G: 3-methoxybenzoic acid H: 2-methoxybenzoic acid I: 2,3-dihydroxybenzoic acid J: 3,5- dihydroxybenzoic acid K: benzoic acid L: p-coumaric acid M: E-cinnamic acid. Colours indicate if no (red), <50% (orange) or >50% label exchange (green) was detected.
(TIF)
Acknowledgments
We thank Prof. M. Karas for MALDI access and Reshma Kadam and Andreas Beust for help during the mutant construction.
Funding Statement
Work in the Bode lab was funded by the Deutsche Forschungsgemeinschaft (DFG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fuchs SW, Bozhüyük KA, Kresovic D, Grundmann F, Dill V, et al. (2013) Formation of 1,3-cyclohexanediones and resorcinols catalyzed by a widely occuring ketosynthase. Angew Chem Int Ed 52: 4108–4112. [DOI] [PubMed] [Google Scholar]
- 2. Kanda N, Ishizaki N, Inoue N, Oshima M, Handa A, et al. (1975) DB-2073, a new alkylresorcinol antibiotic. I. Taxonomy, isolation and characterization. J Antibiot 28: 935–942. [DOI] [PubMed] [Google Scholar]
- 3. Kitahara T, Kanda N (1975) DB-2073, a new alkylresorcinol antibiotic. II. The chemical structure of DB-2073. J Antibiot 28: 943–946. [DOI] [PubMed] [Google Scholar]
- 4. Kato S, Shindo K, Kawai H, Matsuoka M, Mochizuki J (1993) Studies on free radical scavenging substances from microorganisms. III. Isolation and structural elucidation of a novel free radical scavenger, resorstatin. J Antibiot 46: 1024–1026. [DOI] [PubMed] [Google Scholar]
- 5. Joyce SA, Brachmann AO, Glazer I, Lango L, Schwär G, et al. (2008) Bacterial biosynthesis of a multipotent stilbene. Angew Chem Int Ed 47: 1942–1945. [DOI] [PubMed] [Google Scholar]
- 6. Imai S, Fujioka K, Furihata K, Fudo R, Yamanaka S, et al. (1993) Studies on cell growth stimulating substances of low molecular weight. Part 3. Resorcinin, a mammalian cell growth stimulating substance produced by Cytophaga johnsonae . 46: 1319–1322. [DOI] [PubMed] [Google Scholar]
- 7. Achenbach H, Kohl W, Reichenbach H (1979) Investigations on Metabolites of Microorganisms. XX. The Structures of the Flexirubin-type Pigments from Cytophaga johnsonae Cyj1. Chem Ber 112: 1999–2011. [Google Scholar]
- 8. Achenbach H, Kohl W, Reichenbach H (1976) Investigations on Metabolites of Microorganisms. XI. Flexirubin, a novel pigment from Flexibacter elegans . Chem Ber 109: 2490–2502. [Google Scholar]
- 9. Andrewes AG, Jenkins CL, Starr MP, Shepherd J, Hope H (1977) Structure of Xanthomonadin I, A Novel Dibrominated Aryl-Polyene Pigment Produced by the Bacterium Xanthomonas juglandis . Tetrahedron Lett 45: 4023–4024. [Google Scholar]
- 10. Starr MP, Jenkins CL, Bussey LB, Andrewes AG (1977) Chemotaxonomic significance of the xanthomonadins, novel brominated aryl-polyene pigments produced by bacteria of the genus Xanthomonas . Arch Microbiol 113: 1–9. [DOI] [PubMed] [Google Scholar]
- 11. Poplawsky AR, Kawalek MD, Schaad NW (1993) A xanthomonadin-encoding gene cluster for the identification of pathovars of Xanthomonas campestris . Mol Plant Microbe Interact 6: 545–552. [Google Scholar]
- 12. Poplawsky AR, Chun W (1997) pigB determines a diffusible factor needed for extracellular polysaccharide slime and xanthomonadin production in Xanthomonas campestris pv. campestris . J Bacteriol 179: 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goel AK, Rajagopal L, Nagesh N, Sonti RV (2002) Genetic Locus Encoding Functions Involved in Biosynthesis and Outer Membrane Localization of Xanthomonadin in Xanthomonas oryzae pv. oryzae . J Bacteriol 184: 3539–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Y, Qian G, Li Y, Wang Y, Wang Y, et al. (2013) Biosynthetic Mechanism for Sunscreens of the Biocontrol Agent Lysobacter enzymogenes . PLoS One 8: e66633 10.1371/journal.pone.0066633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He YW, Wu J, Zhou L, Yang F, He YQ, et al. (2011) Xanthomonas campestris diffusible factor is 3-hydroxybenzoic acid and is associated with xanthomonadin biosynthesis, cell viability, antioxidant activity, and systemic invasion. Mol Plant Microbe Interact 24: 948–957. [DOI] [PubMed] [Google Scholar]
- 16. Zhou L, Wang JY, Wu J, Wang J, Poplawsky A, et al. (2013) The diffusible factor synthase XanB2 is a bifunctional chorismatase that links the shikimate pathway to ubiquinone and xanthomonadins biosynthetic pathways. Mol Microbiol 87: 80–93. [DOI] [PubMed] [Google Scholar]
- 17. Zhou L, Huang TW, Wang JY, Sun S, Chen G, et al. (2013) The rice bacterial pathogen Xanthomonas oryzae pv. oryzae produces 3-hydroxybenzoic acid and 4-hydroxybenzoic acid via XanB2 for use in xanthomonadin, ubiquinone and exopolysaccharide biosynthesis. Mol Plant Microbe Interact 26: 1239–1248. [DOI] [PubMed] [Google Scholar]
- 18. Jenkins CL, Starr MP (1982) The brominated aryl-polyene (xanthomonadin) pigments of Xanthomonas juglandis protect against photobiological damage. Curr Microbiol 7: 323–326. [Google Scholar]
