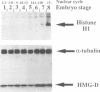
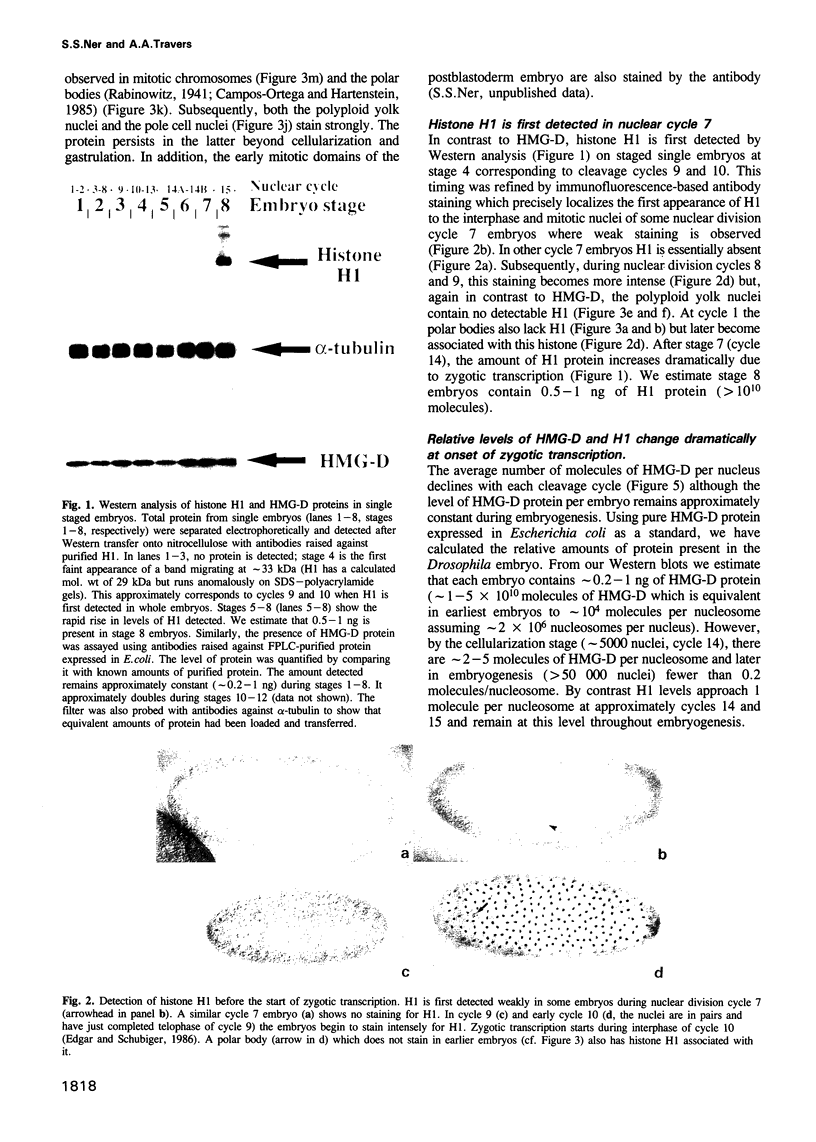
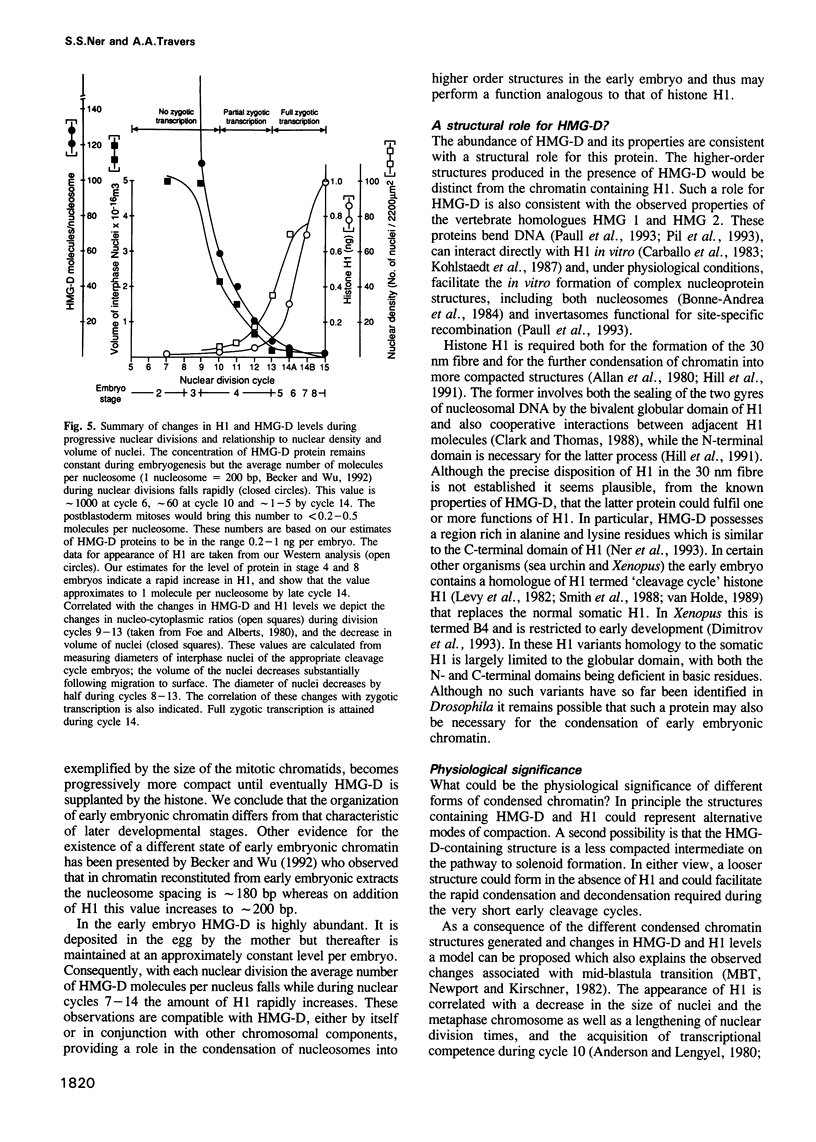
Abstract
We show that HMG-D, an abundant chromosomal protein, is associated with condensed chromatin structures during the first six nuclear cleavage cycles of the developing Drosophila embryo and that histone H1 is absent from these same structures. As H1 accumulates from nuclear division 7 onwards, the nuclei become more compact and transcriptionally active. This compaction is paralleled by a reduction in size of mitotic chromatin. In addition, we find a striking correlation between the switch in HMG-D:H1 ratios and the changes that occur between nuclear cycles 8 and 13 that are collectively termed the mid-blastula transition. This transition is characterized by an increase in the nuclear cycle times, a change in the nucleo-cytoplasmic ratio, and a 5- to 20-fold decrease in nuclear volume. We propose that this is a direct consequence of a re-organization of chromatin from a less condensed state with HMG-D to a more condensed state with H1. We argue that HMG-D, either by itself or in conjunction with other chromosomal proteins, induces a condensed state of chromatin that is distinct from, and less compact than the H1-containing 30 nm fibre and that this state of chromatin could facilitate rapid nuclear cycles.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. The structure of histone H1 and its location in chromatin. Nature. 1980 Dec 25;288(5792):675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- Anderson K. V., Lengyel J. A. Changing rates of histone mRNA synthesis and turnover in Drosophila embryos. Cell. 1980 Oct;21(3):717–727. doi: 10.1016/0092-8674(80)90435-3. [DOI] [PubMed] [Google Scholar]
- Becker P. B., Wu C. Cell-free system for assembly of transcriptionally repressed chromatin from Drosophila embryos. Mol Cell Biol. 1992 May;12(5):2241–2249. doi: 10.1128/mcb.12.5.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M. E., Falciola L., Ferrari S., Lilley D. M. The DNA binding site of HMG1 protein is composed of two similar segments (HMG boxes), both of which have counterparts in other eukaryotic regulatory proteins. EMBO J. 1992 Mar;11(3):1055–1063. doi: 10.1002/j.1460-2075.1992.tb05144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonne-Andrea C., Harper F., Sobczak J., De Recondo A. M. Rat liver HMG1: a physiological nucleosome assembly factor. EMBO J. 1984 May;3(5):1193–1199. doi: 10.1002/j.1460-2075.1984.tb01950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury E. M. Reversible histone modifications and the chromosome cell cycle. Bioessays. 1992 Jan;14(1):9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- Carballo M., Puigdomènech P., Palau J. DNA and histone H1 interact with different domains of HMG 1 and 2 proteins. EMBO J. 1983;2(10):1759–1764. doi: 10.1002/j.1460-2075.1983.tb01654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. J., Thomas J. O. Differences in the binding of H1 variants to DNA. Cooperativity and linker-length related distribution. Eur J Biochem. 1988 Dec 1;178(1):225–233. doi: 10.1111/j.1432-1033.1988.tb14447.x. [DOI] [PubMed] [Google Scholar]
