Abstract
Compelling evidence suggests that the early interaction between porcine circovirus 2 (PCV-2) and the innate immune system is the key event in the pathogenesis of Post-Weaning Multisystemic Wasting Syndrome (PMWS). Furthermore, PCV2 has been detected in bone-marrow samples, potentially enabling an easy spread and reservoir for the virus. To assess the gene-expression differences induced by an in-vitro PCV2b infection in different three different myeloid innate immune cell subsets generated from the same animal, we used the Agilent Porcine Gene Expression Microarray (V2). Alveolar macrophages (AMØs), monocyte-derived dendritic cells (MoDCs) and bone-marrow cells (BMCs) were generated from each animal, and challenged with a UK-isolate of a PCV2 genotype b-strain at a MOI of 0.5. Remarkably, analysis showed a highly distinct and cell-type dependent response to PCV2b challenge. Overall, MoDCs showed the most marked response to PCV2b challenge in vitro and revealed a key role for TNF in the interaction with PCV2b, whereas only few genes were affected in BMCs and AMØs. These observations were further supported by an enrichment of genes in the downstream NF-κB Signalling pathway as well as an up regulation of genes with pro-apoptotic functions post-challenge. PCV2b challenge increases the expression of a large number of immune-related and pro-apoptotic genes mainly in MoDC, which possibly explain the increased inflammation, granulomatous inflammation and lymphocyte depletion seen in PMWS-affected pigs.
Introduction
PCV2, a non-enveloped, single-stranded circular DNA virus, has been recognized as the underlying agent for Post Weaning Multisystemic Wasting Syndrome (PMWS) [1], [2] and is now endemic in most pig-producing countries. Thus far, three genotypes of PCV2 have been described. PCV2a first reported in archived tissue samples, PCV2b, first reported in 2005 in North America and PCV2c recently described in Denmark [3]–[5]. Previous studies did not show differences in virulence of different PCV2 strains [1], [6]; however recent evidence suggests that mutations in circulating PCV2 strains coincided with a dramatic change of pathogenicity and clinical outcome [3], [7], with pigs carrying numerous PCV2 genotypes [8]. The fact that PCV2 has been detected in both PMWS and non-PMWS affected farms and pigs contributed to the notion that different PCV2 strains vary in their pathogenicity [9]. In that context it has to be noted that the dominant PCV2 strain circulating on severely-affected farms in the UK was grouped into genotype PCV2b, as determined in a recent cross-sectional study involving 147 pig farms across England [10].
Whereas the majority of endemic diseases can be controlled by vaccination or eradication of the pathogen, these approaches are less successful in the control of multi-factorial diseases. PMWS represents a typical multi-factorial disease, with pigs between 5 and 12 weeks of age being affected, and which is characterized by increased mortality, weight loss, wasting, dyspnoea and enlarged lymph nodes [8], [11], [12]. Indeed, vaccination against PCV2 seems to be successful in that it is reducing losses on affected farms, but is not inducing sterile immunity and vaccinated pigs still seem to harbour and potentially shed PCV2 [13], [14]. In this respect, the infected cells are of great importance for the distribution of PCV2.
Viruses have evolved complex immune evasion strategies for protection against host immune responses [15], and PCV2-infected pigs seem to harbour the virus in different immune cell subsets [13], [14] without infected cells showing signs of functional differences. Particularly alveolar macrophages (AMØs) and other cell-types of the monocyte/macrophage lineage seem to act as reservoirs and Trojan horse for PCV2, subsequently infecting other immune cells and specifically bone-marrow cells [14], [16]. Lymphocyte depletion, secondary infections with opportunistic pathogens, induction of apoptosis and other changes in immune cell subpopulations and PBMCs are all common characteristics of PMWS in severely affected pigs, strongly suggesting an immunosuppressive status [13], [17], [18]. Indeed, PCV2b-infected plasmacytoid dendritic cells (pDC) were shown to be unresponsive to exposure to further “danger-signals” [19], [20], supporting the notion that PCV2 induces a status of immunsuppression. However, the mechanism by which the immune system is altered remains unclear. A proposed mechanism suggests the presence of immunomodulatory CpG motif in the small circular genome of PCV2 inhibiting critical cytokine secretion [21], [22].
In the last decade, gene expression profiling microarrays have been widely used to reveal the effects of pathogens on host cells and tissues aiming to gain insight into the molecular mechanisms involved in the host-pathogen interactions [23], [24], and a limited number of studies have been performed applying the benefits of microarrays to elucidate the pathogenesis of PCV infection [24]–[27]. In all of these studies, tissues from PCV-infected pigs, rather than specific cellular subsets were analysed. Whereas this allows for a discrimination of genes being differentially expressed in different tissues as a result of the infection, it does not allow for the discrimination of PCV-induced effects in specific cell-types. These ex vivo data showed differential expression of numerous genes in PCV2-infected animals involved in innate immune defence (TLR1, CD14 and CD180), immunosuppressive responses (FGL2 and GPNMB) and pro-inflammatory signals (galectin-3) [24].
Due to the difficulty of reproducing an experimental model of PMWS and lack of advanced pig molecular markers [28], the objective of the present study was to analyse the early molecular mechanisms involved in PCV2b infection of three defined immune cell subsets generated from the same animals, using a genome-wide expression approach. Information obtained from this study will increase our understanding of the features of PCV2 infection during the onset of immune responses in vivo.
Materials and Methods
Ethics Statement
All animal studies were performed according to the regulations and guidance provided under the UK Home Office Animals (Scientific Procedures) Act 1986. Experimental protocols were approved under project licence number PPL 70/7219, as well as the RVC Ethics and Welfare Committee.
Animals
Six crossbred Large White×Landrace pigs (Sus scrofa), free of PCV1, PCV2, swine influenza strains H1N1, H1N3, and H1N5, PRRSV and Mycoplasma hyopneumoniae, as confirmed by qPCR and antibody testing, were sourced from the Animal Health and Veterinary Laboratories Agency (AHVLA) and housed at the pig unit of the Royal Veterinary College. At 6 months of age, all six pigs were humanly euthanized. During housing, PCV2 status of the pigs was repeatedly confirmed by ELISA (antibody) and quantitative real-time PCR on serum. All six pigs remained PCV2 negative throughout.
Generation of immune cell subsets
From each individual animal, the following immune cell subsets were generated:
Porcine alveolar macrophages (AMØs)
Porcine AMØs were obtained from the lungs of each euthanized PCV2-negative pig (n = 6). Briefly, approximately 2×500 ml of sterile PBS (containing 200 µg ml−1 gentamycin and 2.5 µg ml−1 amphotericin B) were delivered into the freshly isolated lungs via tracheal intubation. Following gentle manipulation to detach loosely adherent cells, the aspirate was removed and the resulting cell suspension was placed in a sterile 500 ml bottle. AMØ were recovered following centrifugation (200× g for 10 min at 4°C), washed once and resuspended in growth medium (RPMI 1640 supplemented with 10% (v/v) porcine serum, 200 µg ml−1 Pen/Strep and 0.5 µg ml−1 amphotericin B). AMØs were seeded at 1×106 cells ml−1 and incubated at 37°C/5% CO2 in 6-well plates (Nalgene, USA). The majority of isolated cells expressed the surface antigens CD14 and MHC class II, as analysed by flow-cytometry (data not shown).
Generation of monocyte-derived dendritic cells (MoDCs)
Just before euthanasia, 500 ml of blood was collected from each pig ( = 6) into sterile bottles, containing acid citrate dextrose solution (0.22 M D(+)glucose, 0.2 M sodium citrate and 0.14 M citric acid) at 10% of the total volume. The blood was divided into 50 mL falcon tubes and centrifuged at 1000× g for 25 min. The buffy coat was removed and collected in a sterile glass bottle, diluted 1∶2 (v/v) with calcium/magnesium free PBS (PBSa) at room temperature. Peripheral blood mononuclear cells (PBMCs) were isolated from the buffy coat via density centrifugation over Histopaque (Histopaque 1.077 g ml−1, Sigma, Poole, Dorset, UK) as described previously [29]. After centrifugation (800× g for 25 min at 22°C), the mononuclear cells were recovered from the interface formed between the cell suspension and the Histopaque and washed twice by dilution in cold (4°C) PBS-A 0.03% (w/v) EDTA (PBS/EDTA) and centrifugation (350× g/10 min/4°C). CD14+ monocytes were isolated by positive selection using a MACS system (Miltenyi Biotech, Bergisch Gladbach, Germany), according to the manufacturer's protocol. The cells were cultured for 7 days in DMEM medium with porcine serum (10% v/v), recombinant porcine (rp) granulocyte–macrophage colony-stimulating factor (GM-CSF, 150 ng ml−1) and rp interleukin-4 (IL-4, 100 U ml−1). RpIL-4 and rpGM-CSF were prepared in house, and bioactivity of both cytokines was determined using a TF-1 cell bioassay, with the IL-4 concentration giving half-maximum proliferation being defined as 1 unit. Resulting cells were CD14low, and expressed the DC-SIGN molecule, consistent with the phenotype reported by Huang et al., 2009 [30].
