Abstract
Analysis of ion channel mutants is a widely used approach for dissecting ion channel function and for characterizing the mechanisms of action of channel-directed modulators. Expression of functional potassium channels in potassium-uptake-deficient yeast together with genetic selection approaches offers an unbiased, high-throughput, activity-based readout that can rapidly identify large numbers of active ion channel mutants. Because of the assumption-free nature of the method, detailed biophysical analysis of the functional mutants from such selections can provide new and unexpected insights into both ion channel gating and ion channel modulator mechanisms. Here, we present detailed protocols for generation and identification of functional mutations in potassium channels using yeast selections in the potassium-uptake-deficient strain SGY1528. This approach is applicable for the analysis of structure–function relationships of potassium channels from a wide range of sources including viruses, bacteria, plants, and mammals and can be used as a facile way to probe the interactions between ion channels and small-molecule modulators.
Keywords: SGY1528, Potassium channel, Ion channel, Yeast, Gating
1 Introduction
Accumulation of high concentrations of intracellular potassium is essential for life. In Saccharomyces cerevisiae, two potassium transporters Trk1p and Trk2p serve as the major components of the potassium-uptake system (1, 2) and allow the yeast to grow under low (0.5–1.0 mM) potassium conditions. The double knockout of these transporters, trk1Δtrk2Δ, fails to survive when subjected to low potassium conditions but can be rescued by the heterologous expression of a number of different types of functional potassium channels from viruses (3–5), archaea (6), bacteria (7–9), plants (10–16), and mammals (7, 17–23, 30). This phenotypic rescue is driven by the fact that, unlike animal cells, the membrane potential of yeast is exceptionally negative due to the action of the plasma membrane proton ATPase (24). This situation is enhanced in trk1Δtrk2Δ yeast (25), which have an estimated membrane potential of ~−300 mV (24). Thus, in the presence of a functional, open potassium channel, this negative potential is able to drive intracellular potassium accumulation from the surroundings through the overexpressed channel and allow for rescue of growth. When combined with random or directed mutagenesis protocols, this system can serve as a powerful selection method to identify novel and functional ion channel mutations that provide the starting point for dissection of structure–function relationships. It should be noted that to date, the channels that have been used successfully in this system all have the property of being open under hyperpolarized conditions. For a broader review on different approaches to study ion channel structure and function see ref. 26.
In this chapter, we provide a detailed protocol for generating a random mutagenic library and selection for gain-of-function mutations of a potassium channel in the trk1Δtrk2Δ yeast strain SGY1528 (21) based on a number of studies in which we have used the system to explore both viral and mammalian potassium channel function (4, 5, 18, 19). Additionally, we describe a basic approach towards using the yeast strain to screen for modulator-resistant or modulator-sensitive mutations.
2 Materials
The protocol described here assumes the knowledge of standard molecular biology techniques, E. coli manipulations, and media recipes. For general manipulations with yeast see ref. 27. Use at least ≥98.5% purity-grade chemicals and Milli-Q-grade water.
2.1 Equipment
Tools for colony picking and streaking: while sterile standard pipette tips or microbiological loops can be used for picking and streaking yeast colonies, we routinely use flat toothpicks sterilized in dry conditions in scintillation vials.
Replica plating tool and sterile velveteen squares (Scienceware).
Microtube mixer Tomy MT-360 (Tommytech), or a similar device.
Plasmid DNA isolation kit (Qiagen), or similar.
DNA gel extraction kit (Qiagen), or similar.
0.5 mm acid-washed glass beads, ≥100 μl per sample.
Sterile Whatman paper filter discs (5–10 mm diameter).
0.22 μm sterilization filter units (e.g., Nalgene, cat# 127-0020 or similar).
2.2 Yeast and Plasmids
The protocol describes procedures optimized for the derivative of the Saccharomyces cerevisiae strain W303, SGY1528 (MATa; ade2-1; canl-100; his3-11,15; leu2-3,112; trpl-1; ura3-1; trkl::HIS3; trk2::TRP1) (21). To express potassium channels in yeast, we have been routinely using the multicopy pYes2 (2 μ, URA3, AmpR) shuttle vector (Invitrogen), modified by replacing the galactose-inducible GAL4 promoter with methionine-repressible MET25 promoter (28) as described elsewhere (19). This change allows for an efficient way of controlling channel expression by simply adding or removing methionine from the growth medium. The channel of interest is cloned 5′–3′ between HindIII and XhoI sites of the vector.