- Dimitrov S., Almouzni G., Dasso M., Wolffe A. P. Chromatin transitions during early Xenopus embryogenesis: changes in histone H4 acetylation and in linker histone type. Dev Biol. 1993 Nov;160(1):214–227. doi: 10.1006/dbio.1993.1299. [DOI] [PubMed] [Google Scholar]
- Edgar B. A., Kiehle C. P., Schubiger G. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell. 1986 Jan 31;44(2):365–372. doi: 10.1016/0092-8674(86)90771-3. [DOI] [PubMed] [Google Scholar]
- Edgar B. A., Schubiger G. Parameters controlling transcriptional activation during early Drosophila development. Cell. 1986 Mar 28;44(6):871–877. doi: 10.1016/0092-8674(86)90009-7. [DOI] [PubMed] [Google Scholar]
- Elgin S. C., Hood L. E. Chromosomal proteins of Drosophila embryos. Biochemistry. 1973 Nov 20;12(24):4984–4991. doi: 10.1021/bi00748a026. [DOI] [PubMed] [Google Scholar]
- Fedor M. J. Chromatin structure and gene expression. Curr Opin Cell Biol. 1992 Jun;4(3):436–443. doi: 10.1016/0955-0674(92)90009-2. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V. E., Alberts B. M. Reversible chromosome condensation induced in Drosophila embryos by anoxia: visualization of interphase nuclear organization. J Cell Biol. 1985 May;100(5):1623–1636. doi: 10.1083/jcb.100.5.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V. E., Alberts B. M. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983 May;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- Hill C. S., Rimmer J. M., Green B. N., Finch J. T., Thomas J. O. Histone-DNA interactions and their modulation by phosphorylation of -Ser-Pro-X-Lys/Arg- motifs. EMBO J. 1991 Jul;10(7):1939–1948. doi: 10.1002/j.1460-2075.1991.tb07720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J. B., Pollock J. M., Jr, Rill R. L. Chromatin fractionation procedure that yields nucleosomes containing near-stoichiometric amounts of high mobility group nonhistone chromosomal proteins. Biochemistry. 1979 Aug 21;18(17):3739–3748. doi: 10.1021/bi00584a015. [DOI] [PubMed] [Google Scholar]
- Kohlstaedt L. A., Sung E. C., Fujishige A., Cole R. D. Non-histone chromosomal protein HMG1 modulates the histone H1-induced condensation of DNA. J Biol Chem. 1987 Jan 15;262(2):524–526. [PubMed] [Google Scholar]
- Lawrence P. A., Johnston P. Pattern formation in the Drosophila embryo: allocation of cells to parasegments by even-skipped and fushi tarazu. Development. 1989 Apr;105(4):761–767. doi: 10.1242/dev.105.4.761. [DOI] [PubMed] [Google Scholar]
