Abstract
Inteins are auto-processing domains found in organisms from all domains of life. These proteins carry out a process known as protein splicing, which is a multi-step biochemical reaction comprised of both the cleavage and formation of peptide bonds. While the endogenous substrates of protein splicing are specific essential proteins found in intein-containing host organisms, inteins are also functional in exogenous contexts and can be used to chemically manipulate virtually any polypeptide backbone. Given this, protein chemists have exploited various facets of intein reactivity to modify proteins in myriad ways for both basic biological research as well as potential therapeutic applications. Here, we review the intein field, first focusing on the biological context and phylogenetic diversity of inteins, followed by a description of intein structure and biochemical function. Finally, we discuss prevalent inteinbased technologies, focusing on their applications in chemical biology, followed by persistent caveats of intein chemistry and approaches to alleviate these shortcomings. The findings summarized herein describe two and a half decades of research, leading from a biochemical curiosity to the development of powerful protein engineering tools.
Introduction
The diversity of proteins found in nature is immediately apparent from the vast array of biochemical functions carried out by these proteins to sustain living organisms. To a first approximation, these functions are dictated by a protein’s primary amino acid sequence, which is transcribed and translated from the gene encoding that protein. This sequence carries all of the necessary information for a newly synthesized protein to fold into a well-defined threedimensional structure, and this structure in turn confers its function.1 In reality, however, many proteins require additional factors to fold into their active conformation,2 while others remain intrinsically disordered as a requirement for their function.3 Furthermore, most proteins are matured, activated, inhibited, translocated, and/or degraded through the chemical modification of their side chains and backbones after protein synthesis, adding yet another layer of complexity to their structure and function.4
In many cases, these post-translational modifications (PTMs) expand the chemical and structural repertoire of the canonical twenty amino acids by the addition of new functional groups to their side chains. These enzymatically applied modifications include (but are not limited to) phosphorylation, acetylation, methylation, lipidation, glycosylation, and hydroxylation, and the dynamic interplay between their addition and removal governs biological signaling. In other cases, the primary sequence of a protein is post-translationally altered by the scission of one or more peptide bonds. This “processing” of a polypeptide chain is often carried out enzymatically by proteases and is most commonly utilized for protein degradation. However, it can also serve to activate an enzyme (e.g. the cleavage of prothrombin to yield thrombin), remove a translocation signal (e.g. signal peptide removal during antibody secretion), or mature a protein into a functional state (e.g. the conversion of proinsulin into active insulin). Remarkably, several classes of proteins can modify their own peptide backbones, and these modules are referred to as auto-processing domains.
The capacity for proteases and auto-processing domains to modify polypeptide sequences has garnered tremendous interest, not only for its biological significance, but also for its practical utility. Indeed, biochemists regularly use proteases to remove affinity purification tags from recombinant proteins5 and to process complex biological mixtures for proteomics experiments.6 Proteases are also commonplace in industrial settings, where they are used in detergents and for food production.7 While auto-processing domains are less prevalent in technological settings, some of these proteins are rapidly emerging as powerful tools for chemical biology.8 These useful auto-processing domains comprise a conserved family of proteins known as inteins.
Protein splicing: a wide-spread post-translational modification
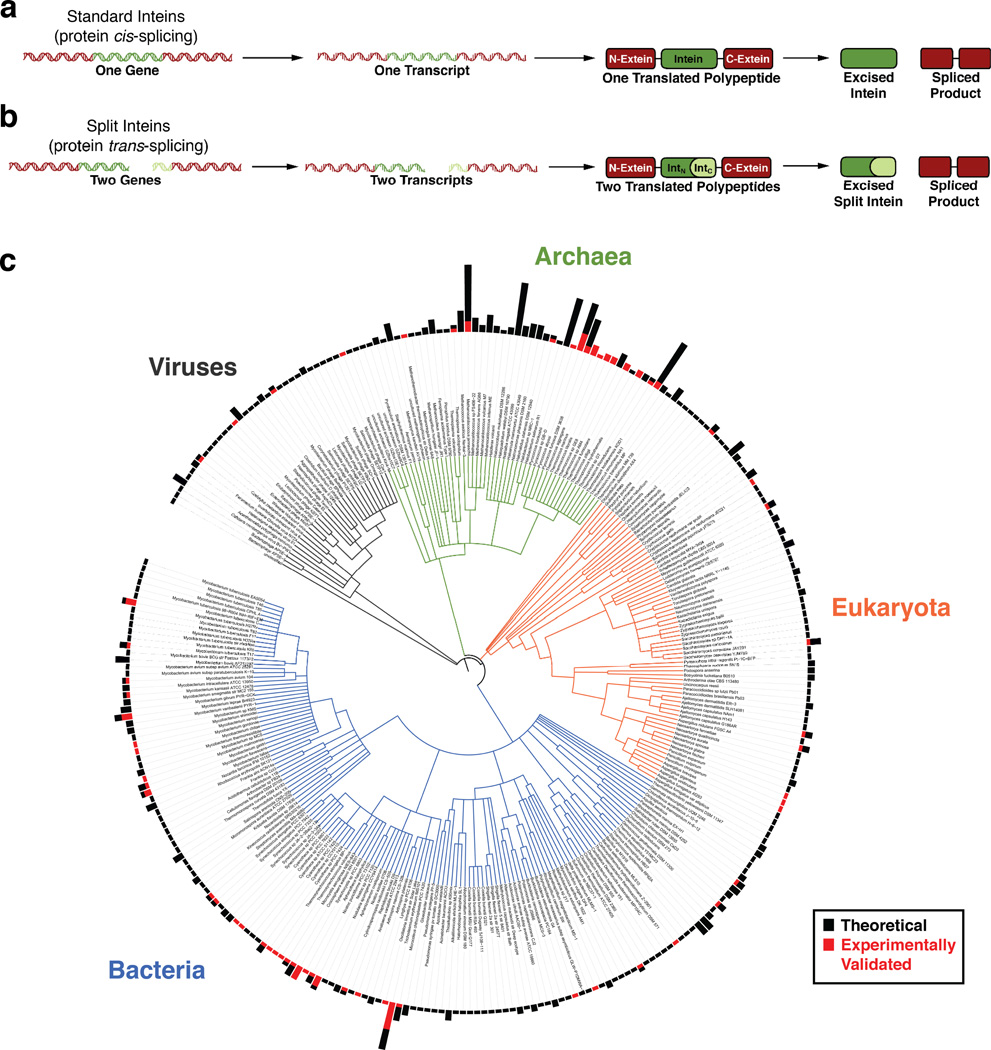
An intein (intervening protein) carries out a unique auto-processing event known as protein splicing in which it excises itself out from a larger precursor polypeptide through the cleavage of two peptide bonds and, in the process, ligates the flanking extein (external protein) sequences through the formation of a new peptide bond. This rearrangement occurs post-translationally (or possibly co-translationally), as intein genes are found embedded in frame within other protein-coding genes (Figure 1a). Furthermore, intein-mediated protein splicing is spontaneous; it requires no external factor or energy source, only the folding of the intein domain.
Figure 1. Protein splicing in nature.
a. Protein cis-splicing by the more prevalent contiguous inteins. b. Protein trans-splicing by the rarer split inteins. IntN refers to the N-intein and IntC refers to the C-intein. c. The phylogenetic distribution of intein-containing organisms. Roughly 300 organisms containing one or more intein are shown in this phylogenetic tree. The bars at the periphery of the tree denote the number of inteins in each organism. The smallest bar indicates one intein, and the largest bar indicates 19 inteins. Black bars indicate inteins identified based on their gene sequence whose splicing capacity has not yet been determined experimentally. Red bars indicate inteins that have been shown experimentally to facilitate protein splicing. Data was extracted from the NEB InBase9, which was last updated in 2010. Several inteins from the recent literature were added to this dataset, but we acknowledge that this tree may not reflect all discovered or characterized inteins. The phylogenetic tree was generated using the Interactive Tree of Life (iTOL) online tool.10 Phylogenetic relationships were automatically inferred by the iTOL software based on the NCBI taxonomic identifiers for each organism.11 Based on this classification system, in some cases different strains of the same organism were combined into one data entry.
The vast phylogeny of inteins
In 1990, the first protein splicing domain was found embedded within the vacuolar proton-translocating ATPase gene in Sacchromyces cerevisiae.12, 13 Since then, taking advantage of several highly conserved sequence motifs in inteins, bioinformatic approaches have identified at least 600 putative intein genes in the genomes of unicellular organisms from all three domains of life (archaea, bacteria, and eukaryota) as well as several viral genomes (Figure 1c).9 Interestingly, this broad distribution of inteins pertains not only to the array of organisms in which they are found, but also the large variety of host genes in which they are embedded. To date, there are at least 70 different intein alleles, distinguished not only by the type of host gene in which the inteins are embedded, but also the integration point within that host gene.9, 14 Furthermore, some proteins have been found containing as many as four inteins embedded at different integration points within their gene, and several organisms have more than one intein (as many as 19 inteins) in their genome (Figure 1c).9
Inteins are typically embedded within essential proteins involved in DNA replication, transcription, and maintenance (e.g. DNA or RNA polymerase subunits, DNA helicases, DNA gyrases, and ribonucleotide reductase) or in other housekeeping genes including essential proteases and metabolic enzymes.14 Within these proteins, the auto-processing domains are commonly inserted at conserved sites in their host that are crucial for host protein function (e.g. ligand binding sites or enzyme active sites).15 Given this, it is largely believed that intein excision is required for host protein function, however no intein has been shown to have a clear regulatory role on its host protein or provide a fitness benefit to the host organism.16, 17 Interestingly, redox-controlled splicing has been shown for some inteins in an exogenous context. It has been proposed that these inteins may inhibit their endogenous host protein’s assembly and function under hyperoxic conditions, however this has not been validated in the native host organism.18 The lack of direct evidence for biologically significant intein function begs the question, “Why do inteins exist?” Thus far, the prevailing view is that inteins are selfish genes with no obvious biological role. Regardless, their mere presence and persistence in microbial genomes is intrinsically fascinating.
