Abstract
Purpose
Subclones bearing the epidermal growth factor receptor (EGFR) T790M mutation concomitant with deletional mutation in exon 19 (Del19) or a point mutation in exon 21 (L858R) have been reported in non-small cell lung cancer patients before any EGFR tyrosine-kinase inhibitor (TKI) treatment. The effect of pretreatment T790M mutation on the survival of patients with both mutations treated with EGFR TKI is still being debated.
Methods
A meta-analysis was undertaken to pool eligible trials to better elucidate whether pretreatment T790M mutation predicts a poorer outcome of EGFR TKI treatment. Hazard ratios and their 95% confidence intervals for progression-free survival (PFS) were extracted and used under the random-effects model.
Results
A total of 246 patients with activating EGFR mutation such as Del19 or L858R participated in four selected trials from 350 articles. The overall incidence of patients with pretreatment T790M mutation was 43.10% (106/246), ranging from 34.88% to 80.00% in the individual trials. The combined hazard ratio for PFS in all four eligible studies was 2.602 (95% confidence interval 1.011–6.695; P=0.047), indicating a shorter PFS in patients who had the T790M mutation before receiving EGFR TKI treatment. Heterogeneity testing indicated significant heterogeneity but the absence of publication bias.
Conclusion
The meta-analysis reported here found that pretreatment T790M mutation had a negative impact on the PFS of non-small cell lung cancer patients with a Del19 or L858R EGFR mutation who received EGFR TKI treatment.
Keywords: EGFR, primary resistance, TKI, Del19, L858R, prediction
Introduction
Epidermal growth factor receptor (EGFR) tyrosine-kinase inhibitor (TKI) has been proven to be an effective first-line treatment for non-small cell lung cancer (NSCLC) patients harboring the activating EGFR mutation of exon 19 deletion or L858R missense mutation,1–5 with a response rate of over 70%, PFS of approximately 9–10 months6 and overall median survival of 30 months.7,8 However, acquired resistance to EGFR TKI therapy almost always develops, even in those who initially had dramatic responses.9 In nearly half of all cases, resistance is caused by a secondary missense T790M mutation.
However, most research has focused on T790M mutation as an acquired resistance after EGFR TKI treatment or on its mechanism in resistance, and the de novo fT790M mutation that existed at baseline before TKI treatment has been ignored in spite of its potential importance. Although always below the detection level of conventional DNA sequencing,10,11 this “baseline T790M” incidence has been reported to be much higher in recent trials using new detection methods. As a result, recently several trials have been published dealing with the predictive effect of pretreatment T790M mutation in activating EGFR mutation-positive patients who received TKI treatment. However, these trials were small in size, used different detection methods, and were unable to establish a definitive relationship between pretreatment T790M and progression-free survival (PFS) in EGFR TKI treatment. As far as we are aware, the metaanalysis presented here is the first major attempt to bridge this knowledge gap by using data pooled from those papers quantitatively, to gain insights into the predictive effect of pretreatment T790M mutation, and whether its detection could be useful in the management of activating EGFR mutation-positive NSCLC patients.
Methods
Eligibility criteria
Prospective studies to be included in this meta-analysis had to meet the following criteria:
Patients enrolled had to have had histologically confirmed stage III or IV NSCLC.
Patients must have had activating EGFR mutations with no EGFR TKI treatment before the study.
Patients had to have been treated with either erlotinib or gefitinib in the study.
T790M had to have been evaluated in the primary lung-cancer tissue of patients before EGFR TKI treatment, without limitation to the detection methods.
Hazard ratios (HRs) and their confidence intervals (CIs) for PFS according to pretreatment T790M mutation status had to have been either reported in the study or calculable by us using data in the original article.
The research had to have been published in English before May 2013.
