Abstract
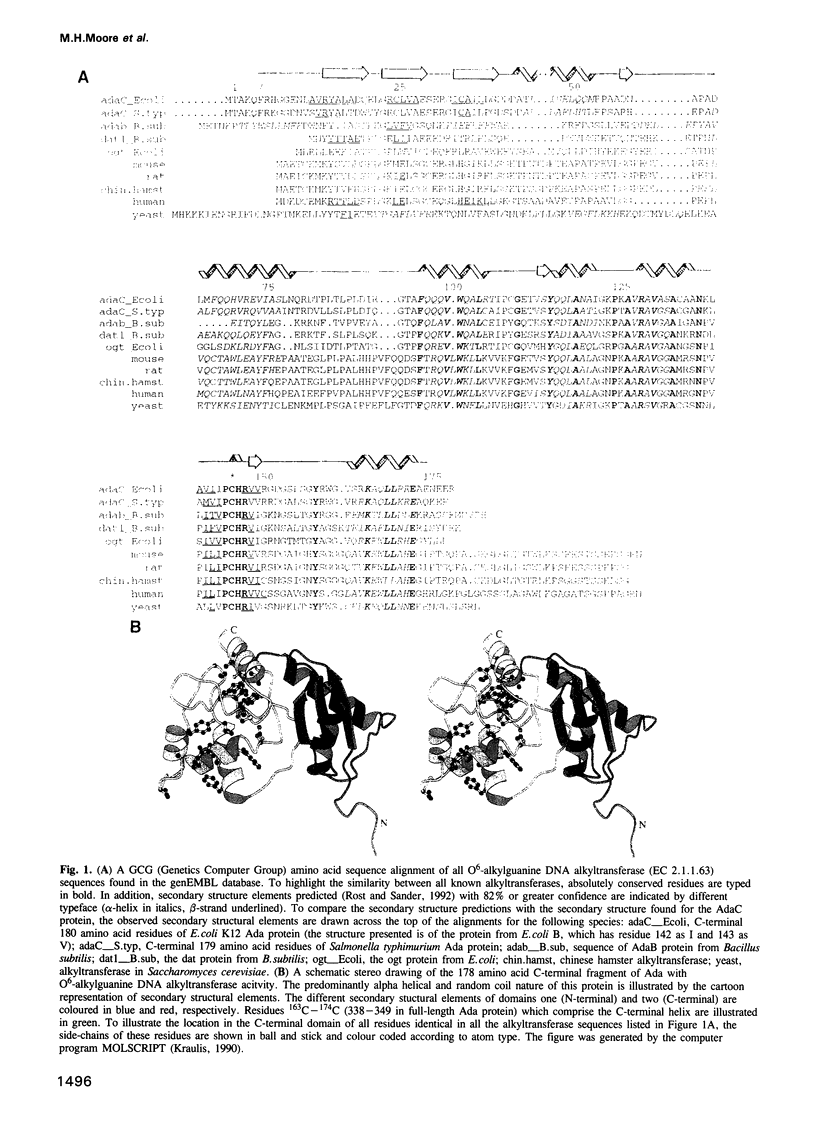
The mutagenic and carcinogenic effects of simple alkylating agents are mainly due to methylation at the O6 position of guanine in DNA. O6-methylguanine directs the incorporation of either thymine or cytosine without blocking DNA replication, resulting in GC to AT transition mutations. In prokaryotic and eukaryotic cells antimutagenic repair is effected by direct reversal of this DNA damage. A suicidal methyltransferase repair protein removes the methyl group from DNA to one of its own cysteine residues. The resulting self-methylation of the active site cysteine renders the protein inactive. Here we report the X-ray structure of the 19 kDa C-terminal domain of the Escherichia coli ada gene product, the prototype of these suicidal methyltransferases. In the crystal structure the active site cysteine is buried. We propose a model for the significant conformational change that the protein must undergo in order to bind DNA and effect methyl transfer.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blow D. M., Birktoft J. J., Hartley B. S. Role of a buried acid group in the mechanism of action of chymotrypsin. Nature. 1969 Jan 25;221(5178):337–340. doi: 10.1038/221337a0. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Costa de Oliveira R., Laval J. The Escherichia coli O6-methylguanine-DNA methyltransferase does not repair promutagenic O6-methylguanine residues when present in Z-DNA. J Biol Chem. 1985 Jul 25;260(15):8711–8715. [PubMed] [Google Scholar]
- Brent T. P., Dolan M. E., Fraenkel-Conrat H., Hall J., Karran P., Laval L., Margison G. P., Montesano R., Pegg A. E., Potter P. M. Repair of O-alkylpyrimidines in mammalian cells: a present consensus. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1759–1762. doi: 10.1073/pnas.85.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Chan C. L., Wu Z., Ciardelli T., Eastman A., Bresnick E. Kinetic and DNA-binding properties of recombinant human O6-methylguanine-DNA methyltransferase. Arch Biochem Biophys. 1993 Jan;300(1):193–200. doi: 10.1006/abbi.1993.1027. [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. IV. Mutagenic specificity in the lacI gene of Escherichia coli. J Mol Biol. 1977 Dec 15;117(3):577–606. doi: 10.1016/0022-2836(77)90059-6. [DOI] [PubMed] [Google Scholar]
- Cowtan K. D., Main P. Improvement of macromolecular electron-density maps by the simultaneous application of real and reciprocal space constraints. Acta Crystallogr D Biol Crystallogr. 1993 Jan 1;49(Pt 1):148–157. doi: 10.1107/S0907444992007698. [DOI] [PubMed] [Google Scholar]
- Demple B., Jacobsson A., Olsson M., Robins P., Lindahl T. Repair of alkylated DNA in Escherichia coli. Physical properties of O6-methylguanine-DNA methyltransferase. J Biol Chem. 1982 Nov 25;257(22):13776–13780. [PubMed] [Google Scholar]