Biological mechanisms of intein spread, persistence, and loss
The broad phylogenetic distribution of inteins suggests that these molecules have ancient origins. Nevertheless, for the reasons outlined below,14 it is clear that their prevalence must not only be due to vertical, but also horizontal gene transmission: (1) Inteins are integrated into a wide variety of host proteins. (2) When an intein exists in one microbial host gene, it is not always found in an orthologous gene in a closely related organism. (3) Inteins are completely absent from multicellular organisms (although some multicellular organisms have homologous domains with related biochemical functions, discussed below in the section “Hint domains: one fold, many functions”). (4) Allelic intein genes typically have higher homology and different codon usage than their host genes. These facts suggest that mechanisms exist to propagate inteins from one host gene and host organism to another, and that inteins can also be lost.
Many inteins have, inserted within their auto-processing domain, another functional module called a homing endonuclease domain (HED). HEDs make double-stranded DNA breaks at specific recognition sequences encoded within intein/HED-free alleles of their own host genes. These breakages initiate a recombination process that results in the integration of an intein/HED gene into a previously intein/HED-free version of that gene (Figure 2a).
Figure 2. Intein spreading and splitting through homing endonucleases.
a. Homing endonuclease activity to convert an intein-free allele into an intein-containing allele. b. Proposed mechanism for intein splitting due to aberrant homing endonuclease invasion followed by chromosomal rearrangements.19
This mechanism likely explains both intra- and inter-species intein spread involving the same host gene and insertion site.14 However, given the sequence specificity of HEDs, it is unlikely that this mechanism allows for intein spread to different insertion sites within the same gene or different genes. Currently, it is unclear how this process occurs, however it is noteworthy that non-allelic inteins have extremely low sequence homology when compared to allelic inteins.14 This suggests that intein spread to a variety of host genes was an ancient process and that non-allelic inteins have diverged since this initial event. The recent discovery of inteins in viral genomes may hold the explanation to the early proliferation of inteins.20
The loss of an intein from a host gene may be driven by negative selection, if the intein is detrimental to host fitness. However, intein loss through gene deletion should be challenging, as it requires precise removal of the intein from within an essential gene. Presumably this process is more likely in diploid or polyploid organisms, where an intein-containing copy of a gene is expendable in the presence of an intein-free copy. Furthermore, the capacity for interchromosomal recombination provides another route to intein loss and may explain the dearth of inteins in multicellular organisms.
The emergence of split inteins
A small fraction (less than 5%) of the identified intein genes encode split inteins.9 Unlike the more common contiguous inteins, these are transcribed and translated as two separate polypeptides, the N-intein and C-intein, each fused to one extein. Upon translation, the intein fragments spontaneously and non-covalently assemble into the canonical intein structure to carry out protein splicing in trans (Figure 1b).
Although several lineages of split inteins independently emerged during evolution, as evidenced by their divergent sequences and their insertion in at least five different host proteins,19, 21, 22 the precise mechanism of intein splitting is not clear. Interestingly, the split site of most split inteins is also the homing endonuclease insertion site within many contiguous inteins. In fact, some split intein genes are separated by an out-of-frame, free-standing HED gene, suggesting that aberrant insertion of an HED into an intein gene could fracture that gene (Figure 2b).19 Oddly, the largest known family of split inteins, found within the DnaE genes of at least 20 cyanobacterial species, has unconserved genomic architecture.21 In different cyanobacterial species, the intein fragments are located in dramatically different regions of the bacterial chromosome, and in some cases, the fragments are encoded on opposite strands as well. This suggests that after an initial fracturing of the intein gene in an early cyanobacterium, the resulting locus was unstable and further rearranged as the organism speciated (Figure 2b).
Split inteins present unique evolutionary challenges and opportunities for their host organisms. On one hand, splitting of an intein gene should be effectively irreversible, and the resulting gene fragments, if still transcribed and translated, have to contend with splicing their exteins in trans. These constraints provide a strong selection filter that would either lead to the termination of that lineage or significant optimization of the newly split intein. On the other hand, these split genes could provide a way to regulate the host gene’s activity. Furthermore, it has been postulated that if an organism contains multiple cross-reactive split inteins, they could serve as a platform for protein evolution through domain shuffling.23 While this proposal is intriguing, thus far no organism has been identified that contains more than one split intein in its genome. On the other hand, a recent biochemical and structural study demonstrated that fragments of split inteins can domain-swap with regions of contiguous inteins. 24 The resulting complexes are active and can yield alternative spliced products distinct from those encoded within a single intein-extein gene fusion. While it is currently not clear if this process occurs in any natural context, several organisms do contain a single split intein and one or more contiguous inteins, and domain swapping between split and contiguous inteins may provide these organisms with evolutionary opportunities through the formation of diverse protein products.
The chemical mechanism of protein splicing
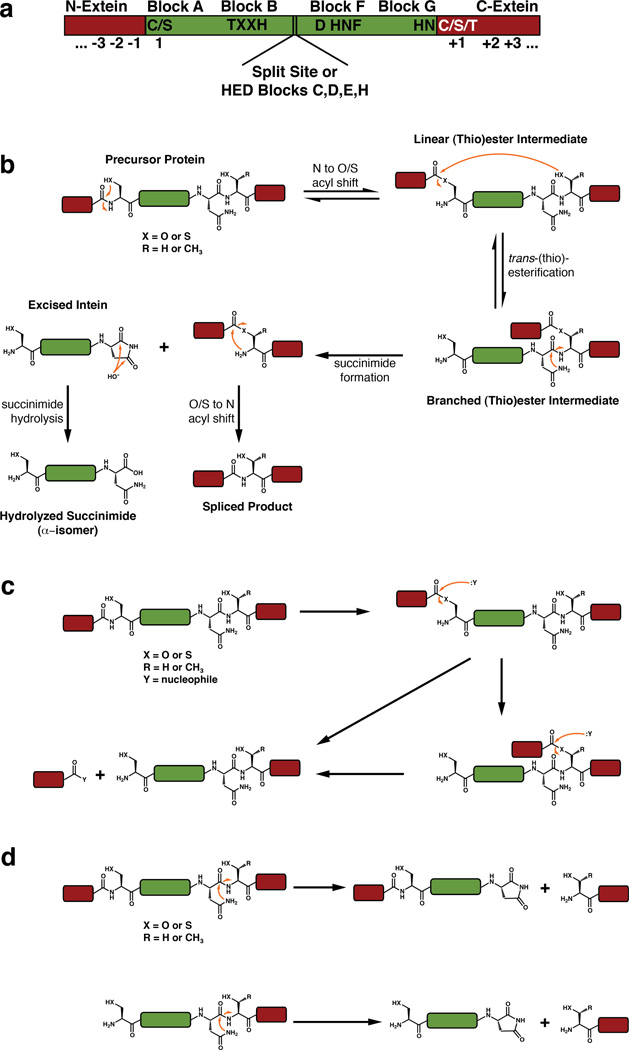
Given that inteins are wide-spread in nature, it is not surprising that non-allelic inteins have low sequence homology (<40%). Despite this fact, inteins are unified by their common biochemical mechanism for protein splicing, which is carried out by several conserved sequence motifs distributed throughout their primary amino acid sequences (Figure 3a).
Figure 3. The chemical mechanism of protein splicing and common side reactions.
a. Conserved sequence motifs that facilitate protein splicing. b. The canonical mechanism of protein splicing. c. N-extein cleavage, also known as N-terminal cleavage, from the linear or branched (thio)ester intermediate. d. C-extein cleavage, also known as C-terminal cleavage, from the precursor protein or the N-extein cleaved protein.
The canonical mechanism and the conserved sequence motifs
The mechanism of protein splicing entails a series of a acyl-transfer reactions that result in the cleavage of two peptide bonds at the intein-extein junctions and the formation of a new peptide bond between the N- and C-exteins (Figure 3b).25 This process is initiated by activation of the peptide bond joining the N-extein and the N-terminus of the intein. Virtually all inteins have a cysteine or serine at their N-terminus (Block A) that attacks the carbonyl carbon of the C-terminal N-extein residue. This N to O/S acyl-shift is facilitated by a conserved threonine and histidine found in Block B (also referred to as the TXXH motif), along with a commonly found aspartate in Block F, and results in the formation of a linear (thio)ester intermediate. Next, this intermediate is subject to trans-(thio)esterification by nucleophilic attack of the first C-extein residue (+1), which is invariably a cysteine, serine, or threonine. The resulting branched (thio)ester intermediate is resolved through a unique transformation: cyclization of the highly conserved C-terminal asparagine of the intein. This process is facilitated by the Block F histidine (found in a highly conserved HNF motif) and the penultimate Block G histidine and may also involve the Block F aspartate. This succinimide formation reaction excises the intein from the reactive complex and leaves behind the exteins attached through a non-peptidic linkage. This structure rapidly rearranges into a stable peptide bond in an intein-independent fashion.26 In addition, the excised intein succinimide will slowly hydrolyze into an α- or β-isomer of the carboxylic acid.
Variations on the canonical mechanism
Some inteins have slightly divergent sequences in the canonical splicing motifs which require alternate mechanisms of splicing. Perhaps the most dramatic of these differences is the lack of a nucleophilic cysteine or serine in Block A. Inteins lacking a Block A nucleophile commonly have an alanine or proline as their N-terminal residue and cannot initiate splicing through the formation of a linear (thio)ester intermediate. Remarkably, these inteins either directly form the typical branched intermediate upon N-terminal activation27 or proceed through a different branched thioester intermediate using an unique cysteine within Block F before forming the canonical branched structure.28 The capacity to bypass the linear (thio)ester intermediate in some inteins is not surprising, as several studies have shown that the N-terminal scissile peptide bond in inteins is destabilized or twisted in the precursor protein.29, 30 Furthermore, the structure of the MjaKlbA intein, which contains an N-terminal alanine, shows that this peptide bond is in a cis conformation,31 which may be unstable and susceptible to nucleophilic attack.
In another surprising variation on the splicing mechanism, a few inteins have been found ending in a glutamine or aspartate, rather than asparagine.32 These inteins could theoretically proceed through the same chemical mechanism, however glutamine cyclization should be less favorable than asparagine cyclization, as it would proceed through a six-membered, rather than five-membered, cyclic intermediate. Cyclization of aspartate would be geometrically similar to asparagine and proceed through the formation of a succinic anhydride rather than a succinimide.