2.3 Yeast Media Components and Transformation Solutions
-Ura/-Met dropout powder: 6.0 g glutamic acid, 2.5 g adenine hemisulfate, 1.2 g arginine, 6.0 g aspartic acid, 1.8 g lysine, 3.0 g phenylalanine, 22.5 g serine, 12.0 g threonine, 2.4 g tryptophane, 1.8 g tyrosine, 9.0 g valine, 1.2 g histidine, 3.4 g leucine. Mix well, and store at room temperature for up to 2 years in moisture-free conditions. This component supplies nucleobases and amino acids that are necessary for yeast growth, except for uracil (for plasmid selection) and methionine (to drive protein expression from the MET25 promoter).
1,000× Vitamin stock solution: 2 mg biotin, 400 ng D-pantothenic acid, 400 mg pyridoxine, 400 mg thiamin, 2 g inositol. Dissolve the components in 1 L water, sterilize through a 0.22 μm filter, and store at −20°C for 2–3 years.
1,000× Trace mineral solution: 50 mg boric acid, 4 mg CuSO4, 10 mg KI, 50 mg FeCl3, 40 mg MnSO4, 90 mg molybdic acid, 40 mg ZnSO4. Dissolve the components in 100 ml water, add 1 ml concentrated HCl, sterilize through a 0.22 μm filter, and store at −20°C for up to 3 years.
Li–TE (lithium–Tris–EDTA): 100 mM LiCl, 10 mM Tris–HCl pH 7.5, 1 mM EDTA. Sterilize through a 0.22 μm filter, store at room temperature.
PEG–TE (polyethylene glycol–Tris–EDTA): 50% (w/v) PEG-3350 (polyethylene glycol with average molecular weight 3,350), 10 mM Tris–HCl pH 7.5, 1 mM EDTA. Sterilize through a 0.22 μm filter, store at room temperature.
2.4 Yeast Media
YPAD/100 K, nonselective medium (liquid): 10 g yeast extract, 20 g dextrose, 20 g peptone, 24 mg adenine hemisulfate, 7.46 g KCl (see Note 1). Dissolve the components in 1 L water, sterilize through a 0.22 μm, and store at room temperature for up to 6 months.
YPAD/100 K, nonselective medium (plates): 10 g yeast extract, 20 g dextrose, 20 g peptone, 20 g Bacto agar, 24 mg adenine hemisulfate, 7.46 g KCl (see Note 1). Mix the components in 1 L water, sterilize by autoclaving, and pour the plates (25–30 ml per plate). Leave the plates at room temperature for 24 h to dry. Store the plates in a plastic bag at 4°C for up to 6 months.
-Ura/-Met 100 K, plasmid-selective synthetic medium (liquid): 1.5 g -Ura/-Met dropout powder, 6.7 g yeast nitrogen base (without amino acids), 20 g dextrose, 7.46 g KCl. Dissolve the components in 1 L water, and bring pH to 6.5 with 1 M Tris (free base) solution. Sterilize the medium through a 0.22 μm filter, and store at room temperature for up to 6 months.
-Ura/-Met 100 K, plasmid-selective synthetic medium (plates): 20 g Bacto agar in 500 ml water prepared separately from 1.5 g -Ura/-Met dropout powder, 6.7 g yeast nitrogen base (without amino acids), 7.46 g KCl, water to 450 ml, to pH 6.5 with 1 M Tris (free base) solution. Autoclave both solutions and combine within 10–15 min after autoclaving to avoid premature solidification of the components. Add 50 ml 40% dextrose (sterilized though a 0.22 μm filter) and pour the plates (25–30 ml per plate). Store the plates in a plastic bag at 4°C for up to 1 year.
-Ura/-Met 1 K and -Ura/-Met 0.5 K test medium (plates): 15 g LE agarose in 500 mg water prepared separately from 2.1 g arginine (free base), 1.5 g dropout powder -Ura/-Met, 10 g dextrose, 1 ml 1,000× trace minerals stock solution, 1 ml 1,000× vitamin stock solution, 1 ml 1 M MgSO4, 0.1 ml 1 M CaCl2, 334 μl or 167 μl 3 M KCl for -Ura/-Met 1 K and -Ura/-Met 0.5 K medium, respectively, water to 500 ml, pH 6.0 with phosphoric acid and sterilized though a 0.22 μm filter. Autoclave the agar and add the solution to autoclaved agar while hot, and pour plates (25–30 ml per plate). Store the plates in a plastic bag for up to 1 year.