Literature search strategy
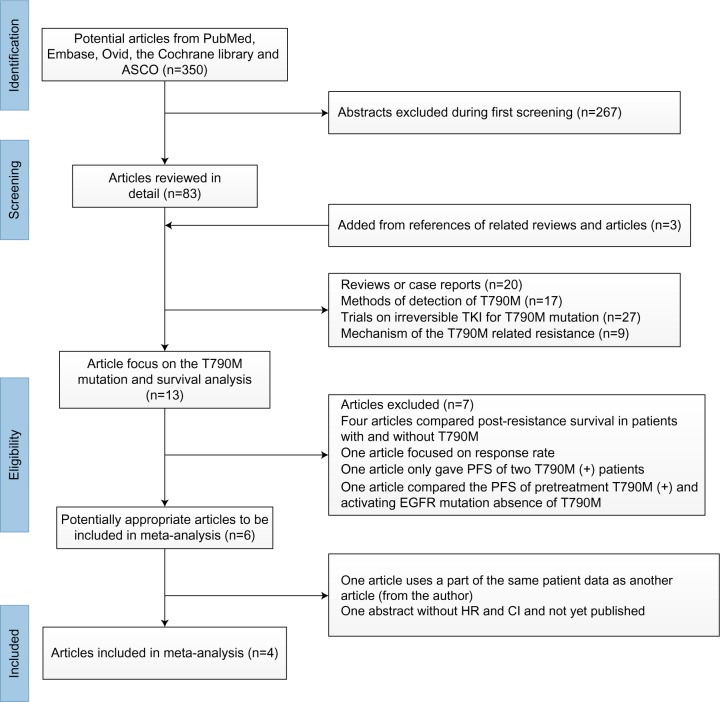
Potential studies were identified by searching PubMed, Embase, Ovid The Cochrane Library, and American Society of Clinical Oncology (ASCO) abstracts. The terms “NSCLC”, “epidermal growth factor receptor”, “T790M”, “PFS”, and “pretreatment”, were used to identify a total of 350 articles once duplicates were discounted. Thirteen potentially eligible trials focusing on the prediction of T790M mutation on survival analysis were identified. Of these, seven studies were excluded: four compared the post-resistance survival in patients with or without T790M mutation;12,13 one trial did not assess the PFS of patients with pretreatment T790M;10 one trial only gave the PFS for two patients with pretreatment T790M mutation and did not have an appropriate control arm;14 and one trial compared the PFS of patients with pretreatment T790M mutation but no activating mutation and patients with activating EGFR mutation.15 Of the remaining six trials, one was an abstract with only the response rate provided, with the full article not yet published;16 one17 used a part of the same patient group as another trial18 – these two trials were excluded; and the remaining four trials were included in the meta-analysis (Figure 1).18–21 The reference lists of all relevant articles were also searched for additional studies.
Figure 1.
Identification process for eligible studies.
Abbreviations: CI, confidence interval; HR, hazard ratio; EGFR, epidermal growth factor receptor; PFS, progression-free survival; TKI, tyrosine-kinase inhibitor.
Data extraction
Information recorded from each trial included study name, year of publication, method of detection, sensitivity, whether the specimens were obtained in surgeries or biopsies, stage when specimens were obtained, histology, and percentage of T790M mutation. We extracted the HRs and the associated 95% CIs for PFS outcomes to assess the effect of pretreatment T790M mutation. These values were obtained directly from three trials.18,20,21 The estimated HR and CI of the first-line group in one trial18 were obtained using the methods proposed by Parmar et al.22 In one article, only survival curves were given.19 We emailed the authors for the HR and CI and were honored to receive also the original PFS for all the patients in that study. Multivariate Cox proportional hazard regression analysis was used to evaluate the HRs and CIs (in Fujita et al’s study,19 only 35 patients with activating EGFR mutations were included).
Information was carefully extracted from all eligible publications independently by two of the authors according to the inclusion criteria presented here. Any disagreement was discussed with the third author until consensus was achieved.
Statistical analysis
HRs and their 95% CIs were used to combine the data. A random effect model by DerSimonian and Laird23 was applied, since this method permits the combined assessment of heterogeneous studies to produce combined HR and 95% CI values. Heterogeneity of the individual HRs was calculated using chi-square tests according to Yusuf et al’s method,24 as well as the heterogeneity tests with I2 and Q statistics. Publication bias was examined by a method described by Begg and Mazumdar25 and Egger et al’s test.26 All P-values presented are two-tailed, with those <0.05 considered statistically significant.
Stata software (v 12.0; StataCorp, College Station, TX, USA) was used to perform most of the data analysis.
Results
Study and patient demographics
A total of 246 patients participated in the four trials (mean 61 per study; range 26–129). All the patients harbored activating EGFR mutations and were treated with gefitinib or erlotinib. All the trials dealt with advanced-stage NSCLC patients. Specimens were all tumor tissue before TKI use, biopsied at an advanced stage in three trials18,20,21 and surgically resected in one trial.19 In all four studies included in this meta-analysis, pretreatment T790M mutation was associated with PFS with EGFR TKI therapy, with three (75%) linking pretreatment T790M with shorter PFS and one (25%) reporting no significant impact on PFS. Table 1 summarizes the main details of the included trials.