Perhaps the most subtle but intriguing deviations from the canonical splicing motifs involve lack of the Block B or penultimate (Block G) histidines.33, 34 For example, in the Thermococcus kodakaraensis CDC21-1 intein and several of its orthologs, the highly conserved Block B histidine, which is required for the initiating N-to-S acyl shift reaction, is a threonine. Intriguingly, this family of inteins has evolved a lysine residue outside of the canonical splicing motifs that compensates for this divergence to yield a functional intein.34 Similarly, in the cyanobacterial split DnaE inteins, the Block G histidine is either a serine or alanine.21 In most inteins, both the Block F and Block G histidines are crucial for resolution of the branched intermediate, as they work in concert to activate the asparagine nucleophile then to protonate the forming amine.26 Currently, it is not clear how these inteins circumvent the need for two histidines, however other proximal amino acids may serve as surrogate general acids or bases during splicing.
Side reactions during protein splicing
Two common side reactions have been observed during experimental analysis of protein splicing reactions.35 The first, N-extein cleavage (also known as N-terminal cleavage) can occur when an external nucleophile, commonly water or a thiol, attacks the linear or branched (thio)ester intermediates to yield a free N-extein (Figure 3c). The second, C-extein cleavage (also known as C-terminal cleavage) can occur from the precursor protein or the N-extein cleaved protein, when the C-terminal asparagine cyclizes in the absence of the branched intermediate structure, releasing the C-extein (Figure 3d). While both of these reactions can be enhanced by mutation of critical intein residues and have been exploited for various technological purposes,35–38 it is not clear whether these reactions are prevalent in an intein’s native environment. Interestingly, however, intein-related auto-processing domains have been found in nature that exploit these side reactions for biochemical outcomes other than protein splicing.
Hint domains: one fold, many functions
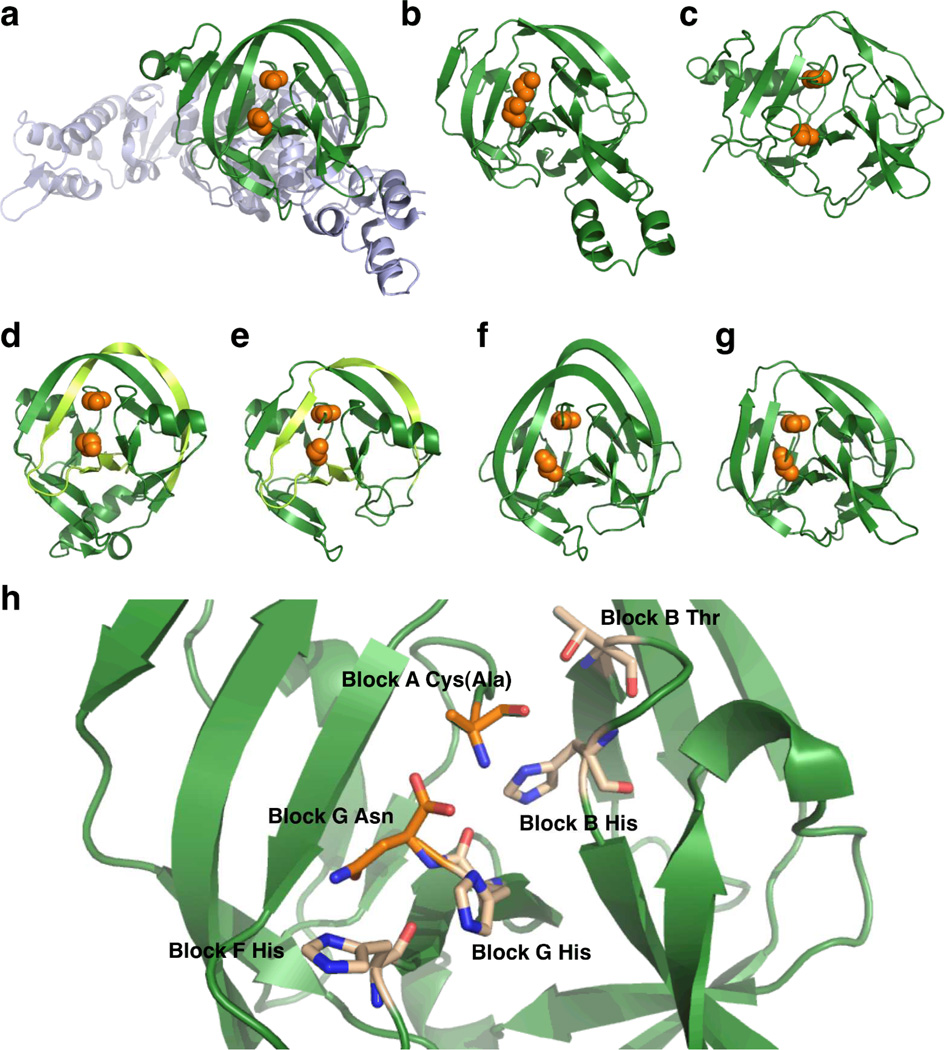
Inteins are actually part of a larger class of proteins known as Hedgehog/intein (Hint) domains. This superfamily is comprised of three members: inteins, bacterial intein-like (BIL) domains, and Hedgehog auto-processing (Hog) domains. While different Hint domains have various insertions, including the large homing endonuclease domains found in some inteins (Figure 4a), all of these proteins have the same core fold comprised primarily of several two-or three-strand β-sheets and loops along with two short α-helices (Figure 4a–g). Interestingly, this conserved horseshoe-like core has a pseudo two-fold symmetry. Given this symmetry, it is believed that the Hint fold likely arose from the gene duplication event of some progenitor protein with unrelated function.39
Figure 4. Comparison of various Hint domain structures.
a. Thermococcus kodakaraensis Pol-2 intein (PDB 2CW7).40 bMycobacterium xenopi GyrA intein (PDB 1AM2).41 c Methanococcus jannaschii KlbA intein (PDB 2JNQ).31 d Synechocystis sp. PCC6803 DnaE split intein (PDB 1ZD7).42 e Nostoc punctiforme DnaE split intein (PDB 2KEQ).43 f Clostridium thermocellum BIL domain 4 (PDB 2LWY).44 gDrosophila melanogaster Hog domain (PDB 1AT0).39 In panels a to g, the Hint domain is shown as green ribbon with the Block A nucleophile and Block G asparagine positions highlighted as orange spheres. In panel a, the homing endonuclease domain is shown in blue. For the split inteins in panels d and e (which are artificially fused in the solved structures), the C-intein region is light green. h. A close up of the MxeGyrA active site, highlighting key residues as sticks. The Block A Cys is mutated to Ala in this structure.
Bacterial intein-like domains
BIL domains can be categorized into two sub-families based on the presence or absence of certain splicing motifs and their resulting biochemical activity.45 A-type BIL domains have all of the conserved splicing motifs, however most lack the obligatory cysteine, serine, or threonine at the +1 residue of the C-extein. As a result, they can sustain N- and C-extein cleavage (Figure 3c,d). While these BILs should not be splicing-competent, some evidence suggests that they can still splice proteins through an alternate mechanism.45, 46 A small subset of BIL domains actually have a serine or threonine at the +1 position, and these BIL domains can indeed carry out protein splicing in addition to N- and C-extein cleavage. B-type BIL domains have an abnormal Block G that is lacking a C-terminal asparagine but contains a conserved cysteine, serine, or threonine two residues upstream of the canonical intein splice junction. Oddly, these BIL domains are capable of both N- and C-extein cleavage (Figure 3c,d), but the latter must proceed through a different mechanism than for inteins and A-type BILs.45
While the biochemical difference between BIL domains and inteins is subtle, their phylogenetic distribution differs dramatically. Unlike inteins, which are embedded within highly conserved sites in essential proteins, BILs are integrated into hyper-variable regions of non-conserved proteins.45 Interestingly, BILs are commonly found attached to secreted proteins, suggesting that they may have a role in protein maturation or translocation. Furthermore, their capacity to activate N-exteins for nucleophilic attack and cleavage without splicing may be utilized for the C-terminal modification of these proteins by any available nucleophile in the cell. Indeed, Nature uses a variation on this mode of post-translational modification in another type of Hint domain, described in the following section.
Hedgehog auto-processing domains
The Hog domain is one of three domains found within the Hedgehog developmental signaling proteins in animals. The N-terminal domain of Hedgehog proteins bear the signaling moiety, the central Hog domain has auto-processing function, and the C-terminal domain binds cholesterol. During the maturation of the Hedgehog signaling proteins, the Hog domain activates the C-terminal amino acid of the signaling region through an N to S acyl shift, analogous to the first step of protein splicing. Following this activation, the cholesterol binding region presents the hydroxyl group of cholesterol as a nucleophile to cleave the signaling domain and tag it with the sterol.47, 48 The resulting product is also palmitoylated near its N-terminus through a more traditional enzymatic mechanism and secreted from cells to act as a morphogen during developmental patterning.
Hog domains are the only known Hint domains in animals, where they have a clear function during development. The lack of inteins or BIL domains in these higher organisms suggests that a single Hint domain existed in the progenetor organism before the emergence of metazoans, and this ancient Hint domain ultimately became the modern Hog protein involved in development. Consistent with this notion, a secreted protein of unknown function discovered in the genome of the choanoflagellate Monosiga ovata was found to be auto-processed by a Hog domain.49 As choanoflagellates and metazoans are believed to have diverged from a common ancestor close to the emergence of multicellularity,50 this protein may provide insights into the evolution of biological function in the Hint superfamily.
The structure of split inteins
Several high-resolution structures of two orthologous cyanobacterial split inteins have been solved. Structures of the DnaE intein from Synechocystis sp. PCC6803 (SspDnaE) were determined using X-ray crystallography (Figure 4d),18, 30, 42 and structures of the DnaE intein from Nostoc punctiforme (NpuDnaE) were determined both by nuclear magnetic resonance (NMR) spectroscopy in solution (Figure 4e)43 and by X-ray crystallography.24 In all cases, the N- and C-intein fragments were recombinantly fused to generate contiguous inteins, and the structures were solved in these fused forms. Regardless, these studies verified that split inteins can adopt the same horseshoe-like fold seen for all other Hint domains. Interestingly, a closer look at the N- and C-intein regions clearly indicates that these fragments are heavily entwined, which would preclude assembly through pre-folded monomers. Furthermore, fluorescence measurements indicate that the SspDnaE intein fragments associate extremely rapidly (kon of ∼107 M−1s−1) and tightly (KD of ∼30 nM).51 The NpuDnaE intein fragments also bind tightly (KD of ∼2 nM)52, albeit with slower association kinetics (kon of 105–106 M−1s−1).53 Additionally, atomic force microscopy measurements on several other split DnaE inteins demonstrate that tight binding is highly conserved in this family.54
This highly efficient binding and the entangled topology seen for artificially fused split inteins raises two important questions: (1) Do split inteins retain this topology when they are actually split? (2) If so, how do split intein fragments efficiently arrive at this structure in trans? Even in the absence of high-resolution structures of truly split inteins, the first question has effectively been resolved. Comparisons of recent NMR data on a contiguous form of the NpuDnaE intein,43 a natively split form,53 and a fragment-swapped homodimer24 indicate that the same fold is clearly adopted when the N- and C-inteins interact in trans.