Generation of bone marrow cells (BMCs)
BMCs were isolated from the sternum of each pig (n = 6) as previously described [31]. Briefly, the sternum was opened and flushed with PBS/0·03% EDTA (w/v) at 37°C. The obtained cell suspension was depleted of erythrocytes and mature granulocytes by centrifugation over Histopaque (1.077 g ml−1, Sigma, Poole, Dorset, UK) at 1000× g for 40 min at room temperature. After a further two PBS/EDTA washes (250× g/10 min/4°C), the cells were cultured for 7 days in αMEM medium with porcine serum (10% v/v), 200 µg ml−1 Pen/Strep, and 0.5 µg ml−1 amphotericin B.
Virus preparation for infection
To avoid potential false-positive results due to LPS contaminations of reagents/media, all substances used were tested for their LPS content using the Endosafe-PTS system (Charles River, Charleston, USA). Samples with an endotoxin content below 0.01 EU ml−1 were considered as LPS-free.
A previously characterized PCV2b isolate from the UK (GenBank accession number JX193799; [10]) was used to generate the virus stock for the experimental infections. The virus was propagated in type-I IFNKO PK15-ALR-NPro cells, free of PCV1 and PCV2. Cells were cultured in MEM containing Earle's salts supplemented with 10% (v/v) tetracycline-free foetal bovine serum (FBS) (Clontech, Saint-Germain-en-Laye, France). To induce the tetracycline-regulated expression of the IFN-KO, tetracycline was added 2 h pre-inoculation. Cell monolayers were inoculated at 50% confluence. After 18 h incubation at 37°C, the inoculum was removed and retained, and the monolayer treated with 300 mM D-glucosamine in Hanks balanced salt solution (HBSS) for 30 min at 37°C. After removal of the glucosamine and subsequent washes with HBSS, cultures were overlaid with retained inoculum. Cultures were overlaid with media 24 h post-inoculation. After a further 3 days incubation media was removed and retained and virus harvested from trypsinised cells by freeze-thawing. The cell lysate was clarified by centrifugation at 1,000× g for 5 min at 4°C and the supernatant added to the retained media. This virus suspension was then concentrated approximately 10-fold using dialysis tubing (Spectra/Por, Biotech Cellulose Ester membrane; Spectrum Europe B.V.) in polyethylene glycol (PEG 12000 flake; Whyte Chemicals Ltd.) at 4°C. Concentrated virus suspension was subsequently dialysed in MEM overnight and aliquoted. PCV2 stocks were titrated on PK15-ALR-NPro cells as described elsewhere [1]. The titre of the virus stock was determined as 106.05 TCID50 ml−1.
In line with the general accepted rules for work with viruses, and similar as described by others [14], [20], [32], supernatant of type-I IFNKO PK15-ALR-NPro cells was used as true mock-infection control to counteract gene expression changes induced by the cell culture supernatant of PK15-ALR-NPro cells rather than being a true result of PCV2b infection.
All three cell types generated from each pig were exposed on the same day to either PCV2b at an MOI of 0.5 or remained uninfected in six-well-plates for 0 h, 1 h and 24 h at 37°C. At the 0 h time-point, cells were overlaid with PCV2b stock at an MOI of 0.5 and immediately washed and lysed; this served as a further control for normal gene expression within the cells. At time-points indicated, cells were lysed in 600 µL GTC buffer (4 M guanidine thiocyanate, 25 mM sodium citrate, 0.5% (w/v) sodium N- lauroylsarcosine, 0.1 M 2-mercaptoethanol, pH 7) after two washes, as required for subsequent RNA extraction.
Assessment of infection by qPCR
The number of PCV2 copies per ng of cDNA isolated from each cell type was determined by comparison to known standards using qPCR. Each sample was measured in triplicate and in a final volume of 20 µl per well in Microamp fast optical 48-microtiter well plates (Applied Biosystems). Each well contained, 2 µl of cDNA (standard or test sample), 10 µl of 2× TaqMan Universal Master Mix II (Applied Biosystems), 50 pmol of each primer (Forward: 5′-GCTCTYTATCGGAGGATTAC-3′, Reverse: 5′-ATAAAAACCATTACGAWGTGATA-3′) (MWG) and 2.5 µM of TaqMan probe (5′FAM-CCATGCCCTGAATTTCCATATGAAAT-3′TAMRA) (Applied Biosystems). The volume of each well was adjusted to 20 µl by addition of nuclease-free water (Sigma-Aldrich). The TaqMan probe and the primers were designed to target a partial (137 bp) sequence of the ORF2 of PCV2 [33]. Standard measurements were performed in a 10-fold serial dilution from 109 PCV2 copies to 0 copies. After plate set up, the qPCR was performed in a StepOne Real-time PCR machine (Applied Biosystems) and StepOne software version 2.2.2 (Applied Biosystems). The cycling conditions were 95°C for 10 mins for polymerase activation followed by 40 cycles of denaturation at 95°C for 15 secs and annealing/extension at 55°C for 1 min. Data was analysed using either StepOne software version 2.2.2 (Applied Biosystems) or Excel 2010 (Microsoft). Ct values of triplicate sample and standard measurements were averaged and this average Ct value was used to calculate the PCV2 ORF2 copy number in samples by comparison to the known standards. The copy number in each sample was then divided by the nanograms of DNA in that sample to give the PCV2 copy number per ng of DNA.
Microarray hybridizations and data analysis
From all cell types generated from each pig, total RNA was extracted at each time point indicated from cells either mock- or PCV2-infected, using the RNeasy kit (Qiagen, Crawley, UK) according to the manufacturer's instructions, including an RNase-free DNase I (Qiagen, Crawley, UK) digestion step at the time-points indicated. RNA integrity and quality was determined using the Agilent 2100 Bioanalyzer (Agilent Technologies, Wokingham, UK). Up to 50 ng of total RNA with RIN values ≥8 was reverse transcribed to cDNA and then transcribed into Cy3-labelled cRNA using the Low Input Quick Amp Gene Expression Labeling Kit (Agilent, Santa Clara, CA) according to the manufacturer's instructions. Hybridization to the Agilent 4×44K Porcine Gene Expression Microarray (V2; Agilent) was performed on a Tecan Hybridization Station HSPro400 (Tecan, Maennersdorf, Switzerland). Microarrays were scanned with an Agilent C Scanner (Agilent Technologies, Wokingham, UK).
Following quality control checks and principal component analysis (PCA), a subset of 12 arrays were removed. A total of 100 arrays were analysed with 38, 21 and 41 arrays representing AMØs, BMCs and MoDCs respectively. Pre-processing steps of background correction and between-array normalization were performed prior to analysis using the software packages R (http://www.r-project.org) and Bioconductor (http://www.bioconductor.org) [34]. Differential expression analysis was performed by the Bioconductor package limma using the functions lmFit and eBayes. With increasing advancements in microarray technology, numerous studies have now used a −1.5≤ fold change (FC) ≥1.5 [35], [36] and have verified the biological significance of these transcripts [37], [38], consequently, in this study differences in gene expression between the treatment groups were considered significant using false discovery rate (FDR)-adjusted P<0.05 [40] and a −1.5≤FC≥1.5 as a cut-off.
Systems Biology Analysis
For further analysis, all differentially expressed genes (P<0.05) were imported into the Ingenuity Systems Pathway Analysis program (IPA; Ingenuity Systems, Redwood City, CA, USA; http://www.ingenuity.com). Canonical pathways analysis identified the pathways from the IPA library of canonical pathways that were most significant to the data set. Transcripts from the data set that met the −1.5≤FC≥1.5 and a FDR-adjusted P<0.05 cut-off and were associated with a canonical pathway in the Ingenuity Knowledge Base were considered for the analysis. The significance of the association between the data set and the canonical pathway is measured by considering: (1) the number of focus genes that participate in that process, (2) the total number of genes that are known to be associated with that process in the selected reference set and lastly, Fischer's exact test is used to calculate a p-value and the corrected p-value, using the Benjamini-Hochberg method [39], is displayed for the Functions and Canonical Pathways (IPA).
Real time quantitative reverse transcription PCR (qRT-PCR) analysis
The significance of differential expression between PCV2b- challenged and unchallenged cells uncovered by Agilent microarray analyses was further validated using real-time PCR. cDNA was prepared from total RNA isolated from each sample analysed in the microarray study using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Life Technologies Corporation, Warrington, UK). cDNA conversions were performed in 20 µl reaction volumes using random primers as per manufacturer's instructions.