Table 1.
Studies selected for meta-analysis
| Study | Year | Number of patients | Method | Sensitivity of method (%) | Specimen | Histology | T790M mutation, n (%) |
|---|---|---|---|---|---|---|---|
| Fujita et al19 | 2012 | 35 | CH | 0.01 | Surgery | AD | 28 (80) |
| Maheswaran et al20 | 2008 | 26 | SARMs assay | 0.1 | Biopsy | AD | 10 (38.46) |
| Rosell et al18 | 2011 | 129 | TaqMan® assay | 0.02 | Biopsy | AD | 45 (34.88) |
| Su et al21 | 2012 | 56 | MALDI-TOF MS | 0.4 | Biopsy | AD (94.3%) SQ (5.7%)* | 23 (41.07) |
Note:
The percentage of 88 patients including 56 activating epidermal growth factor receptor-positive patients.
Abbreviations: AD, adenocarcinoma; CH, colony hybridization; MALDI-TOF MS, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; SARMs, selective androgen-receptor modulators; SQ, squamous-cell carcinoma.
Pretreatment T790M mutation in the four individual studies ranged from 34.88% to 80.00%, while the overall incidence was 43.10% (106/246 patients). Pretreatment T790M mutation was detected by four different methods, modified colony hybridization (CH),19 allele-specific selective androgen-receptor modulators (SARMs) assay,20 highly sensitivity matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS),21 and TaqMan® assay,18 with a sensitivity of 0.01%, 0.1%, 0.4%, and 0.02%, respectively. The pretreatment T790M mutation positivity differed according to the detection method used: 41.01% with MALDI-TOF MS, 38.46% with SARMs assay, 34.88% with TaqMan assay, and 80.00% with CH. Activating EGFR mutation was evaluated by direct sequence in all studies. Three trials examined patients with adenocarcinomas,18–20 while the remaining study21 investigated 94.3% of adenocarcinomas and 5.7% of squamous-cell carcinomas.
Survival
The combined HR for all four eligible studies was 2.602 (95% CI 1.011–6.695; P=0.047), indicating that pretreatment T790M mutation had a negative impact on the PFS of activating EGFR mutation-positive patients treated with EGFR TKI (Figure 2). Heterogeneity was tested (Q=14.65, P=0.002, I2=79.5). There is an important factor that should be taken into account. Was TKI the first-line treatment for the included patients? In fact, all the patients were treated with TKI without any treatment history except in one study,18 in which 64 of 129 patients were second-line TKI treated patients. In this study,18 we combined the HRs of only first-line TKI treated patients. The pooled HR was 1.954 (95% CI 1.068 to 3.574; P=0.03) (Figure 3).
Figure 2.
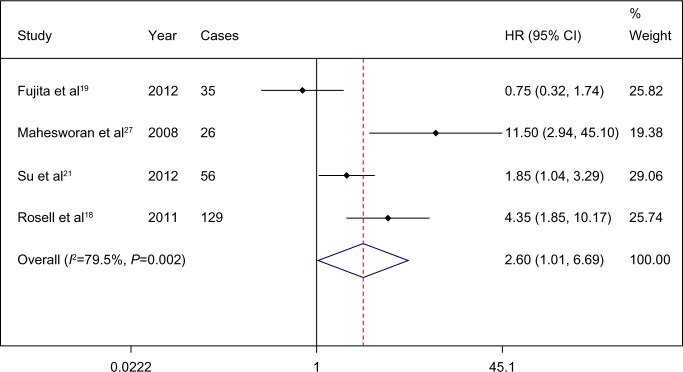
Comparison of progression-free survival between patients with and without a T790M epidermal growth factor receptor (EGFR) mutation prior to treatment with an EGFR tyrosine-kinase inhibitor.
Note: Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; HR, hazard ratio.
Figure 3.