- Levy S., Sures I., Kedes L. The nucleotide and amino acid coding sequence of a gene for H1 histone that interacts with euchromatin. The early embryonic H1 gene of the sea urchin Strongylocentrotus purpuratus. J Biol Chem. 1982 Aug 25;257(16):9438–9443. [PubMed] [Google Scholar]
- Lilley D. M. DNA--protein interactions. HMG has DNA wrapped up. Nature. 1992 May 28;357(6376):282–283. doi: 10.1038/357282a0. [DOI] [PubMed] [Google Scholar]
- Ner S. S., Churchill M. E., Searles M. A., Travers A. A. dHMG-Z, a second HMG-1-related protein in Drosophila melanogaster. Nucleic Acids Res. 1993 Sep 11;21(18):4369–4371. doi: 10.1093/nar/21.18.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ner S. S. HMGs everywhere. Curr Biol. 1992 Apr;2(4):208–210. doi: 10.1016/0960-9822(92)90541-h. [DOI] [PubMed] [Google Scholar]
- Newport J., Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982 Oct;30(3):675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Paull T. T., Haykinson M. J., Johnson R. C. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993 Aug;7(8):1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- Pil P. M., Chow C. S., Lippard S. J. High-mobility-group 1 protein mediates DNA bending as determined by ring closures. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9465–9469. doi: 10.1073/pnas.90.20.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. Y., Allis C. D. Chromatin condensation: does histone H1 dephosphorylation play a role? Trends Biochem Sci. 1992 Mar;17(3):93–98. doi: 10.1016/0968-0004(92)90243-3. [DOI] [PubMed] [Google Scholar]
- Ruddell A., Jacobs-Lorena M. Biphasic pattern of histone gene expression during Drosophila oogenesis. Proc Natl Acad Sci U S A. 1985 May;82(10):3316–3319. doi: 10.1073/pnas.82.10.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröter H., Bode J. The binding sites for large and small high-mobility-group (HMG) proteins. Studies on HMG-nucleosome interactions in vitro. Eur J Biochem. 1982 Oct;127(2):429–436. doi: 10.1111/j.1432-1033.1982.tb06890.x. [DOI] [PubMed] [Google Scholar]
- Smith R. C., Dworkin-Rastl E., Dworkin M. B. Expression of a histone H1-like protein is restricted to early Xenopus development. Genes Dev. 1988 Oct;2(10):1284–1295. doi: 10.1101/gad.2.10.1284. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O. The higher order structure of chromatin and histone H1. J Cell Sci Suppl. 1984;1:1–20. doi: 10.1242/jcs.1984.supplement_1.1. [DOI] [PubMed] [Google Scholar]
- Wagner C. R., Hamana K., Elgin S. C. A high-mobility-group protein and its cDNAs from Drosophila melanogaster. Mol Cell Biol. 1992 May;12(5):1915–1923. doi: 10.1128/mcb.12.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]