Taqman gene expression assays (Applied Biosystems, Life Technologies Corporation, Warrington, UK) were used to determine expression of CXCL2, IL-8, IL-1β, IL-1α, TNF, and PTGES in cDNA samples prepared above according to the manufacturer's instructions. After evaluation of several house-keeping genes according to the MIQE-guidelines [40], glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was chosen as the most stable one with low variation between samples in the microarray analysis. qPCR was carried out in 48-well optical plates (Applied Biosystems, Life Technologies Corporation, Warrington, UK) and each sample was measured in triplicate. Measurements were made using a StepOne qPCR machine (Applied Biosystems, Life Technologies Corporation, Warrington, UK) and StepOne software version 2.1 (Applied Biosystems, Life Technologies Corporation, Warrington, UK). PCR thermal cycling conditions for each amplicon comprised of one cycle at 95°C for 10 mins, followed by 40 cycles at 95°C for 15 seconds and 60°C for 60 seconds.
Changes in gene expression revealed by qPCR were calculated by the 2ΔΔCt method [41]. Normalization was carried out by dividing their relative expression level by the relative expression level of GAPDH.
Results
Confirmation of PCV2b infection in all cell types over time
Quantitative PCR revealed the presence of increasing copy-numbers of PCV2b in all three cell-types, with the highest copy number being detected in alveolar macrophages (Fig. 1a). In addition, specifically AMØs showed substantial cell-morphological changes between 24 h mock-infected (Fig. 1b) and 24 h after infection (Fig. 1d), as well as between 1 h after infection (Fig. 1c) and 24 h after infection (Fig. 1d), as seen by light-microscopy, compared to MoDC infected for the same length of time (Fig. 1 e–f).
Figure 1. Quantitative PCR of PCV2b copy-numbers in immune cell subsets.
Cells were prepared and infected as described, and PCV2 copy numbers analysed at the time-points indicated (Fig. 1a). Effect of PCV2 infection on immune cell subsets was also investigated by light microscopical examination of infected cells (AMØs Fig. 1 c and d: MoDC Fig. 1 f and g) compared to mock-infected cells (Fig. 1 b and e, respectively). Magnification ×40.
PCV2b challenge induces differential gene expression in all three immune cell subsets, with monocyte-derived dendritic cells showing highest differences
Differences in cell-specific gene expression of PCV2b challenged and unchallenged immune cell subsets derived from individual animals (MoDCs, AMØs and BMCs) were assessed using the Agilent Porcine Gene Expression Microarray with a cut-off p-value of p<0.05 and a fold range of −1.5≤FC≥1.5.
Interestingly, some genes were differentially regulated at timepoint 0. We hypothesise at the moment that this is not due to the PCV2 infection, rather than handling of cells (adding virus, washing of cells).
Overall, 92 genes showed differential expression, with the number of differentially expressed genes increasing as infection progresses (Fig. 2). As indicated above, all three immune cell subsets were responsive to PCV2b infection and showed differential gene expression at an early (1 h) and late (24 h) time-point. Interestingly however, substantial differences were seen between the response of the different cell subsets to PCV2b exposure, resulting in a minimal overlap of differentially-expressed transcripts between MoDCs, AMØs and BMCs (Fig. 3). Among the immune cell subset studied, and compared to mock-infection using the cell culture supernatant of PK15 cells as control, MoDCs showed the most marked response to PCV2b challenge with 40 and 30 transcripts differentially-regulated at 1 h and 24 h, respectively (Fig. 2), whereas AMØs hardly reacted to the infection at all, and BMCs showed the greatest number of down-regulated genes (Fig. 2).
Figure 2. Differentially expressed genes in PCV2b-challenged and control immune cell subsets.
Differentially expressed genes at each time-point are shown for the two treatment comparisons (P<0.05, −1.5≤ fold change ≥1.5, N = 6).
Figure 3. Transcriptome differences between immune cell subsets challenged with PCV2b.

Venn diagram of differentially expressed genes after challenge with PCV2b is shown for each immune cell subset (P<0.05, −1.5≤ fold change ≥1.5).
Independent of the cell-type, down-regulation of transcripts appeared only 24 h post PCV2b-challenge, at a time-point when PCV2b replication seemed to drop (see Fig. 1). At the early time point (1 h p.i.), the profiles of gene expression in un-challenged and PCV2b-challenged AMØs and BMCs were very similar, with only one transcript (TNF) was significantly altered in AMØs (Tab. 1). PCV2b challenge, however, had a clear early effect on MoDCs, which showed 40 differentially regulated transcripts (Tab. 1, Fig. 3, Tab. S1, Tab. S2), all of which were upregulated. These transcripts included a plethora of genes involved in inflammation and inflammatory responses, including NF-κB-inducible chemokine and cytokine genes such as the (C-C motif) ligand 20 gene (CCL20), interleukin 1 alpha and beta genes (IL-1α and IL-1β), chemokine (C-X-C motif) ligand 2 (CXCL2), other molecules involved in immune cell trafficking (ADM, CCL4, MAP3K8, NFKBIA, OSM, STAB1, TIMP1 and TNF), antiviral response genes (IRF1), and apoptosis-related genes (HBEGF, IRF1 (includes EG:16362), IER3, JAG1 and TNFAIP3).
Table 1. Annotated transcripts differentially expressed in AMØs, MoDCs and BMCs at 0 h, 1 h and 24 h p.i. following PCV2b infection.
| Cell type | Time-point p.i. | Gene symbol | FC | Gene description |
| AMØs | 0 h | PRDX6 | 2.58 | Peroxiredoxin-6 |
| 1 h | TNF | 2.32 | Tumor necrosis factor | |
| MoDCs | 1 h | IL1B | 6.47 | Interleukin 1-beta |
| CXCL2 | 4.66 | Chemokine (C-X-C motif) ligand 2 | ||
| IL8 | 4.4 | Interleukin 8 | ||
| TNF | 4.17 | Tumor necrosis factor | ||
| IER3 | 3.4 | immediate early response 3 | ||
| CCL4 | 3.37 | Chemokine (C-C motif) ligand 4 | ||
| ADM | 3.27 | adrenomedullin | ||
| IL1A | 3.2 | Interleukin-1 alpha | ||
| MAP3K8 | 2.91 | Mitogen-activated protein kinase 8 | ||
| BTG2 | 2.35 | B-cell translocation gene 2 | ||
| RND3 | 2.29 | rho Family GTPase 3 | ||
| JAG1 | 2.15 | jagged 1 | ||
| TNFAIP3 | 2.14 | tumor necrosis factor, alpha-induced protein 3 | ||
| IRF1 | 2.1 | interferon regulatory factor 1 | ||
| NFKBIA | 2.08 | NF-kappa-B inhibitor alpha | ||
| IRG1 | 2.01 | immune-responsive gene 1 protein homolog | ||
| MFSD2A | 1.98 | Major facilitator superfamily domain-containing protein 2 | ||
| OSM | 1.86 | oncostatin M | ||
| HBEGF | 1.84 | heparin-binding EGF-like growth factor | ||
| CCL20 | 1.82 | chemokine (C-C motif) ligand 20 | ||
| STAB1 | 1.82 | stabilin 1 | ||
| TOB1 | 1.73 | transducer of ERBB2, 1 | ||
| SLC2A6 | 1.71 | solute carrier family 2 (facilitated glucose transporter), member 6 | ||
| CSRNP1 | 1.68 | cysteine-serine-rich nuclear protein 1 | ||
| MoDCs | 24 h | VDAC1P5 | 4.06 | voltage-dependent anion channel 1 pseudogene 5 |
| S100A9 | 3.3 | S100 calcium binding protein A9 | ||
| COX3 | 3.21 | cytochrome oxidase 3 | ||
| ARHGAP25 | 2.85 | Rho GTPase activating protein 25 | ||
| SLC25A6 | 2.71 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 | ||
| RPL32 | 2.46 | ribosomal protein L32 | ||
| MRPL18 | 2.28 | mitochondrial ribosomal protein L18 | ||
| RTN4 | 2.25 | reticulon 4 | ||
| TGH2 | 2.17 | tissue transglutaminase homologue | ||
| PDLIM1 | 2.1 | PDZ and LIM domain 1 | ||
| PLCXD1 | 1.99 | phosphatidylinositol-specific phospholipase C, X domain containing 1 | ||
| CASP10 | 1.78 | caspase 10, apoptosis-related cysteine peptidase | ||
| RANBP1 | 1.77 | RAN binding protein 1 | ||
| VDAC2 | 1.66 | voltage-dependent anion channel 2 | ||
| FCN1 | −1.75 | ficolin (collagen/fibrinogen domain containing) 1 | ||
| BTC | −1.77 | betacellulin | ||
| ITGB5 | −2.03 | integrin, beta 5 | ||
| PIK3IP1 | −2.05 | phosphoinositide-3-kinase interacting protein 1 | ||
| SEPP2 | −2.08 | septin 2 | ||
| RNASE4 | −2.27 | ribonuclease, RNase A family, 4 | ||
| DKK3 | −2.29 | dickkopf 3 homolog | ||
| DUOXA2 | −3 | dual oxidase maturation factor 2 | ||
| BMCs | 0 h | PLDN | 1.56 | pallidin homolog |
| BMCs | 24 h | Es25 | 3.9 | Esterase 25 |
| IGSF8 | 2.78 | immunoglobulin superfamily, member 8 | ||
| PTGES | 2.54 | prostaglandin E synthase | ||
| TMEM237 | −1.6 | transmembrane protein 237 | ||
| MMRN2 | −1.72 | multimerin 2 | ||
| MYL1 | −2.27 | myosin, light chain 1 | ||
| PEG10 | −2.57 | paternally expressed 10 |
A total of 93 transcripts showed differential expression. Annotated transcripts are listed in each immune cell subset from the highest to the lowest fold-change at the different time points p.i.