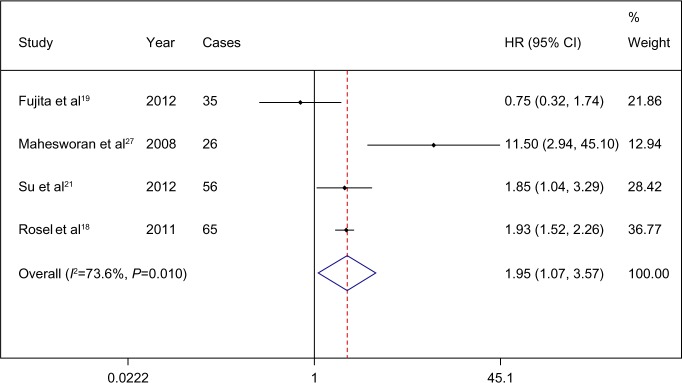
Comparison of progression-free survival between patients with and without a T790M epidermal growth factor receptor (EGFR) mutation prior to first-line treatment with an EGFR tyrosine-kinase inhibitor.
Note: Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; HR, hazard ratio.
Publication bias
To assess publication bias, Begg and Mazumdar’s25 and Egger et al’s26 tests were used, and P-values of 0.308 and 0.424, respectively, were obtained for all four studies; for all three advanced-stage specimen studies, P-values of 0.296 and 0.095, respectively, were obtained. These results suggest an absence of publication bias in all studies.
Discussion
Previously, T790M drug-resistance mutation has generally been considered an acquired mutation, as it has been found in 40% to 50% of patients who had clinical tumor progression after response to TKI.1,2,27 However, in the four studies considered in our meta-analysis and some other studies, T790M mutation was found in more than 30% of patients at baseline, with the highest incidence being 80.00%.19 No association was found between the pretreatment T790M mutation and patient age, size of the tumor, or number of lymph nodes with tumor metastasis.11 Understanding the mechanisms of T790M mutation-related resistance and dealing with the resistance require the correct evaluation of the impact of pretreatment T790M mutation on EGFR TKI treatment in NSCLC patients with activating EGFR mutation.
The present meta-analysis summarized for the first time, as far as we are aware, nearly all of the available research discussing the impact of pretreatment T790M mutation on the survival of NSCLC patients who receive EGFR TKI treatment. The combined analysis of four published studies, which included 246 patients with NSCLC, yielded summary statistics indicating that pretreatment T790M mutation has a negative impact on PFS.
Significant heterogeneity was detected with heterogeneity testing between the four studies in our analysis; the same result was found even after pooling only first-line TKI treatment studies. Fujita et al’s study,19 which showed no significant impact of pretreatment T790M mutation on the PFS of patients receiving EGFR TKI treatment, had the highest incidence of T790M mutation and may have been the source of most of the heterogeneity in the meta-analysis; when this study was excluded, the combined HR was 3.88 (95% CI 1.484–10.184; P=0.006), with inter-study heterogeneity remaining. With the high percentage of T790M mutation detected using the CH method,19 the authors themselves elevated the cutoff sensitivity to 0.5%, resulting in a lower incidence of T790M mutation of 22.9% (calculated using the detailed data obtained from the authors), while the heterogeneity was even more obvious (Q=25.67, P=0.001, I2=88.3). The difference in detection methods could explain the heterogeneity. Unfortunately, since the number of studies focused on the pretreatment T790M issue is limited, and each study in the meta-analysis used a different method of detection, we could not determine which method of detection was the most reliable.
Publication bias25 is a well-known problem in meta-analysis, since positive results tend to be accepted by journals but negative results tend to be rejected or not even submitted by researchers for publication.28 In our analysis, publication bias was not suggested, indicating the summary statistics obtained may approximate the actual average.
Theoretically, pretreatment T790M mutation might have negative impact on PFS due to survival advantage of cells with double mutations in the selective environment of TKI therapy. Some research has shown that the T790M mutant exhibits tyrosine phosphorylation levels comparable to wild-type EGFR, whereas the T790M/L858R double mutant exhibits a substantial increase in phosphorylation, compared with the L858R mutant alone.29 It has also been demonstrated that low percentages of resistant cells in the population (1% and 10%) display similar sensitivity to erlotinib as parental cells (0%),30 whereas sensitivity is reduced when T790M clones made up >25% of the population, which explains why patients whose tumors harbor low levels of T790M can still undergo an objective radiographic response to EGFR TKI treatment,20,31 but relapse. Three studies18,20,21 included in this meta-analysis and two other studies32,33 have all demonstrated a shorter PFS in patients with pretreatment T790M mutation being treated with EGFR TKI. In another part of Maheswaran et al’s study,20 SARMs assay was used for the serial analysis of circulating tumor cells in the blood, and an increased prevalence of the resistance T790M allele within circulating tumor cells over time, during TKI treatment, was shown.