- Demple B. Mutant Escherichia coli Ada proteins simultaneously defective in the repair of O6-methylguanine and in gene activation. Nucleic Acids Res. 1986 Jul 25;14(14):5575–5589. doi: 10.1093/nar/14.14.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B., Sedgwick B., Robins P., Totty N., Waterfield M. D., Lindahl T. Active site and complete sequence of the suicidal methyltransferase that counters alkylation mutagenesis. Proc Natl Acad Sci U S A. 1985 May;82(9):2688–2692. doi: 10.1073/pnas.82.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A. J., Burns P. A., Glickman B. W. N-methyl-N'-nitro-N-nitrosoguanidine induced DNA sequence alteration; non-random components in alkylation mutagenesis. Mutat Res. 1990 Nov-Dec;233(1-2):95–103. doi: 10.1016/0027-5107(90)90154-v. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kalnik M. W., Kouchakdjian M., Li B. F., Swann P. F., Patel D. J. Base pair mismatches and carcinogen-modified bases in DNA: an NMR study of A.C and A.O4meT pairing in dodecanucleotide duplexes. Biochemistry. 1988 Jan 12;27(1):100–108. doi: 10.1021/bi00401a017. [DOI] [PubMed] [Google Scholar]
- Kalnik M. W., Kouchakdjian M., Li B. F., Swann P. F., Patel D. J. Base pair mismatches and carcinogen-modified bases in DNA: an NMR study of G.T and G.O4meT pairing in dodecanucleotide duplexes. Biochemistry. 1988 Jan 12;27(1):108–115. doi: 10.1021/bi00401a018. [DOI] [PubMed] [Google Scholar]
- Katayanagi K., Miyagawa M., Matsushima M., Ishikawa M., Kanaya S., Ikehara M., Matsuzaki T., Morikawa K. Three-dimensional structure of ribonuclease H from E. coli. Nature. 1990 Sep 20;347(6290):306–309. doi: 10.1038/347306a0. [DOI] [PubMed] [Google Scholar]
- Kleene S. J., Toews M. L., Adler J. Isolation of glutamic acid methyl ester from an Escherichia coli membrane protein involved in chemotaxis. J Biol Chem. 1977 May 25;252(10):3214–3218. [PubMed] [Google Scholar]
- Kuo C. F., McRee D. E., Fisher C. L., O'Handley S. F., Cunningham R. P., Tainer J. A. Atomic structure of the DNA repair [4Fe-4S] enzyme endonuclease III. Science. 1992 Oct 16;258(5081):434–440. doi: 10.1126/science.1411536. [DOI] [PubMed] [Google Scholar]
- Leonard G. A., Thomson J., Watson W. P., Brown T. High-resolution structure of a mutagenic lesion in DNA. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9573–9576. doi: 10.1073/pnas.87.24.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Demple B., Robins P. Suicide inactivation of the E. coli O6-methylguanine-DNA methyltransferase. EMBO J. 1982;1(11):1359–1363. doi: 10.1002/j.1460-2075.1982.tb01323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Sedgwick B., Sekiguchi M., Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- Moody P. C., Demple B. Crystallization of O6-methylguanine-DNA methyltransferase from Escherichia coli. J Mol Biol. 1988 Apr 20;200(4):751–752. doi: 10.1016/0022-2836(88)90488-3. [DOI] [PubMed] [Google Scholar]