At the late time-point, comparison of PCV2b-challenged immune cell subsets and non-challenged control cells revealed a total of 49 differentially expressed genes. Among the 30 genes identified in MoDCs, 19 showed upregulation whereas 11 genes were down-regulated (Tab. 1, Fig. 2, Tab. S2). The main transcripts with increased expression in MoDCs were VDAC1P5 (also known as VDAC5P or VDAC3, a voltage-dependent anion-selective channel protein) the myeloid-related protein S100A9 (potent chemotactic factor for immune cells) and COX3 (involved in prostaglandin biosynthesis). Other upregulated transcripts included genes with apoptotic function (CASP10, RANBP1, VDAC2 and SLC25A6). Down-regulated transcripts with a FC≤−1.5 included ITGB5, PIK3IP1, SEPP2, RNASE4, DKK3 and DUOXA2.
Only seven genes were significantly upregulated in BMCs, whereas 12 transcripts were down-regulated. Two of the top-ranking genes (ID BW961486 and 13183) could not be annotated, however, IGSF8, a member of the immunoglobulin protein superfamily functioning in cell migration and viral infection, and PTGES, an inflammatory mediator, showed upregulation in BMCs 24 h after PCV2b-challenge (Tab. 1, Tab. S3). Four of the 12 transcripts downregulated in PCV2b-challenged BMCs were annotated and included the transmembrane protein 237 (TMEM237), MMRN2, MYL1 and PEG10, a paternally-expressed negative regulator of TGFβ receptor signalling (Tab. 1, Tab. S3).
To corroborate these findings, quantitative real-time PCR analysis was performed for several key pro-inflammatory mediators, these included CXCL2, IL-8, IL-1β, IL-1α, TNF and PTGNES (Tab. 2). In general, the direction of upregulation correlated well between the two platforms; however, the magnitude of fold change was generally higher in the quantitative PCR approach.
Table 2. Validation of microarray data by qRT-PCR.
| Gene name | TaqMan® Gene Expression Assay | Microarray FC | qRT qPCR FC |
| CXCL2 | Ss03378360_u1 | 4.66 | 10.22 |
| IL-8 | Ss03392437_m1 | 4.40 | 17.58 |
| IL-1β | Ss03393804_m1 | 6.47 | 191.21 |
| IL-1α | Ss03391335_m1 | 3.20 | 18.09 |
| TNF | Ss03391318_g1 | 4.17 | 8.32 |
| PTGNES | Ss03392129_m1 | 2.54 | 11.92 |
PCV2b infection in MoDC promotes upregulation of genes with granulocyte infiltration and apoptotic function
Ingenuity Systems Pathway Analysis (IPA) allowed the identification of canonical pathways and functional processes affected by PCV2b challenge. Pathways that were differentially affected between PCV2-b challenged and non-challenged MoDCS at 1 h p.i. primarily involved the inflammatory responses and related functional groups including NF-κB Signalling (1st ranked; P = 3.61E-09) with seven out of the 175 genes present in this pathway significantly affected, Role of proinflammatory hypercytokinemia/hyperchemokinemia (associated with Influenza pathology; Suppl. Fig. 1), Innate and Adaptive Immune Cell Communication (3rd ranked), PPAR-, IL-6- (4th, 5th respectively) and Communication between Innate and Adaptive Immune Cells (P = 2.25E-07) (Tabl. S1, Tab. S4).
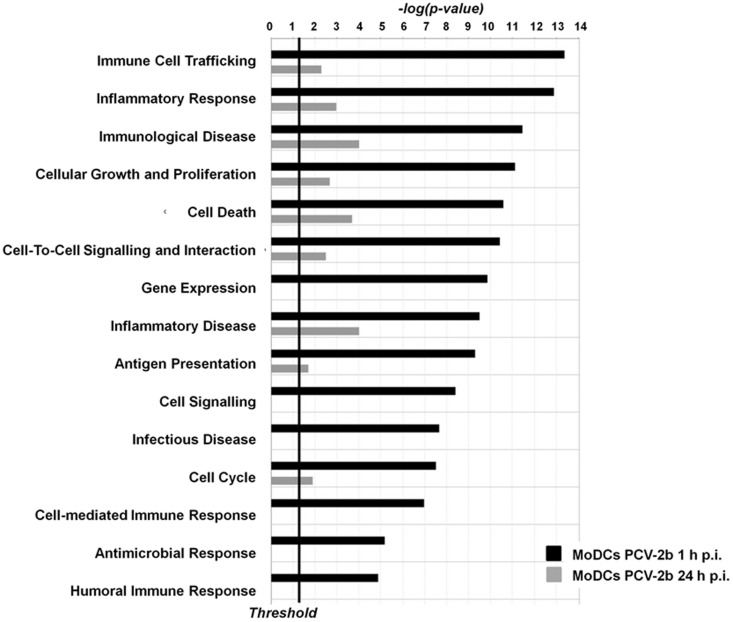
Significant upregulation of five genes within proinflammatory hypercytokinemia/hyperchemokinemia pathway (Fig. S1) indicated a strong pro-inflammatory response associated with PCV2b challenge in MoDCs leading to the recruitment of immune cells to the site of infection. As unique histopathological characteristic of PMWS-affected pigs is excessive granulomatous inflammation in the lymphoid tissues, genes with granulocyte/leukocyte infiltration function were depicted separately in Tab. 3. All but three of the genes (ADM, TNFAIP3 and TIMP1) showed a predicted increasing effect facilitating granulocyte/leukocyte infiltration. Indeed, functional analysis of differentially-expressed transcripts revealed an enrichment of genes associated with immune cell trafficking, inflammatory responses and immunological disease (Fig. 4, Tab. S1).
Table 3. Downstream effect analysis of upregulated genes involved in cell migration and infiltration in MoDCs 1.
| ID | FC | Effect of Gene on Granulocyte/Leucocyte Infiltration |
| IL1B | 6.470 | Increase |
| CXCL2 | 4.660 | Increase |
| IL8 | 4.400 | Increase |
| TNF | 4.170 | Increase |
| CCL4 | 3.370 | Increase |
| ADM | 3.270 | Decrease |
| IL1A | 3.200 | Increase |
| TNFAIP3 | 2.140 | Decrease |
| NFKBIA | 2.080 | Increase |
| OSM | 1.860 | Increase |
| TIMP1 | 1.770 | Decrease |
Figure 4. Biological process analysis of genes differentially expressed in MoDCs at 1 h and 24 h post PCV2b challenge.
Differentially expressed genes (P<0.05) were imported into IPA Ingenuity software to determine significantly enriched biological processes. Data represent the distribution in cell function categories of statistically significantly enriched biological processes (P<0.05) at 0 h and 24 h post PCV2b challenge.
Even with the large number of genes differentially expressed, only 3 canonical pathways reached significance of p<0.05 at 24 h p.i., including Role of IL-17A in Psoriasis (p = 0.0136), RAN Signalling (p = 0.0178) and Role of RIG1-like Receptors in Antiviral Innate Immunity (p = 0.0454; Tab. S1, Tab. S3), an important pathway involved in the host recognition of viral PAMPs. Interestingly, none of the top ranking canonical pathways identified by IPA were similar to those identified at the 1 h time-point.
Functional analysis revealed an enrichment of genes involved in Cell Death, with largely increasing effects at both time-points (Fig. 4, Tab. 4). PCV2-induced apoptosis may contribute to limiting the host response [42], this may be particularly important during the acute phase of infection when viral replication takes place [43]. An enrichment of differentially expressed genes with functions in apoptosis, both pro- and anti-apoptotic, were identified in MoDC at both time-points with a shift towards a more pro-apoptotic state at the later time-point where six out of nine genes have a predicted increasing effect (Tab. 4). Further network analyses depicting the top-networks of interacting genes for MoDC 1 h and 24 h after PCV2b infection are shown in figures S3 and S4.