- 19. Rajagopal L, Sundari CS, Balasubramanian D, Sonti RV (1997) The bacterial pigment xanthomonadin offers protection against photodamage. FEBS Lett 415: 125–128. [DOI] [PubMed] [Google Scholar]
- 20. Poplawsky AR, Urban SC, Chun W (2000) Biological role of xanthomonadin pigments in Xanthomonas campestris pv. campestris. Appl Environ Microbiol 66: 5123–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reinhold B, Hurek T, Niemann EG, Fendrik I (1986) Close association of azospirillum and diazotrophic rods with different root zones of kallar grass. Appl Environ Microbiol 52: 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Studier FW (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41: 207–234. [DOI] [PubMed] [Google Scholar]
- 23. Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, et al. (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41: D808–D815 10.1093/nar/gks1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reimer D, Pos KM, Thines M, Grün P, Bode HB (2011) A natural prodrug activation mechanism in nonribosomal peptide synthesis. Nat Chem Biol 7: 888–890. [DOI] [PubMed] [Google Scholar]
- 25. Fuchs SW, Sachs CC, Kegler C, Nollmann FI, Karas M, et al. (2012) Neutral loss fragmentation pattern based screening for arginine-rich natural products in Xenorhabdus and Photorhabdus . Anal Chem 84: 6948–6955. [DOI] [PubMed] [Google Scholar]
- 26. Phelan VV, Du Y, McLean JA, Bachmann BO (2009) Adenylation enzyme characterization using gamma -(18)O(4)-ATP pyrophosphate exchange. Chem Biol 16: 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: A laboratory manual, 2nd edn. New York, USA: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 28. Krause A, Bischoff B, Miché L, Battistoni F, Reinhold-Hurek B (2011) Exploring the function of alcohol dehydrogeanses during the endophytic life of Azoarcus sp. strain BH72. Mol. Plant-Microbe Interact 24: 1325–1332. [DOI] [PubMed] [Google Scholar]
- 29. Böhm M, Hurek T, Reinhold-Hurek B (2007) Twitching motility is essential for endophytic rice colonization by the N2-fixing endophyte Azoarcus sp. strain BH72. Mol. Plant-Microbe Interact 20: 526–533. [DOI] [PubMed] [Google Scholar]
- 30. Fautz E, Reichenbach H (1980) A simple test for flexirubin-type pigments. FEMS Microbiol Lett 8: 87–91. [Google Scholar]
- 31. Schöner TA, Fuchs SW, Schönau C, Bode HB (2013) Initiation of the flexirubin biosynthesis in Chitinophaga pinensis . Microb Biotechnol 10.1111/1751-7915.12110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Andexer JN, Kendrew SG, Nur-e-Alam M, Lazos O, Foster TA, et al. (2011) Biosynthesis of the immunosuppressants FK506, FK520, and rapamycin involves a previously undescribed family of enzymes acting on chorismate. Proc Natl Acad Sci U S A 108: 4776–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Juneja P, Hubrich F, Diederichs K, Welte W, Andexer JN (2013) Mechanistic Implications for the Chorismatase FkbO Based on the Crystal Structure. J Mol Biol 10.1016/j.jmb.2013.09.006 [DOI] [PubMed] [Google Scholar]
- 34. Siebert M, Bechthold A, Melzer M, May U, Berger U, et al. (1992) Ubiquinone biosynthesis. Cloning of the genes coding for chorismate pyruvate-lyase and 4-hydroxybenzoate octaprenyl transferase from Escherichia coli . FEBS Lett 307: 347–350. [DOI] [PubMed] [Google Scholar]
- 35. Achenbach H, Kohl W (1979) Investigations on Metabolites of Microorganisms, XVIII. The Mass Spectrometric Fragmentation of Flexirubin-type Pigments. Chem Ber 112: 209–217. [Google Scholar]
- 36. Jenkins CL, Starr MP (1982) The Pigment of Xanthomonas populi is a Nonbrominated Aryl-Heptaene Belonging to Xanthomonadin Pigment Group 11. Curr Microbiol 7: 195–198. [Google Scholar]
- 37. Jenkins CL, Starr MP (1985) Formation of halogenated aryl-polyene (Xanthomonadin) pigments by the type and other yellow-pigmented strains of Xanthomonas maltophilia . Ann Inst Pasteur Microbiol 136B: 257–264. [DOI] [PubMed] [Google Scholar]
- 38. Dianese JC, Schaad NW (1982) Isolation and Characterization of Inner and Outer Membranes of Xanthomonas campestris pv. campestris . Phytopathology 72: 1284–1289. [Google Scholar]
- 39. Krause A, Ramakumar A, Bartels D, Battistoni F, Bekel T, et al. (2006) Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nat Biotechnol 24: 1385–1391. [DOI] [PubMed] [Google Scholar]
- 40. Han JI, Choi HK, Lee SW, Orwin PM, Kim J, et al. (2011) Complete genome sequence of the metabolically versatile plant growth-promoting endophyte Variovorax paradoxus S110. J Bacteriol 193: 1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nanda AK, Andrio E, Marino D, Pauly N, Dunand C (2010) Reactive oxygen species during plant-microorganism early interactions. J Integr Plant Biol 52: 195–204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Predicted gene clusters for arcuflavine-like biosynthesis in Dechloromonas aromatica RCB.