- Morikawa K., Matsumoto O., Tsujimoto M., Katayanagi K., Ariyoshi M., Doi T., Ikehara M., Inaoka T., Ohtsuka E. X-ray structure of T4 endonuclease V: an excision repair enzyme specific for a pyrimidine dimer. Science. 1992 Apr 24;256(5056):523–526. doi: 10.1126/science.1575827. [DOI] [PubMed] [Google Scholar]
- Olsson M., Lindahl T. Repair of alkylated DNA in Escherichia coli. Methyl group transfer from O6-methylguanine to a protein cysteine residue. J Biol Chem. 1980 Nov 25;255(22):10569–10571. [PubMed] [Google Scholar]
- Patel D. J., Shapiro L., Kozlowski S. A., Gaffney B. L., Jones R. A. Structural studies of the O6meG.C interaction in the d(C-G-C-G-A-A-T-T-C-O6meG-C-G) duplex. Biochemistry. 1986 Mar 11;25(5):1027–1036. doi: 10.1021/bi00353a012. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Shapiro L., Kozlowski S. A., Gaffney B. L., Jones R. A. Structural studies of the O6meG.T interaction in the d(C-G-T-G-A-A-T-T-C-O6meG-C-G) duplex. Biochemistry. 1986 Mar 11;25(5):1036–1042. doi: 10.1021/bi00353a013. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. Jury returns on structure prediction. Nature. 1992 Dec 10;360(6404):540–540. doi: 10.1038/360540b0. [DOI] [PubMed] [Google Scholar]
- Schendel P. F., Robins P. E. Repair of O6-methylguanine in adapted Escherichia coli. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6017–6020. doi: 10.1073/pnas.75.12.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe J. W., Travers A. A. What is evolution playing at? Curr Biol. 1993 Sep 1;3(9):628–630. doi: 10.1016/0960-9822(93)90016-h. [DOI] [PubMed] [Google Scholar]
- Sedgwick B. In vitro proteolytic cleavage of the Escherichia coli Ada protein by the ompT gene product. J Bacteriol. 1989 Apr;171(4):2249–2251. doi: 10.1128/jb.171.4.2249-2251.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick B., Robins P., Totty N., Lindahl T. Functional domains and methyl acceptor sites of the Escherichia coli ada protein. J Biol Chem. 1988 Mar 25;263(9):4430–4433. [PubMed] [Google Scholar]
- Spratt T. E., de los Santos H. Reaction of O6-alkylguanine-DNA alkyltransferase with O6-methylguanine analogues: evidence that the oxygen of O6-methylguanine is protonated by the protein to effect methyl transfer. Biochemistry. 1992 Apr 14;31(14):3688–3694. doi: 10.1021/bi00129a018. [DOI] [PubMed] [Google Scholar]
- Takahashi M., Sakumi K., Sekiguchi M. Interaction of Ada protein with DNA examined by fluorescence anisotropy of the protein. Biochemistry. 1990 Apr 10;29(14):3431–3436. doi: 10.1021/bi00466a002. [DOI] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Demple B., Li B., Lindahl T. Induction of resistance to alkylating agents in E. coli: the ada+ gene product serves both as a regulatory protein and as an enzyme for repair of mutagenic damage. EMBO J. 1984 Sep;3(9):2151–2157. doi: 10.1002/j.1460-2075.1984.tb02105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Hendrickson W. A., Crouch R. J., Satow Y. Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein. Science. 1990 Sep 21;249(4975):1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]