Table 4. Downstream effect analysis of differentially-expressed genes involved in apoptosis in MoDCs 1 h and 24 h post PCV2b challenge.
| Cell Type | ID | FC | Predicted Effect of Gene on Apoptosis |
| MoDCs 1 h p.i. | |||
| IL1B | 6.470 | Increased | |
| IL8 | 4.400 | Increased | |
| TNF | 4.170 | Increased | |
| IER3 | 3.400 | Increased | |
| IL1A | 3.200 | Increased | |
| MAP3K8 | 2.910 | Increased | |
| IRF1 | 2.100 | Increased | |
| NFKBIA | 2.080 | Increased | |
| OSM | 1.860 | Increased | |
| HBEGF | 1.840 | Affected | |
| CXCL2 | 4.660 | Decreased | |
| CCL4 | 3.370 | Decreased | |
| ADM | 3.270 | Decreased | |
| BTG2 | 2.350 | Decreased | |
| RND3 | 2.290 | Decreased | |
| JAG1 | 2.150 | Decreased | |
| TNFAIP3 | 2.140 | Decreased | |
| TIMP1 | 1.770 | Decreased | |
| INSR | 1.600 | Decreased | |
| MoDCs 24 h p.i. | |||
| S100A9 | 3.3 | Increase | |
| SLC25A6 | 2.71 | Increase | |
| RTN4 | 2.25 | Increase | |
| CASP10 | 1.78 | Increase | |
| RANBP1 | 1.77 | Affects | |
| VDAC2 | 1.66 | Increase | |
| BTC | −1.77 | Increase | |
| ITGB5 | −2.03 | Decrease | |
| PIK3IP1 | −2.05 | Decrease | |
| DKK3 | −2.29 | Decrease |
Functional pathway analysis reveals significant activation of cellular development pathways in PCV2b infected BMCs
While no differential gene expression was detected at one hour post infection, 18 genes showed differential expression at 24 h with seven genes significantly upregulated.
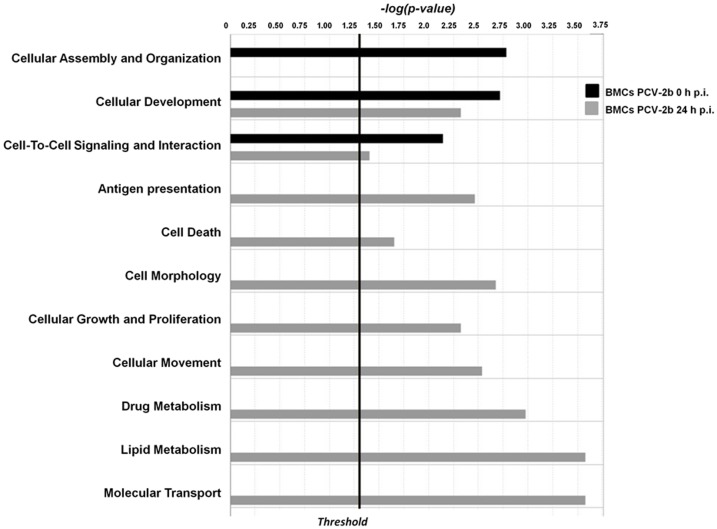
To gain a further insight into the potential mechanisms of the effects of PCV2b challenge on BMCs, the identified transcripts were mapped to networks available in the Ingenuity database. Three networks were identified by scores between three and 13. The scores take the number of focus molecules and the size of the network into account to estimate the relevance of this network. In accordance with the enrichment of genes with tissue development functions (Fig. 5), the highest-scoring network (Network Score 13) revealed a significant link with connective tissue development and function, embryonic and organ development (Fig. S2, Suppl. Fig. 5, Tab. S3).
Figure 5. Biological process analysis of genes differentially expressed in BMCs at 1 h and 24 h post PCV2b challenge.
Differentially expressed genes (P<0.05) were imported into IPA Ingenuity software to determine significantly enriched biological processes. Data represent the distribution in cell function categories of statistically significantly enriched biological processes (P<0.05) at 0 h and 24 h post PCV2b challenge.
Among the top ranking canonical pathways identified by IPA, several impact on immune response, including Eicosanoid Signalling (1st ranked), important for its contribution to the inflammatory response in a variety of diseases (such as arthritis and asthma) and CXCR4 Signalling pathway (9th ranked) (Tab. S3). Prostaglandin E Synthase (PTGES; FC = 2.54) was central to most pathways and functions affected and has been shown to be induced by IL1- and NFκB during inflammatory conditions [44]. Prostaglandin E Synthase converts prostaglandin endoperoxide H2 (PGH2) to prostaglandin E2 (PGE2), which is a potent immunoregulatory lipid mediator with key roles in the regulation of virus replication and modulation of inflammatory responses following infection [45]. Indeed, PGE2 has not only been shown to downregulate IL-12 and IL-23 production, two of the major driving cytokines for a Th1 response [46], but also leads to the development of myeloid-derived suppressor cells [47] and has dampening effects on MoDC maturation [48].
Discussion
PCV-2 is the causative agent of PMWS, a widespread complex multifactorial disease with recognized immunosuppressive characteristics [18]. Compelling evidence suggests that the intricate interaction between the host immune system to the presence of PCV2 and the ability of PCV2 to interfere with immune defence is a key event in the pathological outcome of PMWS. While productive replication of PCV-2 is most likely restricted to epithelial and endothelial cells [32], [49], the virus has been shown to infect and persist in cells of the immune system, including MØ, DC and lymphocytes [14], [50]–[52]. The current study assessed the early molecular mechanisms involved in PCV2 infection of immune cell subsets using a genome-wide expression approach. To aid to the clinical significance of this study, a pathogenic PCV2b strain currently circulating in the UK was used.
Similar as described in earlier publication [14], [53], PCV2b seems to readily infection of all three cell types in the absence of clear viral replication. It is also interesting to mention that there does not seem to be a correlation between viral copy numbers in different cell-types and changes in gene expression levels as analysed by microarray. Agilent microarray analysis of the transcriptome differences between challenged and non-challenged cells revealed a reduced host response of AMØs and BMCs to PCV2b-challenge in vitro at both early and later time-points. This result was particularly surprising considering AMØs are among the first innate immune cell to encounter PCV2 in the lung [54] and function in phagocytic clearance, inflammatory reactions and tissue homeostasis [55], but may also be partially affected by the experimental set-up in which cells were exposed to PCV2b at 0 h, and virus was washed off again straight away. However, this early immunological ignorance of AMØs towards PCV2b infection has been observed by others [53]. Indeed, a recent study utilising a proteomics approach of AMØs infected with PCV2 revealed no obvious changes at 24 h p.i., but resulted only in 9 up-regulated and 12 down-regulated proteins at 48 h p.i. [56], which included cytoskeleton proteins, macromolecular biosynthesis-associated proteins, stress response proteins, signal transduction proteins, energy metabolism, and ubiquitin proteasome pathway-associated proteins. Moreover, nine corresponding genes of the differentially expressed proteins were quantified by real time RT-PCR to examine the transcriptional profiles. Thus, our data regarding gene-expression differences in AMØs fit very well to these data described by proteome-analysis. A decreased immune recognition and response to PCV2b may be a genuine feature of these cells. During the writing of this manuscript, results of a study were published [57] showing that PCV2 infection of AMØs resulted in differential regulation of far more genes than described here or in Cheng et al [56]. Whereas the differences between these results are not completely clear, they can potentially attributed to using a different mock-infection (supernatant of PK15 cells here versus media) or the MOI and virus strain used (UK strain versus Chinese strain, MOI of 0.5 versus MOI of 1). Indeed, several publications have shown that PCV2 isolates from China show a rearranged genotype [58]–[62]. Further explanation for the differences in results could be attributed to the treatment of cells (frozen-thawed versus fresh in the current study) as well as the testing for LPS (all reagents tested for LPS in the current study), which may also explain the TLR activation described in the recent publication [57]. Thus, whereas both studies identified the same central molecules, the resulting differences may be attributed to how the AMØs were treated rather than virus specific differences.
AMØs (24 h) and MoDCs (1 h) had increased gene expression levels of TNF which also represented the only transcript common to two of the three immune cells. TNF is produced by activated MØs in response to microbial/viral stimuli and regulates a wide range of biological activities, including cell differentiation, proliferation and death, as well as inflammatory responses and innate and adaptive immune responses [63]–[65]. An increase in TNF expression in AMØs and MoDCs in response to viral triggers agrees well with previous in vivo studies. Indeed, Kim et al. (2006) [66] showed excessive production of TNFá by porcine parvovirus (PPV) in a dual infection model in vivo, while PCV-2-challenge alone did not induce TNFá to the same extent. Similar results were obtained in a PRRSV/PCV-2 dual infection model of AMØs in vitro [67], [68]. Interestingly, in this model the excessive production of TNF by the co-infecting pathogen potentiated PCV-2-induced PMWS. Signalling through TNF activates various downstream cascades, including activation of transcription factor NF-κB and pro-apoptotic pro-caspases 8 and 10 [63]. In support of this is the enrichment of genes in the NF-κB Signalling pathway in MoDCs 1 h post PCV2b-challenge (Suppl. Tab. 1), the up-regulation of caspase 10 and an enrichment of genes with mainly pro-apoptotic functions 24 h post-challenge. Thereby, our findings further demonstrate a clear role for TNF in the response to PCV-2 challenge in AMØs and MoDCs, substantiating a role of this key cytokine in the aetiology of PMWS.