The latter question of association mechanism has largely been addressed as well through bioinformatic and biophysical studies. Several groups have noted the dramatic charge segregation between N- and C-inteins in the DnaE intein family,51, 55 and our bioinformatic analyses indicate that this charge segregation is generally absent in contiguous inteins.52 Furthermore, mutation of specific charged residues at the fragment interface in the NpuDnaE intein significantly perturbs fragment association, validating the importance of electrostatic interactions for binding.52 Biophysical analyses of the isolated SspDnaE intein fragments suggest that these fragments are at least partly disordered in isolation,56 so their binding must proceed through a disorder-to-order transition. Consistent, with this observation, we recently characterized the structures of the isolated NpuDnaE N- and C-intein fragments using NMR spectroscopy and protein engineering. We found that the C-intein is completely disordered, while the N-intein has a bipartite structure comprised of a well-folded region and an equal-length disordered segment. Using isolated segments of the N-intein, we showed that the C-intein preferentially engages the disordered region of the N-intein through electrostatic interactions to form a compact, native-like binding/folding intermediate. This intermediate then collapses onto the folded portion of the N-intein to yield the native trans-splicing complex.52
Applications of protein splicing
In their native context, inteins facilitate both the cleavage and formation of peptide bonds. Early studies on inteins demonstrated that they could also carry out protein splicing in exogenous contexts, often between polypeptides unrelated to the endogenous host protein.57–59 In fact, the only absolute sequence requirement for intein-mediated splicing outside of the intein itself is the presence of a cysteine, serine, or threonine at the first residue of the C-extein (the +1 position). As such, these molecules should be ideal tools for protein chemistry and engineering. Indeed, intein chemistry is exploited in a variety of powerful technological applications centered around the cleavage and/or formation of peptide bonds.8
Tagless protein purification
The side reactions of protein splicing, N- and C-extein cleavage, can both be enhanced by the introduction of specific point mutations in the conserved splicing motifs. For example, mutation of the Block A nucleophile from cysteine/serine to alanine precludes the formation of the linear and branched (thio)ester intermediates. In this context, many inteins have a basal level of asparagine cyclization and thus C-extein cleavage which can be inhibited at slightly acidic pH.38, 60 Alternatively, mutation of the Block G asparagine to alanine precludes the irreversible branch resolution step, resulting in a trapped equilibrium between the precursor amide and the two (thio)ester intermediates. The (thio)esters can be treated with base to induce N-extein cleavage by hydrolysis.36, 60 These mutant inteins can be used to obtain recombinant proteins without affinity purification tags for use in biochemical studies. Specifically, a protein of interest is fused to the N- or C-terminus of an intein bearing the appropriate mutations, and an affinity purification tag is fused to the other terminus of the intein. After affinity enrichment on a solid support, the pH of the system can be raised to induce N- or C-terminal cleavage, resulting in the release of an untagged protein.60
In vitro protein semi-synthesis
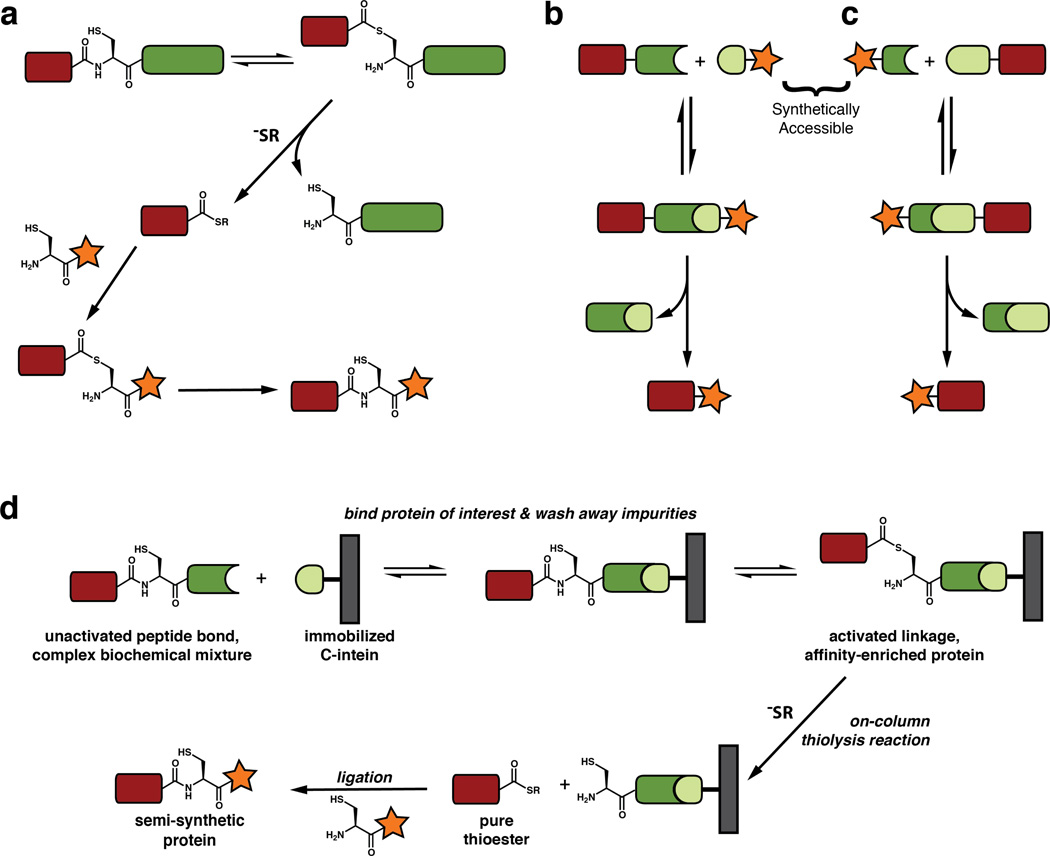
N-extein cleavage from a mutant intein can be induced not only by hydrolysis but also through attack by nucleophiles other than water.36 Cleavage using small-molecule alkyl or aryl thiols will result in a protein of interest bearing an α-thioester, rather than a free carboxylic acid (Figure 5a). In the well-known Native Chemical Ligation (NCL) reaction, a peptide C-terminal thioester can be condensed with a 1,2-aminothiol moiety (such as a polypeptide containing an N-terminal cysteine) to form a native peptide bond.61 While NCL allows for the total chemical synthesis of small proteins through the ligation of synthetic peptides, the use of inteins to generate protein thioesters recombinantly, allows for this reaction to be applied to significantly larger macromolecules (Figure 5a).37 This semi-synthetic iteration of NCL, referred to as Expressed Protein Ligation (EPL), has been used to site-specifically incorporate a wide array of chemical moieties into proteins that are typically inaccessible through purely recombinant methods. These include post-translational modifications, unnatural amino acids, backbone modifications, photochemical cross-linkers, biophysical probes, and imaging probes.8 Finally, it is noteworthy that while intein-mediated EPL traditionally involves the production of the thioester fragment recombinantly, this fragment can also be generated synthetically, and the N-terminal cysteine-containing protein can be made recombinantly, exposing the cysteine either using proteolysis or an intein tag capable of C-extein cleavage.62
Figure 5. In vitro protein semi-synthesis.
a. Expressed Protein Ligation (EPL). A C-terminal thioester is generated by cleavage from the intein followed by condensation with an N-terminal cysteine-containing peptide or protein. b. Semi-synthesis by protein trans-splicing (PTS) with a synthetically accessible C-intein. c. Semi-synthesis by protein trans-splicing (PTS) with a synthetically accessible N-intein. d. Affinity capture and protein modification using streamlined EPL.
Protein semi-synthesis by EPL typically employs contiguous inteins that are mutated to prevent protein splicing and generate a reactive handle for fragment condensation. Split inteins offer an alternative approach to protein semi-synthesis, as the complete protein trans-splicing (PTS) reaction is effectively a fragment condensation reaction mediated by the complementary N- and C-intein fragments (Figure 5b,c). The rates of standard chemical ligation reactions, including EPL, are strongly concentration-dependent, as they rely on the random collision of peptide fragments.63 As such, EPL reactions are often carried out under denaturing conditions, where large proteins or insoluble protein fragments can be solubilized at high concentrations. By contrast, PTS by naturally occurring split inteins is facilitated by a tight protein–protein interaction and thus shows low concentration dependence.51 This makes split inteins attractive tools for protein semi-synthesis of challenging substrates where sample concentration, solubility, and the ability to refold the ligation product are limiting factors.64
Importantly, the smaller C-intein fragment of naturally split inteins is typically between 30 and 40 amino acids, making it synthetically accessible through solid-phase peptide synthesis. However the addition of any “cargo” for protein labeling puts this fragment nearly out of synthetic reach. To address this issue, several groups have engineered artificially split inteins with non-canonical split sites as close as 6 residues from the intein C-terminus (Figure 5b)43, 65, 66 or 11 residues from the N-terminus (Figure 5c).67 While these constructs often have lower splicing efficiencies and binding affinities than naturally split inteins, at least one fragment is synthetically accessible, providing a means to modify the N- or C-terminus of a protein using PTS.68
Recently, we exploited both the binding properties and reactivity of naturally split inteins to expedite protein semi-synthesis using Expressed Protein Ligation,69 taking advantage of the highly efficient N-terminal peptide bond activation observed for the split DnaE inteins70 and the tight binding of DnaE fragments (Figure 5d).52 In this streamlined iteration of EPL, a protein of interest is expressed as a C-terminal fusion to a DnaE N-intein, while a C-intein is covalently immobilized on a solid support. A complex biological mixture containing the fusion is passed over the C-intein column to capture the desired protein. After washing away impurities, the protein of interest can be cleaved off of the immobilized split intein using a small molecule thiol to yield a highly pure product bearing a C-terminal α-thioester. Importantly, this product can be directly condensed with a 1,2-aminothiol (e.g. an N-terminal cysteine-containing peptide) to yield a semi-synthetic protein. This updated EPL protocol has several advantages over traditional EPL: (1) the use of highly active DnaE inteins can yield thioesters more rapidly than other commonly used inteins, (2) since the intein is split, there is no premature cleavage from the N-intein prior to binding the immobilized C-intein (e.g. during protein expression), and (3) the high affinity of the split intein fragments allows for protein isolation and modification directly a heterogeneous source (e.g. a cell lysate). Notably, this technique is applicable to challenging substrates including poorly behaved protein fragments and therapeutically relevant monoclonal antibodies.