In contrast, Fujita et al19 reported a nonsignificant and even positive correlation between presence of T790M mutation before EGFR TKI treatment and PFS (when the patients were divided with a cutoff sensitivity of 0.5%). Why the predictive role in this trial was found to be positive, in contrast to the results in most of the other trials, is unknown. However, the stage when the specimens were obtained (during curative surgeries) and the treatment received (surgery) are factors that may account for this discrepancy. In contrast, a longer survival time after TKI resistance in patients with T790M mutation was observed in several studies.13 So T790M mutation might be a positive prognostic factor for overall survival but a negative predictive factor for PFS in patients with activating EGFR mutation. There is no reasonable explanation for this as yet. It seems the beginning of T790M mutation could be heterogeneity, and the function as well as incidence of the T790M mutation could be different in different stages, which might also be influenced by treatment such as surgery. It becomes more apparent that the mechanisms underlying acquired TKI resistance are more complex than expected, and multiple factors are involved. The selection advantage is far from enough to elucidate the mechanism. In addition to the documented changes, such as secondary T790M mutation, altered EGFR trafficking, and Met gene amplification,1–3,6 EGFR genetic heterogeneity might also contribute to acquired TKI resistance through the selection of preexisting resistant wild clones, at least in a proportion of NSCLCs.
The lack of an established therapeutic option for NSCLC patients who have progressive disease after EGFR TKI failure poses a great challenge to physicians in terms of how best to manage this growing group of lung-cancer patients. Second-generation TKIs that irreversibly inhibit the EGFR TKI domain may offer the potential for overcoming the resistance of T790M mutation to erlotinib and gefitinib. In the present meta-analysis, we demonstrated the negative impact of pretreatment T790M mutation on the PFS of NSCLC patients under EGFR TKI treatment. Thus, the detection of a minor clone harboring pretreatment drug-resistant EGFR T790M mutation by a suitable method may be useful in predicting the clinical outcome of patients and in choosing appropriate cancer therapies for them.
Conclusion
In our meta-analysis, we found that pretreatment T790M mutation has a negative impact on the PFS of activating EGFR mutation NSCLC patients being treated with EGFR TKI. Although a shorter PFS was observed in several studies in these patients, since the best detection method and the presence of T790M mutation are still in debate, the possible prediction role and function of pretreatment T790M requires further studies to demonstrate.
Acknowledgments
This work was supported by a grant from Wu Jie Ping-Roche Medical Special Foundation (320.6750.1388).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Balak MN, Gong Y, Riely GJ, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12(21):6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 2.Kosaka T, Yatabe Y, Endoh H, et al. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin Cancer Res. 2006;12(19):5764–5769. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schiller JH, Harrington D, Belani CP, et al. Eastern Cooperative Oncology Group. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346(2):92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 5.Scagliotti GV, De Marinis F, Rinaldi M, et al. Italian Lung Cancer Project Phase III randomized trial comparing three platinum-based doublets in advanced non-small-cell lung cancer. J Clin Oncol. 2002;20(21):4285–4291. doi: 10.1200/JCO.2002.02.068. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352(8):786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 7.Mitsudomi T, Morita S, Yatabe Y, et al. West Japan Oncology Group Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11(2):121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 8.Rosell R, Moran T, Queralt C, et al. Spanish Lung Cancer Group Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361(10):958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 9.Morita S, Okamoto I, Kobayashi K, et al. Combined survival analysis of prospective clinical trials of gefitinib for non-small cell lung cancer with EGFR mutations. Clin Cancer Res. 2009;15(13):4493–4498. doi: 10.1158/1078-0432.CCR-09-0391. [DOI] [PubMed] [Google Scholar]