Further functional analysis of MoDCs revealed an enrichment of genes involved in apoptosis with largely increasing effects at both time-points (Tab. 4). The role of apoptosis in the development of PMWS remains under investigation. While T- and B-lymphocyte depletion in lymphoid tissues, lymphopenia in peripheral blood and a reduction of B cells is a hallmark in pigs that progress to develop clinical signs of PMWS [69], this decline may similarly result from reduced proliferation rather than increased apoptosis [43]. However, it has been shown that ORF3 facilitates PCV-2-induced apoptosis in vitro and in vivo [70]. This study has identified an increased expression of more than 15 pro-apoptotic genes at both time-points suggesting a potential role for apoptosis in the pathogenesis of PMWS and characteristic lesions.
Significant upregulation of five genes within the ‘Proinflammatory Hypercytokinemia/hyperchemokinemia’ pathway (Fig. S1) indicated a strong pro-inflammatory response associated with PCV2b challenge in MoDCs, which resemble inflammatory DC in vivo [71], leading to the recruitment of immune cells to the site of infection. Indeed, a unique histopathological characteristic of PMWS-affected pigs is excessive granulomatous inflammation in lymphoid and other tissues [72]. Pathogenesis of these immune mediated lesions includes the recruitment of circulating monocytes to the site of inflammation [73]. This is facilitated by signalling through NFκB, p38, or MAPKs-mediated regulation of pro-inflammatory cytokine expression (IL1-β, TNF and IL-6) together with chemokines and cell adhesion proteins [74]. This was further supported by the enrichment of differentially-expressed transcripts with biological functions in immune cell trafficking and cell-to-cell signalling and the PCV2b-induced stimulation of pro-inflammatory cytokines and chemoattractants as seen in MoDC are similar to those seen in subclinically PCV2 infected pigs [25].
Antiviral immune responses in vivo are mediated by a variety of cell types, thus, it is important to understand the dynamic interaction between the immune cell subsets and viral pathogens, including PCV2. Our present study has described the transcriptional response of three immune cell subsets challenged with the dominant UK PCV2b strain currently circulating on severely PMWS-affected farms.
Analysis of the differentially-expressed genes showed a distinct cell-type dependent response to PCV2b challenge and identified several key molecules, functions and pathways involved in the host response to PCV2b. The better understanding of these mechanisms could provide new approaches to PCV2 infection and the development of novel immunotherapies or vaccines for the treatment of PMWS.
Supporting Information
Pathway ‘Role of Hypercytokinemia/hyperchemokinemia in the Pathogenesis of Influenza’. Pathway analysis with the IPA software allowed identification of pathways that were differentially expressed between PCV-2b-challenged and unchallenged MoDCs 1 h post infection. The ‘Role of Hypercytokinemia/hyperchemokinemia in the Pathogenesis of Influenza’ pathway was one of the most affected pathways (P = 5.21E-09), with five out of the 44 genes present significantly affected. Significant upregulation of five genes within this pathway indicated a strong pro-inflammatory response associated with PCV-2b challenge in MoDCs.
(TIF)
Network Analysis of MoDC 1 h p.i. Analysis with the IPA software identified transcripts that could be mapped to networks available in the Ingenuity database. In MoDC 1 h p.i., four networks were identified with the highest ranking network revealing a significant link with Cell-To-Cell Signaling and Interaction, Cellular Growth and Proliferation, Renal and Urological System Development and Function.
(TIF)
Network Analysis of MoDC 24 h p.i. Analysis with the IPA software identified transcripts that could be mapped to networks available in the Ingenuity database. In MoDC 24 h p.i., only two networks were identified with the highest ranking network revealing a significant link with Cell Death and Survival, Cellular Development, Hematological System Development and Function.
(TIF)
Network Analysis of BMCs 24 h p.i. Analysis with the IPA software identified transcripts that could be mapped to networks available in the Ingenuity database. In BMCs 24 h p.i., the highest ranking network revealed a significant link with Cellular Development, Hematological System Development and Function, Hematopoiesis.
(TIF)
Pathway analysis of genes differentially expressed in between PCV-2b-challenged and non-challenged immune cell subsets. Differentially expressed genes (P<0.05) were imported into the Ingenuity Pathways Analysis (IPA) software. Canonical pathways analysis identified the pathways from the IPA library of canonical pathways that were most significant to the data set. Molecules from the data set that met the −1.5≤ fold change ≥1.5 and a FDR-adjusted P<0.05 cutoff and were associated with a canonical pathway in the Ingenuity Knowledge Base were considered for the analysis.
(DOCX)
Gene list of DE genes grouped by IPA based on gene function in MoDCs after 1 h and 24 h p.i. Genes in red and bold represent significantly upregulated transcripts expressed in this study. Genes in green and bold represent significantly downregulated transcripts expressed in this study. A network score of >2 was considered significant (p<0.01).
(DOCX)
Gene list of DE genes grouped by IPA based on gene function in BMCs after 24 h p.i. Genes in green and bold represent significantly downregulated transcripts expressed in this study. Genes in red and bold represent significantly upregulated transcripts expressed in this study. A network score of >2 was considered significant (p<0.01).
(DOCX)
Pathway analysis of genes differentially expressed in between PCV2b-challenged and non-challenged immune cell subsets. Differentially expressed genes (P<0.05) were imported into the Ingenuity Pathways Analysis (IPA) software. Canonical pathways analysis identified the pathways from the IPA library of canonical pathways that were most significant to the data set. Molecules from the data set that met the −1.5≤ fold change ≥1.5 and a FDR-adjusted P<0.05 cutoff and were associated with a canonical pathway in the Ingenuity Knowledge Base were considered for the analysis.
(DOCX)
Acknowledgments
We thank Christopher Browne and Ao Zho for help with the experimental animals, and Anna La Rocca for provision of the PK15-ALR-NPro cells. This manuscript is publication No PB_00579 of the RVC.
Funding Statement
This work was funded by a grant (BB/FO18394/1) from the BBSRC ‘Combating Endemic Diseases of Farmed Animals for Sustainability’ (CEDFAS) initiative, with contributions from BPEX, Biobest Laboratories and Zoetis Animal Health. The funder Zoetis Animal Health provided support in the form of salaries for author TK, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of this author are articulated in the ‘author contributions’ section. All other funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Allan GM, Mc NF, Meehan BM, Kennedy S, Mackie DP, et al. (1999) Isolation and characterisation of circoviruses from pigs with wasting syndromes in Spain, Denmark and Northern Ireland. VetMicrobiol 66: 115–123. [DOI] [PubMed] [Google Scholar]
- 2. Bolin SR, Stoffregen WC, Nayar GP, Hamel AL (2001) Postweaning multisystemic wasting syndrome induced after experimental inoculation of cesarean-derived, colostrum-deprived piglets with type 2 porcine circovirus. J VetDiagnInvest 13: 185–194. [DOI] [PubMed] [Google Scholar]
- 3. Cheung AK, Lager KM, Kohutyuk OI, Vincent AL, Henry SC, et al. (2007) Detection of two porcine circovirus type 2 genotypic groups in United States swine herds. ArchVirol 152: 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dupont K, Nielsen EO, Baekbo P, Larsen LE (2008) Genomic analysis of PCV2 isolates from Danish archives and a current PMWS case-control study supports a shift in genotypes with time. VetMicrobiol 128: 56–64. [DOI] [PubMed] [Google Scholar]
- 5. Jacobsen B, Krueger L, Seeliger F, Bruegmann M, Segales J, et al. (2009) Retrospective study on the occurrence of porcine circovirus 2 infection and associated entities in Northern Germany. VetMicrobiol 138: 27–33. [DOI] [PubMed] [Google Scholar]
- 6. de BC, Beven V, Bigarre L, Thiery R, Rose N, et al. (2004) Molecular characterization of Porcine circovirus type 2 isolates from post-weaning multisystemic wasting syndrome-affected and non-affected pigs. J GenVirol 85: 293–304. [DOI] [PubMed] [Google Scholar]
- 7. Carman S, Cai HY, DeLay J, Youssef SA, McEwen BJ, et al. (2008) The emergence of a new strain of porcine circovirus-2 in Ontario and Quebec swine and its association with severe porcine circovirus associated disease–2004–2006. CanJ VetRes 72: 259–268. [PMC free article] [PubMed] [Google Scholar]