Segmental isotopic labeling
Both EPL and PTS are useful intein-based tools for in vitro protein semi-synthesis. One of the most important applications of these techniques is the segmental isotopic labeling of proteins for NMR spectroscopy studies. NMR is commonly used to characterize the structure and dynamics of proteins, however this approach requires stable enrichment of NMR-active 15N, 13C, and/or 2H nuclei in the protein of interest, as the natural abundance of these heavy isotopes is too low for practical utility. Standardized techniques now exist to incorporate these isotopes uniformly into the backbone and side chains of recombinant proteins by growing bacterial cells in an isotopically enriched medium.71 However, for large proteins (>150 amino acids) and proteins with highly degenerate sequences, significant peak overlap in uniformly labeled samples makes unambiguous interpretation of the spectra challenging.
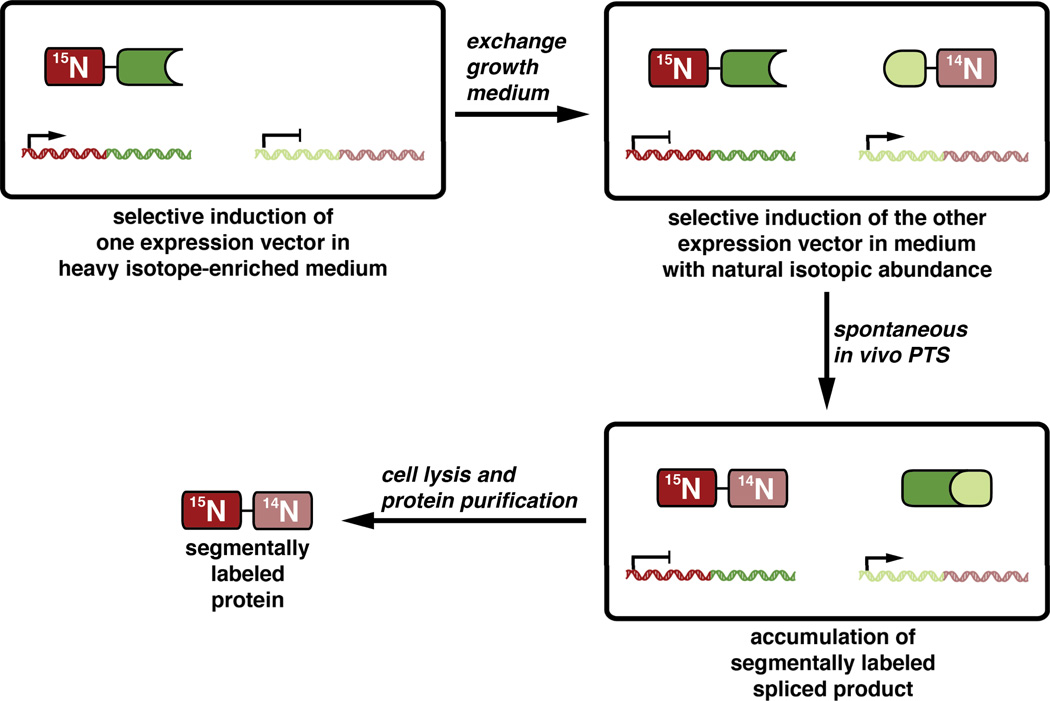
Using either EPL72 or PTS,73 protein fragments with different isotope labeling schemes can be ligated to generate segmentally labeled full-length protein samples with dramatically simplified NMR spectra. Indeed, this approach has been used to aid in the analysis of a wide variety of interesting proteins.74, 75 Two advantages of using PTS with naturally split inteins in this application are that segmentally labeled proteins can be assembled in vitro under non-denaturing conditions or in vivo through the sequential expression of each fragment in the same cell culture with an intermediate exchange of the medium to include or exclude stable isotope sources (Figure 6).76
Figure 6. Segmental isotopic labeling using split inteinsin vivo.
Protein and peptide cyclization
Both EPL and PTS have also been used to generate cyclic polypeptides. Using EPL, a protein can be cyclized when its N-terminus contains an exposed cysteine and its C-terminus is activated through an intein tag to yield an α-thioester. The activated C-terminus can then be directly attacked by the N-terminal cysteine to yield a cyclic thioester, which will rearrange into the more stable peptide bond (Figure 7a).77 The resulting cyclic proteins should have increased thermodynamic and in vivo stability, which in turn can improve their biological function, making this an intriguing approach for enhancing the efficacy of protein-based therapeutics.
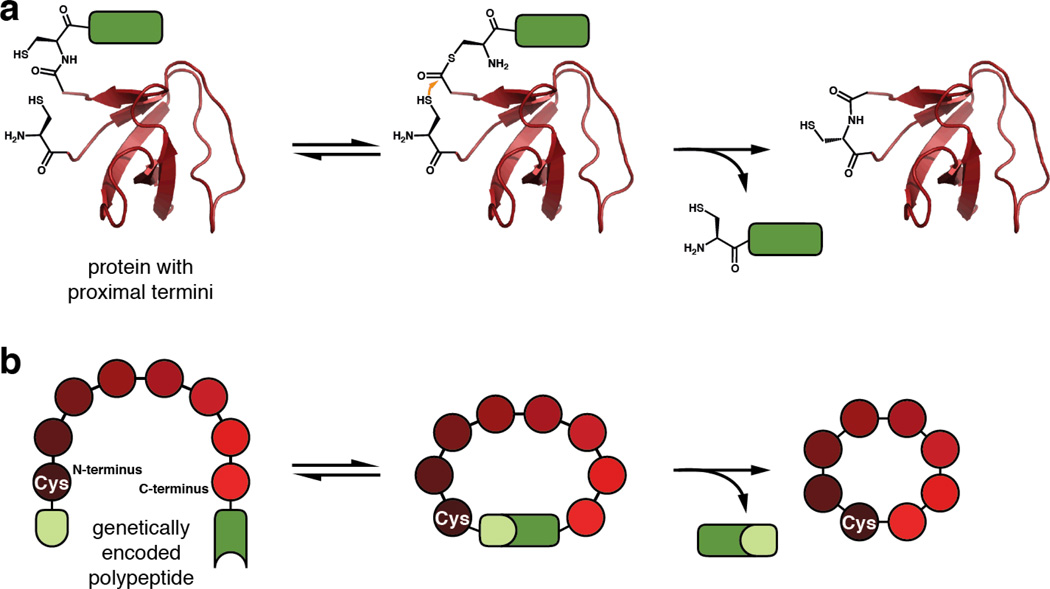
Figure 7. Protein and peptide cyclization.
a. Cyclization of a protein using EPL. The rendering is based on the N-terminal SH3 domain of c-Crk-II (PDB 1M30), which has been head-to-tail cyclized using this method.77 b. Split Intein-mediated Circular Ligation Of Peptides and ProteinS (SICLOPPS).
The efficiency of protein cyclization by EPL is largely dependent on the proximity of the N- and C-termini in the folded state.78 As an alternative, split inteins can be used to force the termini together through the association of the intein fragments. To this end, a technique known as Split Intein-mediated Circular Ligation Of Peptides and ProteinS (SICLOPPS) was developed.79 By inverting the order of intein fragments around a polypeptide of interest, a target sequence can be head-to-tail cyclized (Figure 7b). This reaction results in the formation of a native peptide bond upon excision of the N- and C-inteins, leaving behind a single cysteine residue. The power of this technology is two-fold. First, like EPL, it allows for the production of cyclic proteins with enhanced biophysical and biological properties conferred by their augmented stability.77 Perhaps more significantly, it provides a method to generate large libraries of genetically encoded cyclic peptides for rapid screening. Using SICLOPPS, researchers have discovered methyltransferase inhibitors,80 protease inhibitors,81 modulators of protein–protein interactions,82 and molecules that reduce the cellular pathology of Parkinson’s disease.83
Conditional protein splicing
Conditional protein splicing (CPS), the activation or inhibition of protein splicing by an extrinsic modulator, is perhaps the most intriguing application of inteins. The basic premise of CPS is that inteins control the primary sequence, and thus function, of the proteins they are splicing, so controlling intein activity would provide a way to “turn on” any protein at will, even in vivo. Current CPS systems come in three flavors: (1) Contiguous inteins have been fused to ligand binding domains that can allosterically modulate protein splicing in response to a small molecule (Figure 8a).84, 85 (2) Both contiguous and naturally split inteins have been chemically caged at or near active-site residues to control splicing or cleavage in response to light or proteolysis (Figure 8b).86–88 (3) Artificially split inteins, which cannot spontaneously associate, have been fused to heterodimerization domains to reconstitute their splicing activity in response to small molecules or light (Figure 8c).89–91
Figure 8. Conditional protein splicing.
a. Intein activation through conformational change induced by ligand binding to a fused ligand binding domain. b. Intein activation through deprotection of a photo-caged active site residue. c. Activation of an artificially split intein through chemically induced dimerization.
The post-translational activation of protein function in response to these exogenous factors should be faster than traditional molecular biology techniques involving inducible promoters, tunable in a dose-dependent manner, and portable, as inteins can splice in a wide variety of protein contexts. For example, a photochemically caged intein has been used to generate an allosteric activator of prothrombin in human blood plasma,88 and an intein controlled by chemically induced dimerization has been used to activate firefly luciferase in live fruit-flies.92 Given these facts, the CPS systems described above are promising tools for cell biology and for the development of “smart” protein therapeutics that are only activated at the appropriate site of action.