- 10.Inukai M, Toyooka S, Ito S, et al. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res. 2006;66(16):7854–7858. doi: 10.1158/0008-5472.CAN-06-1951. [DOI] [PubMed] [Google Scholar]
- 11.Oh JE, An CH, Yoo NJ, Lee SH. Detection of low-level EGFR T790M mutation in lung cancer tissues. APMIS. 2011;119(7):403–411. doi: 10.1111/j.1600-0463.2011.02738.x. [DOI] [PubMed] [Google Scholar]
- 12.Oxnard GR, Arcila ME, Sima CS, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res. 2011;17(6):1616–1622. doi: 10.1158/1078-0432.CCR-10-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uramoto H, Yamada T, Yano S, Kondo N, Hasegawa S, Tanaka F. Prognostic value of acquired resistance-related molecules in Japanese patients with NSCLC treated with an EGFR-TKI. Anticancer Res. 2012;32(9):3785–3790. [PubMed] [Google Scholar]
- 14.Wu JY, Yu CJ, Chang YC, Yang CH, Shih JY, Yang PC. Effectiveness of tyrosine kinase inhibitors on “uncommon” epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res. 2011;17(11):3812–3821. doi: 10.1158/1078-0432.CCR-10-3408. [DOI] [PubMed] [Google Scholar]
- 15.Mack PC, Holland WS, Burich RA, et al. EGFR mutations detected in plasma are associated with patient outcomes in erlotinib plus docetaxel-treated non-small cell lung cancer. J Thorac Oncol. 2009;4(12):1466–1472. doi: 10.1097/JTO.0b013e3181bbf239. [DOI] [PubMed] [Google Scholar]
- 16.Rosell R, Molina-Vila MA, Taron M, et al. EGFR compound mutants and survival on erlotinib in non-small cell lung cancer (NSCLC) patients (p) in the EURTAC study. J Clin Oncol. 2012;30(Suppl 15):7522. [Google Scholar]
- 17.Karachaliou N, Costa C, Gimenez-Capitan A, et al. Spanish Lung Cancer Group BRCA1, LMO4, and CtIP mRNA expression in erlotinib-treated non-small-cell lung cancer patients with EGFR mutations. J Thorac Oncol. 2013;8(3):295–300. doi: 10.1097/JTO.0b013e31827db621. [DOI] [PubMed] [Google Scholar]
- 18.Rosell R, Molina MA, Costa C, et al. Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res. 2011;17(5):1160–1168. doi: 10.1158/1078-0432.CCR-10-2158. [DOI] [PubMed] [Google Scholar]
- 19.Fujita Y, Suda K, Kimura H, et al. Highly sensitive detection of EGFR T790M mutation using colony hybridization predicts favorable prognosis of patients with lung cancer harboring activating EGFR mutation. J Thorac Oncol. 2012;7(11):1640–1644. doi: 10.1097/JTO.0b013e3182653d7f. [DOI] [PubMed] [Google Scholar]
- 20.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359(4):366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su KY, Chen HY, Li KC, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol. 2012;30(4):433–440. doi: 10.1200/JCO.2011.38.3224. [DOI] [PubMed] [Google Scholar]
- 22.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–2834. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 23.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 24.Yusuf S, Peto R, Lewis J, Collins R, Sleight P. Beta blockade during and after myocardial infarction: an overview of the randomized trials. Prog Cardiovasc Dis. 1985;27(5):335–371. doi: 10.1016/s0033-0620(85)80003-7. [DOI] [PubMed] [Google Scholar]
- 25.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. [PubMed] [Google Scholar]
- 26.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molina-Vila MA, Bertran-Alamillo J, Reguart N, et al. A sensitive method for detecting EGFR mutations in non-small cell lung cancer samples with few tumor cells. J Thorac Oncol. 2008;3(11):1224–1235. doi: 10.1097/JTO.0b013e318189f579. [DOI] [PubMed] [Google Scholar]
- 28.Earleywine M. The file drawer problem in the meta-analysis of subjective responses to alcohol. Am J Psychiatry. 1993;150(9):1435–1436. doi: 10.1176/ajp.150.9.1435c. [DOI] [PubMed] [Google Scholar]
- 29.Mulloy R, Ferrand A, Kim Y, et al. Epidermal growth factor receptor mutants from human lung cancers exhibit enhanced catalytic activity and increased sensitivity to gefitinib. Cancer Res. 2007;67(5):2325–2330. doi: 10.1158/0008-5472.CAN-06-4293. [DOI] [PubMed] [Google Scholar]
- 30.Chmielecki J, Foo J, Oxnard GR, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med. 2011;3(90):90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mok TS. Living with imperfection. J Clin Oncol. 2010;28(2):191–192. doi: 10.1200/JCO.2009.25.8574. [DOI] [PubMed] [Google Scholar]
- 32.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351(8):781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 33.Whitcombe D, Theaker J, Guy SP, Brown T, Little S. Detection of PCR products using self-probing amplicons and fluorescence. Nat Biotechnol. 1999;17(8):804–807. doi: 10.1038/11751. [DOI] [PubMed] [Google Scholar]