- 8. Grau-Roma L, Crisci E, Sibila M, Lopez-Soria S, Nofrarias M, et al. (2008) A proposal on porcine circovirus type 2 (PCV2) genotype definition and their relation with postweaning multisystemic wasting syndrome (PMWS) occurrence. VetMicrobiol 128: 23–35. [DOI] [PubMed] [Google Scholar]
- 9. Larochelle R, Magar R, D'Allaire S (2003) Comparative serologic and virologic study of commercial swine herds with and without postweaning multisystemic wasting syndrome. CanJ VetRes 67: 114–120. [PMC free article] [PubMed] [Google Scholar]
- 10. Wieland B, Werling D, Nevel A, Rycroft A, Demmers TG, et al. (2012) Porcine circovirus type 2 infection before and during an outbreak of postweaning multisystemic wasting syndrome on a pig farm in the UK. VetRec 170: 596. [DOI] [PubMed] [Google Scholar]
- 11. Gillespie J, Opriessnig T, Meng XJ, Pelzer K, Buechner-Maxwell V (2009) Porcine circovirus type 2 and porcine circovirus-associated disease. J VetInternMed 23: 1151–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Segales J, Olvera A, Grau-Roma L, Charreyre C, Nauwynck H, et al. (2008) PCV-2 genotype definition and nomenclature. VetRec 162: 867–868. [DOI] [PubMed] [Google Scholar]
- 13. Chang HW, Jeng CR, Lin TL, Liu JJ, Chiou MT, et al. (2006) Immunopathological effects of porcine circovirus type 2 (PCV2) on swine alveolar macrophages by in vitro inoculation. VetImmunol Immunopathol 110: 207–219. [DOI] [PubMed] [Google Scholar]
- 14. Vincent IE, Carrasco CP, Herrmann B, Meehan BM, Allan GM, et al. (2003) Dendritic cells harbor infectious porcine circovirus type 2 in the absence of apparent cell modulation or replication of the virus. J Virol 77: 13288–13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garcia-Sastre A, Biron CA (2006) Type 1 interferons and the virus-host relationship: a lesson in detente. Science 312: 879–882. [DOI] [PubMed] [Google Scholar]
- 16. Hansen MS, Segales J, Fernandes LT, Grau-Roma L, Bille-Hansen V, et al. (2013) Detection of Porcine Circovirus Type 2 and Viral Replication by In Situ Hybridization in Primary Lymphoid Organs From Naturally and Experimentally Infected Pigs. Vet Pathol [DOI] [PubMed] [Google Scholar]
- 17. Chae JS, Choi KS (2011) Proinflammatory cytokine expression in the lung of pigs with porcine circovirus type 2-associated respiratory disease. ResVetSci 90: 321–323. [DOI] [PubMed] [Google Scholar]
- 18. Segales J, Domingo M, Chianini F, Majo N, Dominguez J, et al. (2004) Immunosuppression in postweaning multisystemic wasting syndrome affected pigs. VetMicrobiol 98: 151–158. [DOI] [PubMed] [Google Scholar]
- 19. Vincent IE, Balmelli C, Meehan B, Allan G, Summerfield A, et al. (2007) Silencing of natural interferon producing cell activation by porcine circovirus type 2 DNA. Immunology 120: 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vincent IE, Carrasco CP, Guzylack-Piriou L, Herrmann B, McNeilly F, et al. (2005) Subset-dependent modulation of dendritic cell activity by circovirus type 2. Immunology 115: 388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Balmelli C, Steiner E, Moulin H, Peduto N, Herrmann B, et al. (2011) Porcine circovirus type 2 DNA influences cytoskeleton rearrangements in plasmacytoid and monocyte-derived dendritic cells. Immunology 132: 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hasslung FC, Berg M, Allan GM, Meehan BM, McNeilly F, et al. (2003) Identification of a sequence from the genome of porcine circovirus type 2 with an inhibitory effect on IFN-alpha production by porcine PBMCs. J GenVirol 84: 2937–2945. [DOI] [PubMed] [Google Scholar]
- 23. Fink J, Gu F, Ling L, Tolfvenstam T, Olfat F, et al. (2007) Host gene expression profiling of dengue virus infection in cell lines and patients. PLoS Negl Trop Dis 1: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee G, Han D, Song JY, Lee YS, Kang KS, et al. (2010) Genomic expression profiling in lymph nodes with lymphoid depletion from porcine circovirus 2-infected pigs. J GenVirol 91: 2585–2591. [DOI] [PubMed] [Google Scholar]
- 25. Fernandes LT, Tomas A, Bensaid A, Perez-Enciso M, Sibila M, et al. (2009) Exploratory study on the transcriptional profile of pigs subclinically infected with porcine circovirus type 2. Anim Biotechnol 20: 96–109. [DOI] [PubMed] [Google Scholar]
- 26. Fernandes LT, Tomas A, Bensaid A, Sibila M, Sanchez A, et al. (2012) Microarray analysis of mediastinal lymph node of pigs naturally affected by postweaning multisystemic wasting syndrome. Virus Res 165: 134–142. [DOI] [PubMed] [Google Scholar]
- 27. Tomas A, Fernandes LT, Sanchez A, Segales J (2010) Time course differential gene expression in response to porcine circovirus type 2 subclinical infection. Vet Res 41: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tomas A, Fernandes LT, Valero O, Segales J (2008) A meta-analysis on experimental infections with porcine circovirus type 2 (PCV2). Vet Microbiol 132: 260–273. [DOI] [PubMed] [Google Scholar]
- 29. Carrasco CP, Rigden RC, Schaffner R, Gerber H, Neuhaus V, et al. (2001) Porcine dendritic cells generated in vitro: morphological, phenotypic and functional properties. Immunology 104: 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang YW, Dryman BA, Li W, Meng XJ (2009) Porcine DC-SIGN: molecular cloning, gene structure, tissue distribution and binding characteristics. Dev Comp Immunol 33: 464–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Summerfield A, McCullough KC (1997) Porcine bone marrow myeloid cells: phenotype and adhesion molecule expression. J Leukoc Biol 62: 176–185. [DOI] [PubMed] [Google Scholar]
- 32. Steiner E, Balmelli C, Herrmann B, Summerfield A, McCullough K (2008) Porcine circovirus type 2 displays pluripotency in cell targeting. Virology 378: 311–322. [DOI] [PubMed] [Google Scholar]
- 33. Grierson SS, King DP, Sandvik T, Hicks D, Spencer Y, et al. (2004) Detection and genetic typing of type 2 porcine circoviruses in archived pig tissues from the UK. Arch Virol 149: 1171–1183. [DOI] [PubMed] [Google Scholar]
- 34. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Coussens PM, Colvin CJ, Wiersma K, Abouzied A, Sipkovsky S (2002) Gene expression profiling of peripheral blood mononuclear cells from cattle infected with Mycobacterium paratuberculosis. InfectImmun 70: 5494–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yuan L, Hillman JD, Progulske-Fox A (2005) Microarray analysis of quorum-sensing-regulated genes in Porphyromonas gingivalis. InfectImmun 73: 4146–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, et al. (2000) Functional discovery via a compendium of expression profiles. Cell 102: 109–126. [DOI] [PubMed] [Google Scholar]
- 38. Yang IV, Chen E, Hasseman JP, Liang W, Frank BC, et al. (2002) Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol 3: research0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benjamini Y, Hochberg Y (1995) Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. JRoyStatistSocSerB 57 57: 289–300. [Google Scholar]
- 40. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. ClinChem 55: 611–622. [DOI] [PubMed] [Google Scholar]
- 41. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 42. O'Brien V (1998) Viruses and apoptosis. J GenVirol 79 Pt 8: 1833–1845. [DOI] [PubMed] [Google Scholar]
- 43. Resendes AR, Majo N, Segales J, Espadamala J, Mateu E, et al. (2004) Apoptosis in normal lymphoid organs from healthy normal, conventional pigs at different ages detected by TUNEL and cleaved caspase-3 immunohistochemistry in paraffin-embedded tissues. VetImmunol Immunopathol 99: 203–213. [DOI] [PubMed] [Google Scholar]
- 44. Dubois RN, Abramson SB, Crofford L, Gupta RA, Simon LS, et al. (1998) Cyclooxygenase in biology and disease. FASEB J 12: 1063–1073. [PubMed] [Google Scholar]
- 45. Steer SA, Corbett JA (2003) The role and regulation of COX-2 during viral infection. Viral Immunol 16: 447–460. [DOI] [PubMed] [Google Scholar]
- 46. Kalim KW, Groettrup M (2013) Prostaglandin E2 inhibits IL-23 and IL-12 production by human monocytes through down-regulation of their common p40 subunit. MolImmunol 53: 274–282. [DOI] [PubMed] [Google Scholar]
- 47. Obermajer N, Kalinski P (2012) Generation of myeloid-derived suppressor cells using prostaglandin E2. TransplantRes 1: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bruckner M, Dickel D, Singer E, Legler DF (2012) Distinct modulation of chemokine expression patterns in human monocyte-derived dendritic cells by prostaglandin E(2). Cell Immunol 276: 52–58. [DOI] [PubMed] [Google Scholar]
- 49. Perez-Martin E, Rovira A, Calsamiglia M, Mankertz A, Rodriguez F, et al. (2007) A new method to identify cell types that support porcine circovirus type 2 replication in formalin-fixed, paraffin-embedded swine tissues. J VirolMethods 146: 86–95. [DOI] [PubMed] [Google Scholar]
- 50. Allan GM, Ellis JA (2000) Porcine circoviruses: a review. J VetDiagnInvest 12: 3–14. [DOI] [PubMed] [Google Scholar]