In vivo protein semi-synthesis
The most significant advantage of PTS-based protein semi-synthesis over EPL with contiguous inteins is that it can be readily applied in vivo. In purified systems, reaction specificity is governed simply by the functional groups present in the molecules of interest, but living systems are heterogeneous and chemically complex, making orthogonal chemistry more challenging.93 Split inteins overcome this problem by acting as ligation auxiliaries that engender virtually absolute specificity to the ligation reaction of interest. Indeed PTS has been used in vivo to site-specifically label proteins with synthetic probes.94, 95 In these studies, a fluorophore was attached to a synthetically accessible DnaE C-intein, and, using a protein transduction system, the construct was readily delivered into cells expressing the DnaE N-intein fused to a target protein of interest. The intein fragments readily associated and spliced, thereby site-specifically labeling proteins in living cells.
Protein trans-splicing has also been utilized for other cell-based applications. In one study, researchers sought to address the size-limitation of adeno-associated viral vectors (ADVVs) used to deliver therapeutic genes. To overcome this caveat, they split the genetic cargo into two fragments fused to SspDnaE intein genes, packaged within two different ADVVs, and demonstrated that the desired therapeutic gene product could be assembled in vivo by PTS.96 In another report, PTS was used to enhance the tissue specificity of Cre recombinase, an enzyme commonly used for genetic engineering of eukaryotic cells and organisms. Here, the authors engineered DNA encoding two split intein-fused inactive fragments of Cre recombinase under the control of different enhancer elements, and they generated transgenic mice incorporating this DNA into their genome. Then, they demonstrated that Cre recombinase was only assembled and active in cells where both enhancers were utilized to activate gene transcription. Using this strategy, they were able to genetically modify specific cell types at the intersectional domains of different enhancer pairs in transgenic mice.97 These examples demonstrate the broad utility of split inteins in complex biological systems.
Caveats of protein splicing
While numerous intein-based technologies have been developed and widely-used, the full scope of their application is limited by two common properties of most inteins: (1) slow splicing and cleavage reactions and (2) dependence on local extein sequence composition.
Reaction kinetics and yield
Most commonly used inteins carry out splicing reactions slowly, on the order of several minutes to hours (Table 1). For example, the contiguous MxeGyrA intein, most commonly used to generate protein thioesters for EPL, carries out the overall splicing reaction with a half-life of ten hours at 25 °C.26 The first-discovered split intein, SspDnaE intein, has a half-life of 75 minutes at 30 °C.70 Furthermore, at 37 °C, the Ssp intein has an increased rate of N-extein cleavage, lowering the overall yield of the splicing reaction.70 Recently, however, two groups demonstrated that the related NpuDnaE split intein could carry out protein trans-splicing with extremely high yields in vivo98 and a half-life of 63 seconds in vitro,99 both at 37 °C. This anomalous splicing efficiency prompted the intriguing questions: “Is the Npu intein an outlier?” and “How can this intein splice so rapidly?”
Table 1.
In vitro splicing kinetics and yields for several inteins.a
| Intein Name |
Type | Temperature (°C) |
ksplice (s−1) | t1/2b | Splicing Yield | Reference |
|---|---|---|---|---|---|---|
| MxeGyrA | contiguous | 25 | 1.9 × 10−5 | 10 h | >90% c | 26 |
| PabPolIII | contiguous | 70 | 1.6 × 10−5 | 12 h | 74% d | 101 |
| MjaKlbA | contiguous | 42 | 2.2 × 10−3 | 5 m | 64% d | 102 |
| SspDnaB | artificially split | 25 | 9.9 × 10−4 | 12 m | 32–56% e | 103 |
| SceVMA | artificially split | 25 | 1.2 × 10−3 | 10 m | 67–73% e | 103 |
| SspDnaE | naturally split | 37 | 1.5 × 10−4 | 76 m | <50% c | 70 |
| NpuDnaE | naturally split | 37 | 3.7 × 10−2 | 19 s | >90% f | 70 |
| AvaDnaE | naturally split | 37 | 3.1 × 10−2 | 23 s | >90% f | 70 |
| CraDnaE | naturally split | 37 | 1.2 × 10−2 | 58 s | >90% f | 70 |
| CspDnaE | naturally split | 37 | 1.8 × 10−2 | 39 s | >90% f | 70 |
| CwaDnaE | naturally split | 37 | 5.0 × 10−3 | 140 s | 80–90% f | 70 |
| MchtDNaE | naturally split | 37 | 2.4 × 10−2 | 29 s | >90% f | 70 |
| OliDnaE | naturally split | 37 | 1.6 × 10−2 | 44 s | >90% f | 70 |
| TerDnaE | naturally split | 37 | 8.5 × 10−3 | 82 s | >90% f | 70 |
| gp41-1 | naturally split | 45 | 1.8 × 10−1 | 4 s | 85–95% d | 100 |
| gp41-8 | naturally split | 37 | 4.5 × 10−2 | 15 s | 85–95% d | 100 |
| NrdJ-1 | naturally split | 37 | 9.8 × 10−2 | 7 s | 85–95% d | 100 |
| IMPDH-1 | naturally split | 37 | 8.7 × 10−2 | 8 s | 90–95% d | 100 |
The entries in this table represent one reported set of rates and yields for each intein, typically under conditions of optimal pH, temperature, and extein context. Thus, they roughly reflect the maximum potential of each intein. We note that several of these parameters have also been measured for these inteins under sub-optimal conditions and are reported in the primary literature.
Calculated from reported first-order rate constants
Estimated based on visual inspection of raw data
Reported yield
Estimated from reported error bars at reaction endpoint
Based on consumption of the limiting fragment (with no side products observed)
To address this question, we compared the splicing activities of 18 split DnaE inteins, including Ssp and Npu, under identical conditions in E. coli. Interestingly, we found that more than half of these had splicing efficiencies on par with Npu, and these highly active inteins carried out splicing in vitro in tens of seconds to a few minutes, like Npu (Table 1).70 Additionally, our in vivo dataset allowed us to identify sequence elements that differentiate fast and slow inteins, providing the first insight into the molecular determinants for efficient splicing. In a related study, another group characterized several non-DnaE split inteins identified through metagenomic sequencing and found four new inteins with splicing kinetics similar to or faster than the DnaE inteins (Table 1).100 Collectively, these discoveries of ultrafast DnaE and non-DnaE inteins suggest that rapid protein splicing is far more prevalent than previously imagined.
Extein dependence
Even the NpuDnaE intein, with its remarkable splicing kinetics, suffers from another pervasive functional caveat of all inteins: reaction kinetics and overall yield are highly dependent on the identity of extein residues surrounding the splice junctions (Figure 3, positions −3, −2, −1, +2, and +3). Typically, inteins have the fastest reaction rates and least side reactions when embedded between the local extein residues similar to those found in their endogenous host protein. Deviation at the local N-extein residues (especially the −1 position) can have a profound effect on the initial N to O/S acyl shift reaction,60 which has practical implications for thioester formation during EPL. Deviation at the local C-extein residues (especially the +2 C-extein position) has been shown to dramatically reduce the overall splicing rate and yield for all of the split DnaE inteins, including Npu.70, 98, 104–107 This strong extein dependence often necessitates that native extein residues be added to a target protein during protein semi-synthesis and segmental isotope labeling when employing protein trans-splicing.108
Improving intein-based technologies
While the aforementioned caveats apparently call into question the broad utility of inteins for protein chemistry and engineering, the steady increase in intein technology-related papers published since the discovery of protein splicing argues that these shortcomings have not inhibited practical intein use. Furthermore, substantial efforts have been made to overcome the deficiencies of currently used inteins. For example, several groups have applied directed evolution techniques to improve splicing and cleavage rates,109 pH sensitivity,38 tolerance to non-native extein residues,105, 110 temperature dependence,105 and even conditionality in response to a small molecule.85
In addition, the rigorous characterization of intein structure and reactivity has opened avenues for the rational design of more useful inteins. Indeed, elucidation of the protein splicing mechanism guided researchers towards the first EPL platforms,36, 37 which have had a profound impact on protein chemistry. Furthermore, an understanding of intein secondary structure topology and backbone dynamics has aided in the identification of non-canonical split sites to make synthetically accessible split intein fragments.43, 65–67 Structural studies have also aided in addressing the extein-dependence problem. For example, recent NMR and crystal structures of the Pyrococcus horikoshii RadA intein revealed interactions that inhibit splicing in the presence of certain amino acids at the −1 N-extein position.111 Based on this structural data, specific mutations were introduced within the intein to alleviate this inhibition, thereby rationally designing a more promiscuous PhoRadA intein. Similarly, we recently carried out a detailed kinetic and structural analysis of C-extein dependence in the NpuDnaE split intein, which demonstrated that this intein requires a bulky amino acid at the +2 position to constrain active site motions.107 In the absence of a bulky +2 amino acid, reaction kinetics are roughly 100-fold slower, and active site residues are highly dynamic and poorly poised for splicing. Interestingly, we found that this excessive flexibility could be reduced by mutation of a proximal active site loop, which in turn improved splicing kinetics with the unfavorable extein. These successes strongly indicate that basic intein research can fuel the development and improvement of useful intein based technologies.
Conclusions and Outlook
Inteins are a curious class of auto-processing domains that are adept at breaking and making peptide bonds. Their ancient origins and pervasiveness in nature suggest that these proteins once had an important biological function. While only remnants of this natural function are evident now in intein-related proteins, the utility of inteins in a technological setting is undeniable. These proteins have been used in countless ways as protein engineering tools for both basic and applied research. Naturally split inteins are particularly intriguing, both from a basic biochemical/biophysical standpoint, as well as for their applications. Their capacity to carry out protein spicing in trans can provide additional benefits when applied to each of the techniques discussed above. Furthermore, the discovery of several fast trans-splicing inteins, coupled with strategies to make inteins splice in a traceless manner, should vastly improve intein-based technologies and make them more applicable to biological systems.
The characterization of these new, ultrafast inteins also brings to light a major deficiency in the intein field: there are several hundred known inteins, and while this number is rapidly increasing as new microbial genomes are sequenced, only a small fraction (less than 15%) have been experimentally characterized in any way (Figure 1c). Future efforts should focus on rigorously characterizing inteins as they are discovered with the goal of finding more robust tools for chemical biology.