(DOCX)
Predicted gene clusters for arcuflavin-like biosynthesis in Variovorax paradoxus S110.
(DOCX)
Predicted gene clusters for arcuflavin-like biosynthesis in Sideroxydans lithotrophicus ES-1.
(DOCX)
Gene cluster for xanthomonadin biosynthesis in Xanthomonas campestris pv. campestris ATCC33913.
(DOCX)
Strains and plasmids used in this work.
(DOCX)
Primers and PCR products used in this study.
(DOCX)
Predicted gene clusters encoding polyene-associated biosynthesis and DAR-biosynthesis proteins.
(DOCX)
Phenotype of Azoarcus sp. BH72 wildtype and mutant strains. Growth and pigmentation of wildtype Azoarcus sp. BH72 and insertional mutants on VM-agar (with ethanol) plate (left) or a kanamycin containing VM-agar (with ethanol) plate (right).
(TIF)
HPLC-MS-analysis and mass spectra of 8 from Azoarcus sp. BH72. A: HPLC-MS-analysis of wildtype 8 (I) and after feeding with 4-fluoro-3-hydroxybenzoic acid (4F-3HBA) (II) in raw extracts. Traces show EIC of m/z 607.5 [M-H+] (orange), m/z 625.5 [M-H+] (green) and UV at 420 nm (blue). Chromatograms are drawn to the same scale. B: MALDI-iontrap-MS2 of wildtype 8 (I), m/z 611.4 [M]+• (II) and m/z 626.4 [M]+• (III) from feeding experiments with d3-methionine or 4F-3HBA, respectively. C: MALDI-iontrap-MS3 mass spectra of the polyene-fragments m/z 317.3 (I), m/z 320.2 (II) and m/z 335.3 (III), that were obtained from the precursors m/z 608.4 [M]+•(I), m/z 611.4 [M]+• (II) and m/z 626.4 [M]+•(III). Mass shifts mentioned in the text are indicated.
(TIF)
GC-MS analysis of E. coli cultures overexpressing arcB and in vitro assays with purified ArcB. A: Mass spectra of 3-hydroxybenzoic acid (3-HBA) standard (I) and the compounds identified at min 18.5 (II) and min 19.7 (III) in E. coli cultures overexpressing arcB. B: Total ion chromatograms of 4-hydroxybenzoic acid (4-HBA) standard (I) and 3-HBA standard (II) and chorismic acid containing in vitro assays incubated with (III) or without (IV) ArcB and stopped after 10 (blue) or 60 min (red) incubation, respectively.
(TIF)
SDS-PAGE analysis of purified Azoarcus proteins. M: PageRuler™ Unstained protein ladder (Fermentas). Lane 1–8: Fractions of ArcB purification with the purified protein in Lane 8 (expected size 35,24 kDA) Lane 10: Purified ArcT (expected size: 47,1 kDa). Stars indicate protein bands with the expected size.
(TIF)
MALDI-MS analysis of [γ-18O4]-ATP containing assays with ArcT and various substrates. A mass shift from m/z 514 to m/z 506 indicates the exchange of the ATP-label and therefore activity of ArcT with the substrate. Substrates: A: none B: 3-hydroxybenzoic acid C: 4-fluoro-3-hydroxybenzoic acid D: 4-fluorobenzoic acid E: 4-hydroxybenzoic acid F: 4-methoxybenzoic acid G: 3-methoxybenzoic acid H: 2-methoxybenzoic acid I: 2,3-dihydroxybenzoic acid J: 3,5- dihydroxybenzoic acid K: benzoic acid L: p-coumaric acid M: E-cinnamic acid. Colours indicate if no (red), <50% (orange) or >50% label exchange (green) was detected.
(TIF)