- 51. Rosell C, Segales J, Plana-Duran J, Balasch M, Rodriguez-Arrioja GM, et al. (1999) Pathological, immunohistochemical, and in-situ hybridization studies of natural cases of postweaning multisystemic wasting syndrome (PMWS) in pigs. J Comp Pathol 120: 59–78. [DOI] [PubMed] [Google Scholar]
- 52. Sorden SD, Harms PA, Nawagitgul P, Cavanaugh D, Paul PS (1999) Development of a polyclonal-antibody-based immunohistochemical method for the detection of type 2 porcine circovirus in formalin-fixed, paraffin-embedded tissue. J VetDiagnInvest 11: 528–530. [DOI] [PubMed] [Google Scholar]
- 53. Gilpin DF, McCullough K, Meehan BM, McNeilly F, McNair I, et al. (2003) In vitro studies on the infection and replication of porcine circovirus type 2 in cells of the porcine immune system. Vet Immunol Immunopathol 94: 149–161. [DOI] [PubMed] [Google Scholar]
- 54. Gordon SB, Read RC (2002) Macrophage defences against respiratory tract infections. BrMed Bull 61: 45–61. [DOI] [PubMed] [Google Scholar]
- 55. Serhan CN, Brain SD, Buckley CD, Gilroy DW, Haslett C, et al. (2007) Resolution of inflammation: state of the art, definitions and terms. FASEB J 21: 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cheng S, Zhang M, Li W, Wang Y, Liu Y, et al. (2012) Proteomic analysis of porcine alveolar macrophages infected with porcine circovirus type 2. J Proteomics 75: 3258–3269. [DOI] [PubMed] [Google Scholar]
- 57. Li W, Liu S, Wang Y, Deng F, Yan W, et al. (2013) Transcription analysis of the porcine alveolar macrophage response to porcine circovirus type 2. BMC Genomics 14: 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Huang Z, Su S, Wei CY, Xie JX, Zhu WJ, et al. (2012) Complete genome sequence of a novel field strain of rearranged porcine circovirus type 2 in southern China. J Virol 86: 10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li B, Ma J, Liu Y, Wen L, Yu Z, et al. (2012) Complete genome sequence of a highly prevalent porcine circovirus 2 isolated from piglet stool samples in China. J Virol 86: 4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lin SQ, Shen JG, Gao FL, Cai W, Huang Z, et al. (2012) Complete genome sequence of narcissus late season yellows virus infecting Chinese narcissus in China. Arch Virol 157: 1821–1824. [DOI] [PubMed] [Google Scholar]
- 61. Wen L, He K, Ni Y, Zhang X, Li B, et al. (2012) Complete genome sequence of the rearranged porcine circovirus type 2. J Virol 86: 5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wen L, He K, Yu Z, Mao A, Ni Y, et al. (2012) Complete genome sequence of a novel porcine circovirus-like agent. J Virol 86: 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Benedict CA (2003) Viruses and the TNF-related cytokines, an evolving battle. Cytokine Growth Factor Rev 14: 349–357. [DOI] [PubMed] [Google Scholar]
- 64. Rahman MM, McFadden G (2006) Modulation of tumor necrosis factor by microbial pathogens. PLoS Pathog 2: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yarilina A, Park-Min KH, Antoniv T, Hu X, Ivashkiv LB (2008) TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat Immunol 9: 378–387. [DOI] [PubMed] [Google Scholar]
- 66. Kim J, Ha Y, Chae C (2006) Potentiation of porcine circovirus 2-induced postweaning multisystemic wasting syndrome by porcine parvovirus is associated with excessive production of tumor necrosis factor-alpha. Vet Pathol 43: 718–725. [DOI] [PubMed] [Google Scholar]
- 67. Chang HW, Jeng CR, Liu JJ, Lin TL, Chang CC, et al. (2005) Reduction of porcine reproductive and respiratory syndrome virus (PRRSV) infection in swine alveolar macrophages by porcine circovirus 2 (PCV2)-induced interferon-alpha. Vet Microbiol 108: 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sinha A, Lin K, Hemann M, Shen H, Beach NM, et al. (2012) ORF1 but not ORF2 dependent differences are important for in vitro replication of PCV2 in porcine alveolar macrophages singularly or coinfected with PRRSV. Vet Microbiol 158: 95–103. [DOI] [PubMed] [Google Scholar]
- 69. Harding JC (2004) The clinical expression and emergence of porcine circovirus 2. Vet Microbiol 98: 131–135. [DOI] [PubMed] [Google Scholar]
- 70. Liu J, Zhu Y, Chen I, Lau J, He F, et al. (2007) The ORF3 protein of porcine circovirus type 2 interacts with porcine ubiquitin E3 ligase Pirh2 and facilitates p53 expression in viral infection. J Virol 81: 9560–9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Segura E, Touzot M, Bohineust A, Cappuccio A, Chiocchia G, et al. (2013) Human inflammatory dendritic cells induce Th17 cell differentiation. Immunity 38: 336–348. [DOI] [PubMed] [Google Scholar]
- 72. Krakowka S, Ellis JA, McNeilly F, Gilpin D, Meehan B, et al. (2002) Immunologic features of porcine circovirus type 2 infection. Viral Immunol 15: 567–582. [DOI] [PubMed] [Google Scholar]
- 73. Sibille Y, Reynolds HY (1990) Macrophages and polymorphonuclear neutrophils in lung defense and injury. Am Rev Respir Dis 141: 471–501. [DOI] [PubMed] [Google Scholar]
- 74. Beck IM, Vanden Berghe W, Vermeulen L, Yamamoto KR, Haegeman G, et al. (2009) Crosstalk in inflammation: the interplay of glucocorticoid receptor-based mechanisms and kinases and phosphatases. Endocr Rev 30: 830–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pathway ‘Role of Hypercytokinemia/hyperchemokinemia in the Pathogenesis of Influenza’. Pathway analysis with the IPA software allowed identification of pathways that were differentially expressed between PCV-2b-challenged and unchallenged MoDCs 1 h post infection. The ‘Role of Hypercytokinemia/hyperchemokinemia in the Pathogenesis of Influenza’ pathway was one of the most affected pathways (P = 5.21E-09), with five out of the 44 genes present significantly affected. Significant upregulation of five genes within this pathway indicated a strong pro-inflammatory response associated with PCV-2b challenge in MoDCs.
(TIF)
Network Analysis of MoDC 1 h p.i. Analysis with the IPA software identified transcripts that could be mapped to networks available in the Ingenuity database. In MoDC 1 h p.i., four networks were identified with the highest ranking network revealing a significant link with Cell-To-Cell Signaling and Interaction, Cellular Growth and Proliferation, Renal and Urological System Development and Function.
(TIF)
Network Analysis of MoDC 24 h p.i. Analysis with the IPA software identified transcripts that could be mapped to networks available in the Ingenuity database. In MoDC 24 h p.i., only two networks were identified with the highest ranking network revealing a significant link with Cell Death and Survival, Cellular Development, Hematological System Development and Function.
(TIF)
Network Analysis of BMCs 24 h p.i. Analysis with the IPA software identified transcripts that could be mapped to networks available in the Ingenuity database. In BMCs 24 h p.i., the highest ranking network revealed a significant link with Cellular Development, Hematological System Development and Function, Hematopoiesis.
(TIF)
Pathway analysis of genes differentially expressed in between PCV-2b-challenged and non-challenged immune cell subsets. Differentially expressed genes (P<0.05) were imported into the Ingenuity Pathways Analysis (IPA) software. Canonical pathways analysis identified the pathways from the IPA library of canonical pathways that were most significant to the data set. Molecules from the data set that met the −1.5≤ fold change ≥1.5 and a FDR-adjusted P<0.05 cutoff and were associated with a canonical pathway in the Ingenuity Knowledge Base were considered for the analysis.
(DOCX)
Gene list of DE genes grouped by IPA based on gene function in MoDCs after 1 h and 24 h p.i. Genes in red and bold represent significantly upregulated transcripts expressed in this study. Genes in green and bold represent significantly downregulated transcripts expressed in this study. A network score of >2 was considered significant (p<0.01).
(DOCX)
Gene list of DE genes grouped by IPA based on gene function in BMCs after 24 h p.i. Genes in green and bold represent significantly downregulated transcripts expressed in this study. Genes in red and bold represent significantly upregulated transcripts expressed in this study. A network score of >2 was considered significant (p<0.01).
(DOCX)
Pathway analysis of genes differentially expressed in between PCV2b-challenged and non-challenged immune cell subsets. Differentially expressed genes (P<0.05) were imported into the Ingenuity Pathways Analysis (IPA) software. Canonical pathways analysis identified the pathways from the IPA library of canonical pathways that were most significant to the data set. Molecules from the data set that met the −1.5≤ fold change ≥1.5 and a FDR-adjusted P<0.05 cutoff and were associated with a canonical pathway in the Ingenuity Knowledge Base were considered for the analysis.
(DOCX)