Intein-based technologies have largely been bolstered by mechanistic investigations into their structure and activity. In the future, these basic efforts could lead to the design of a truly universal intein: one that is high-yielding with rapid splicing or cleavage kinetics in the context of any extein sequence, can readily splice in trans, has a synthetically accessible fragment, and can even be controlled by an exogenous stimulus. Realistically, such a design may never be attained. However, in striving for such a lofty goal, we will presumably continue to learn more about the intricacies of protein splicing and possibly discover new inteins with interesting biochemical and perhaps biological functions.
Acknowledgements
The authors thank the members of the Muir laboratory for many valuable discussions. Some of the work discussed in this perspective was carried out in the authors’ laboratory and was supported by the U.S. National Institutes of Health (NIH grant GM086868).
References
- 1.Anfinsen CB. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Hartl FU, Hayer-Hartl M. Nat. Struct. Mol. Biol. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 3.Uversky VN, Gillespie JR, Fink AL. Protein Struct. Funct. Genet. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 4.Walsh CT, Garneau-Tsodikova S, Gatto GJ. Angew. Chem. Int. Ed. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 5.Waugh DS. Protein Expr. Purif. 2011;80:283–293. doi: 10.1016/j.pep.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlosser A, Vanselow JT, Kramer A. Anal. Chem. 2005;77:5243–5250. doi: 10.1021/ac050232m. [DOI] [PubMed] [Google Scholar]
- 7.Rao MB, Tanksale AM, Ghatge MS, Deshpande VV. Microbiol. Mol. Biol. Rev. 1998;62:597–635. doi: 10.1128/mmbr.62.3.597-635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vila-Perelló M, Muir TW. Cell. 2010;143:191–200. doi: 10.1016/j.cell.2010.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perler FB. Nucleic Acids Res. 2002;30:383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Letunic I, Bork P. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 11.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Mizrachi I, Ostell J, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Yaschenko E, Ye J. Nucleic Acids Res. 2009;37:D5–15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirata R, Ohsumk Y, Nakano A, Kawasaki H, Suzuki K, Anraku Y. J. Biol. Chem. 1990;265:6726–6733. [PubMed] [Google Scholar]
- 13.Kane PM, Yamashiro CT, Wolczyk DF, Neff N, Goebl M, Stevens TH. Science. 1990;250:651–657. doi: 10.1126/science.2146742. [DOI] [PubMed] [Google Scholar]
- 14.Pietrokovski S. Trends Genet. 2001;17:465–472. doi: 10.1016/s0168-9525(01)02365-4. [DOI] [PubMed] [Google Scholar]
- 15.Dalgaard JZ, Moser MJ, Hughey R, Mian IS. J. Comput. Biol. 1997;4:193–214. doi: 10.1089/cmb.1997.4.193. [DOI] [PubMed] [Google Scholar]
- 16.Frischkorn K, Sander P, Scholz M, Teschner K, Prammananan T, Böttger EC. Mol. Microbiol. 1998;29:1203–1214. doi: 10.1046/j.1365-2958.1998.01003.x. [DOI] [PubMed] [Google Scholar]
- 17.Papavinasasundaram KG, Colston MJ, Davis EO. Mol. Microbiol. 1998;30:525–534. doi: 10.1046/j.1365-2958.1998.01083.x. [DOI] [PubMed] [Google Scholar]
- 18.Callahan BP, Topilina NI, Stanger MJ, Van Roey P, Belfort M. Nat. Struct. Mol. Biol. 2011 doi: 10.1038/nsmb.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dassa B, London N, Stoddard BL, Schueler-Furman O, Pietrokovski S. Nucleic Acids Res. 2009;37:2560–2573. doi: 10.1093/nar/gkp095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogata H, Raoult D, Claverie J-M. Virol. J. 2005;2:8. doi: 10.1186/1743-422X-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caspi J, Amitai G, Belenkiy O, Pietrokovski S. Mol. Microbiol. 2003;50:1569–1577. doi: 10.1046/j.1365-2958.2003.03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi JJ, Nam KH, Min B, Kim S-J, Söll D, Kwon S-T. J. Mol. Biol. 2006;356:1093–1106. doi: 10.1016/j.jmb.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 23.Perler FB. Trends Biochem. Sci. 1999;24:209–211. doi: 10.1016/s0968-0004(99)01403-6. [DOI] [PubMed] [Google Scholar]
- 24.Aranko AS, Oeemig JS, Kajander T, Iwaï H. Nat. Chem. Biol. 2013;9:616–622. doi: 10.1038/nchembio.1320. [DOI] [PubMed] [Google Scholar]
- 25.Volkmann G, Mootz HD. Cell. Mol. Life Sci. 2012 doi: 10.1007/s00018-012-1120-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frutos S, Goger M, Giovani B, Cowburn D, Muir TW. Nat. Chem. Biol. 2010;6:527. doi: 10.1038/nchembio.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Southworth MW, Benner J, Perler FB. EMBO J. 2000;19:5019–5026. doi: 10.1093/emboj/19.18.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tori K, Dassa B, Johnson MA, Southworth MW, Brace LE, Ishino Y, Pietrokovski S, Perler FB. J. Biol. Chem. 2010;285:2515–2526. doi: 10.1074/jbc.M109.069567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romanelli A, Shekhtman A, Cowburn D, Muir TW. Proc. Natl. Acad. Sci. USA. 2004;101:6397–6402. doi: 10.1073/pnas.0306616101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dearden AK, Callahan B, Roey PV, Li Z, Kumar U, Belfort M, Nayak SK. Protein Sci. 2013;22:557–563. doi: 10.1002/pro.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson MA, Southworth MW, Herrmann T, Brace L, Perler FB, Wüthrich K. Protein Sci. 2007;16:1316–1328. doi: 10.1110/ps.072816707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amitai G, Dassa B, Pietrokovski S. J. Biol. Chem. 2004;279:3121. doi: 10.1074/jbc.M311343200. [DOI] [PubMed] [Google Scholar]
- 33.Chen L, Benner J, Perler FB. J. Biol. Chem. 2000;275:20431–20435. doi: 10.1074/jbc.M000178200. [DOI] [PubMed] [Google Scholar]
- 34.Tori K, Cheriyan M, Pedamallu CS, Contreras MA, Perler FB. Biochemistry. 2012;51:2496–2505. doi: 10.1021/bi201840k. [DOI] [PubMed] [Google Scholar]
- 35.Chong S, Shao Y, Paulus H, Benner J, Perler FB, Xu MQ. J. Biol. Chem. 1996;271:22159–22168. doi: 10.1074/jbc.271.36.22159. [DOI] [PubMed] [Google Scholar]
- 36.Chong S, Williams KS, Wotkowicz C, Xu MQ. J. Biol. Chem. 1998;273:10567–10577. doi: 10.1074/jbc.273.17.10567. [DOI] [PubMed] [Google Scholar]
- 37.Muir TW, Sondhi D, Cole PA. Proc. Natl. Acad. Sci. USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood DW, Wu W, Belfort G, Derbyshire V, Belfort M. Nat. Biotechnol. 1999;17:889–892. doi: 10.1038/12879. [DOI] [PubMed] [Google Scholar]
- 39.Hall TM, Porter JA, Young KE, Koonin EV, Beachy PA, Leahy DJ. Cell. 1997;91:85–97. doi: 10.1016/s0092-8674(01)80011-8. [DOI] [PubMed] [Google Scholar]
- 40.Matsumura H, Takahashi H, Inoue T, Yamamoto T, Hashimoto H, Nishioka M, Fujiwara S, Takagi M, Imanaka T, Kai Y. Proteins. 2006;63:711–715. doi: 10.1002/prot.20858. [DOI] [PubMed] [Google Scholar]
- 41.Klabunde T, Sharma S, Telenti A, Jacobs WR, Sacchettini JC. Nat. Struct. Biol. 1998;5:31–36. doi: 10.1038/nsb0198-31. [DOI] [PubMed] [Google Scholar]
- 42.Sun P, Ye S, Ferrandon S, Evans TC, Xu M-Q, Rao Z. J. Mol. Biol. 2005;353:1093–1105. doi: 10.1016/j.jmb.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 43.Oeemig JS, Aranko AS, Djupsjöbacka J, Heinämäki K, Iwaï H. FEBS Lett. 2009;583:1451–1456. doi: 10.1016/j.febslet.2009.03.058. [DOI] [PubMed] [Google Scholar]
- 44.Aranko AS, Oeemig JS, Iwaï H. FEBS J. 2013;280:3256–3269. doi: 10.1111/febs.12307. [DOI] [PubMed] [Google Scholar]
- 45.Amitai G, Belenkiy O, Dassa B, Shainskaya A, Pietrokovski S. Mol. Microbiol. 2003;47:61–73. doi: 10.1046/j.1365-2958.2003.03283.x. [DOI] [PubMed] [Google Scholar]
- 46.Dassa B, Haviv H, Amitai G, Pietrokovski S. J. Biol. Chem. 2004;279:32001–32007. doi: 10.1074/jbc.M404562200. [DOI] [PubMed] [Google Scholar]
- 47.Lee JJ, Ekker SC, von Kessler DP, Porter JA, Sun BI, Beachy PA. Science. 1994;266:1528–1537. doi: 10.1126/science.7985023. [DOI] [PubMed] [Google Scholar]
- 48.Koonin EV. Trends Biochem. Sci. 1995;20:141–142. doi: 10.1016/s0968-0004(00)88989-6. [DOI] [PubMed] [Google Scholar]
- 49.Snell EA, Brooke NM, Taylor WR, Casane D, Philippe H, Holland PWH. Proc. Biol. Sci. 2006;273:401–407. doi: 10.1098/rspb.2005.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.King N, Carroll SB. Proc. Natl. Acad. Sci. USA. 2001;98:15032–15037. doi: 10.1073/pnas.261477698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi J, Muir TW. J. Am. Chem. Soc. 2005;127:6198–6206. doi: 10.1021/ja042287w. [DOI] [PubMed] [Google Scholar]
- 52.Shah NH, Vila-Perelló M, Muir TW. Angew. Chem. Int. Ed. 2011;50:6511–6515. doi: 10.1002/anie.201102909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah NH, Eryilmaz E, Cowburn D, Muir TW. J. Am. Chem. Soc. 2013 doi: 10.1021/ja401015p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sorci M, Dassa B, Liu H, Anand G, Dutta AK, Pietrokovski S, Belfort M, Belfort G. Anal. Chem. 2013;85:6080–6088. doi: 10.1021/ac400949t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dassa B, Amitai G, Caspi J, Schueler-Furman O, Pietrokovski S. Biochemistry. 2007;46:322–330. doi: 10.1021/bi0611762. [DOI] [PubMed] [Google Scholar]
- 56.Zheng Y, Wu Q, Wang C, Xu M-Q, Liu Y. Biosci. Rep. 2012;32:433–442. doi: 10.1042/BSR20120049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis EO, Jenner PJ, Brooks PC, Colston MJ, Sedgwick SG. Cell. 1992;71:201–210. doi: 10.1016/0092-8674(92)90349-h. [DOI] [PubMed] [Google Scholar]
- 58.Xu MQ, Southworth MW, Mersha FB, Hornstra LJ, Perler FB. Cell. 1993;75:1371–1377. doi: 10.1016/0092-8674(93)90623-x. [DOI] [PubMed] [Google Scholar]
- 59.Cooper AA, Chen YJ, Lindorfer MA, Stevens TH. EMBO J. 1993;12:2575–2583. doi: 10.1002/j.1460-2075.1993.tb05913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Southworth M, Amaya K, Evans T, Xu M, Perler F. BioTechniques. 1999;27:110–120. doi: 10.2144/99271st04. [DOI] [PubMed] [Google Scholar]
- 61.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 62.Muir TW. Annu. Rev. Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 63.Dawson P, Churchill M, Ghadiri M, Kent S. J. Am. Chem. Soc. 1997;119:4325–4329. [Google Scholar]
- 64.Mootz HD. ChemBioChem. 2009;10:2579–2589. doi: 10.1002/cbic.200900370. [DOI] [PubMed] [Google Scholar]
- 65.Appleby JH, Zhou K, Volkmann G, Liu X-Q. J. Biol. Chem. 2009;284:6194–6199. doi: 10.1074/jbc.M805474200. [DOI] [PubMed] [Google Scholar]
- 66.Aranko AS, Züger S, Buchinger E, Iwaï H. PLoS ONE. 2009;4:e5185. doi: 10.1371/journal.pone.0005185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ludwig C, Pfeiff M, Linne U, Mootz HD. Angew. Chem. Int. Ed. 2006;45:5218–5221. doi: 10.1002/anie.200600570. [DOI] [PubMed] [Google Scholar]
- 68.Ludwig C, Schwarzer D, Mootz HD. J. Biol. Chem. 2008;283:25264–25272. doi: 10.1074/jbc.M802972200. [DOI] [PubMed] [Google Scholar]
- 69.Vila-Perelló M, Liu Z, Shah NH, Willis JA, Idoyaga J, Muir TW. J. Am. Chem. Soc. 2013;135:286–292. doi: 10.1021/ja309126m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shah NH, Dann GP, Vila-Perelló M, Liu Z, Muir TW. J. Am. Chem. Soc. 2012;134:11338–11341. doi: 10.1021/ja303226x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jansson M, Li YC, Jendeberg L, Anderson S, Montelione GT, Nilsson B. J. Biomol. NMR. 1996;7:131–141. doi: 10.1007/BF00203823. [DOI] [PubMed] [Google Scholar]
- 72.Xu R, Ayers B, Cowburn D, Muir TW. Proc. Natl. Acad. Sci. USA. 1999;96:388–393. doi: 10.1073/pnas.96.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamazaki T, Otomo T, Oda N, Kyogoku Y, Uegaki K, Ito N, Ishino Y, Nakamura H. J. Am. Chem. Soc. 1998;120:5591–5592. [Google Scholar]
- 74.Liu D, Xu R, Cowburn D. Meth. Enzymol. 2009;462:151–175. doi: 10.1016/S0076-6879(09)62008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Volkmann G, Iwaï H. Mol. Biosyst. 2010;6:2110–2121. doi: 10.1039/c0mb00034e. [DOI] [PubMed] [Google Scholar]
- 76.Züger S, Iwai H. Nat. Biotechnol. 2005;23:736–740. doi: 10.1038/nbt1097. [DOI] [PubMed] [Google Scholar]
- 77.Camarero J, Muir T. J. Am. Chem. Soc. 1999;121:5597–5598. [Google Scholar]
- 78.Camarero J, Pavel J, Muir TW. Angew. Chem. Int. Ed. 1998;37:347–349. doi: 10.1002/(SICI)1521-3773(19980216)37:3<347::AID-ANIE347>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 79.Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ. Proc. Natl. Acad. Sci. USA. 1999;96:13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Naumann TA, Tavassoli A, Benkovic SJ. ChemBioChem. 2008;9:194–197. doi: 10.1002/cbic.200700561. [DOI] [PubMed] [Google Scholar]
- 81.Young TS, Young DD, Ahmad I, Louis JM, Benkovic SJ, Schultz PG. Proc. Natl. Acad. Sci. USA. 2011;108:11052–11056. doi: 10.1073/pnas.1108045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tavassoli A, Lu Q, Gam J, Pan H, Benkovic SJ, Cohen SN. ACS Chem. Biol. 2008;3:757–764. doi: 10.1021/cb800193n. [DOI] [PubMed] [Google Scholar]
- 83.Kritzer JA, Hamamichi S, McCaffery JM, Santagata S, Naumann TA, Caldwell KA, Caldwell GA, Lindquist S. Nat. Chem. Biol. 2009;5:655–663. doi: 10.1038/nchembio.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Skretas G, Wood DW. Protein Sci. 2005;14:523–532. doi: 10.1110/ps.04996905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Buskirk AR, Ong Y-C, Gartner ZJ, Liu DR. Proc. Natl. Acad. Sci. USA. 2004;101:10505–10510. doi: 10.1073/pnas.0402762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cook SN, Jack WE, Xiong X, Danley LE, Ellman JA, Schultz PG, Noren CJ. Angew. Chem. Int. Ed. 1995;34:1629–1630. [Google Scholar]
- 87.Vila-Perelló M, Hori Y, Ribó M, Muir TW. Angew. Chem. Int. Ed. 2008;47:7764–7767. doi: 10.1002/anie.200802502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Binschik J, Zettler J, Mootz HD. Angew. Chem. Int. Ed. 2011;50:3249–3252. doi: 10.1002/anie.201007078. [DOI] [PubMed] [Google Scholar]
- 89.Mootz HD, Muir TW. J. Am. Chem. Soc. 2002;124:9044–9045. doi: 10.1021/ja026769o. [DOI] [PubMed] [Google Scholar]
- 90.Mootz HD, Blum ES, Tyszkiewicz AB, Muir TW. J. Am. Chem. Soc. 2003;125:10561–10569. doi: 10.1021/ja0362813. [DOI] [PubMed] [Google Scholar]
- 91.Tyszkiewicz AB, Muir TW. Nat. Methods. 2008;5:303–305. doi: 10.1038/nmeth.1189. [DOI] [PubMed] [Google Scholar]
- 92.Schwartz EC, Saez L, Young MW, Muir TW. Nat. Chem. Biol. 2007;3:50–54. doi: 10.1038/nchembio832. [DOI] [PubMed] [Google Scholar]
- 93.Prescher JA, Bertozzi CR. Nat. Chem. Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 94.Giriat I, Muir TW. J. Am. Chem. Soc. 2003;125:7180–7181. doi: 10.1021/ja034736i. [DOI] [PubMed] [Google Scholar]
- 95.Borra R, Dong D, Elnagar AY, Woldemariam GA, Camarero JA. J. Am. Chem. Soc. 2012;134:6344–6353. doi: 10.1021/ja300209u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li J, Sun W, Wang B, Xiao X, Liu X-Q. Hum. Gene Ther. 2008;19:958. doi: 10.1089/hum.2008.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang P, Chen T, Sakurai K, Han B-X, He Z, Feng G, Wang F. Sci. Rep. 2012;2:497. doi: 10.1038/srep00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Iwai H, Züger S, Jin J, Tam P-H. FEBS Lett. 2006;580:1853–1858. doi: 10.1016/j.febslet.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 99.Zettler J, Schütz V, Mootz HD. FEBS Lett. 2009;583:909–914. doi: 10.1016/j.febslet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 100.Carvajal-Vallejos P, Pallissé R, Mootz HD, Schmidt SR. J. Biol. Chem. 2012;287:28686–28696. doi: 10.1074/jbc.M112.372680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mills KV, Manning JS, Garcia AM, Wuerdeman LA. J. Biol. Chem. 2004;279:20685–20691. doi: 10.1074/jbc.M400887200. [DOI] [PubMed] [Google Scholar]
- 102.Saleh L, Southworth MW, Considine N, O'Neill C, Benner J, Bollinger JM, Perler FB. Biochemistry. 2011;50:10576–10589. doi: 10.1021/bi200810j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brenzel S, Kurpiers T, Mootz HD. Biochemistry. 2006;45:1571–1578. doi: 10.1021/bi051697+. [DOI] [PubMed] [Google Scholar]
- 104.Amitai G, Callahan BP, Stanger MJ, Belfort G, Belfort M. Proc. Natl. Acad. Sci. USA. 2009;106:11005–11010. doi: 10.1073/pnas.0904366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lockless SW, Muir TW. Proc. Natl. Acad. Sci. USA. 2009;106:10999–11004. doi: 10.1073/pnas.0902964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cheriyan M, Pedamallu CS, Tori K, Perler F. J. Biol. Chem. 2013;288:6202–6211. doi: 10.1074/jbc.M112.433094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shah NH, Eryilmaz E, Cowburn D, Muir TW. J. Am. Chem. Soc. 2013;135:5839–5847. doi: 10.1021/ja401015p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Muona M, Aranko AS, Raulinaitis V, Iwaï H. Nat. Protoc. 2010;5:574–587. doi: 10.1038/nprot.2009.240. [DOI] [PubMed] [Google Scholar]
- 109.Hiraga K, Soga I, Dansereau JT, Pereira B, Derbyshire V, Du Z, Wang C, Van Roey P, Belfort G, Belfort M. J. Mol. Biol. 2009;393:1106–1117. doi: 10.1016/j.jmb.2009.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Appleby-Tagoe JH, Thiel IV, Wang Y, Wang Y, Mootz HD, Liu X-Q. J. Biol. Chem. 2011;286:34440–34447. doi: 10.1074/jbc.M111.277350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Oeemig JS, Zhou D, Kajander T, Wlodawer A, Iwaï H. J. Mol. Biol. 2012;421:85–99. doi: 10.1016/j.jmb.2012.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]