2.5 Molecular Biology Solutions
10× Error-prone Taq buffer: 100 mM Tris–HCl pH 8.3, 500 mM KCl, 70 mM MgCl2, 5 mM MnCl2, 0.1% gelatin.
10× Error-prone dNTP stock solution: 5 mM dATP, 5 mM dGTP, 25 mM dCTP, 25 mM dTTP in Milli-Q-grade water. Aliquot by 15–20 μl and store at −20°C for up to 1 year. Avoid freeze–thaw cycles.
3 Methods
3.1 Preparing Yeast Stock for Long-Term Storage
To prepare SGY1528 yeast stock for long-term storage, streak the cells onto YPAD/100 K plates and grow at 30°C until pinhead-sized individual colonies appear (see Note 2).
Pick 3–5 healthy colonies, inoculate 5 ml YPAD/100 K liquid, and grow in a ventilated tube overnight at 30°C at constant shaking at ~225 rpm (rotation per minute).
Collect the cells by centrifugation for 10 min at 4,000 × g; discard supernatant.
Resuspend the cells in 2.5 ml sterile 30% glycerol.
Aliquot in 0.5 ml samples using screw-cap or regular Eppendorf tubes.
Flash-freeze in liquid nitrogen and store at −80°C for up to 3 years.
3.2 Library Generation Using Taq Polymerase
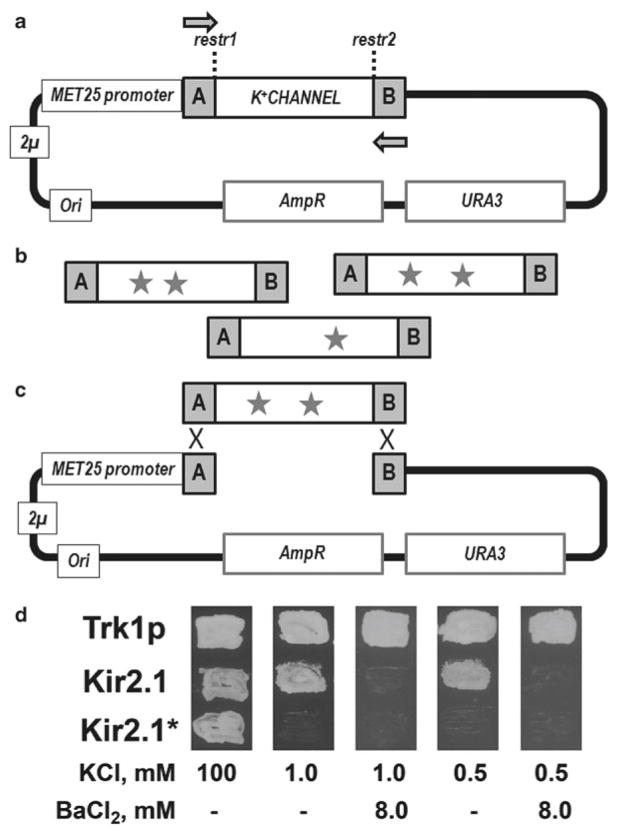
For an excellent collection of various methods for mutagenic library construction, see ref. 29. Here, we present a Taq polymerase-based protocol to generate random library of a full-length gene. To increase the rate of erroneous nucleotide incorporation, PCR is done under error-prone conditions. Of note, the mutations that appear during the initial PCR cycles will be overrepresented in the pool of the mutated PCR products at the end of the reaction. To account for this, we run at least four independent identical PCRs, and combine the PCR products. This protocol generates 1–5 nucleotide substitutions per 1 kb of gene length. The library is generated in a PCR using 40–45 nucleotide long primers that match the regions immediately flanking the targeted gene. Incorporation of the PCR product into the plasmid occurs by homologous recombination between the sites flanking the targeted gene on the PCR product, and the linearized vector backbone (Fig. 1).
Fig. 1.
Library generation and functional expression of a potassium channel in yeast. (a) A scheme of the multicopy pYES2-based vector carrying yeast and bacterial origins of replication (2 μ and Ori, respectively), yeast and bacterial selection markers (AmpR and URA3, respectively), and a potassium channel gene (K+ CHANNEL) under the control of the methionine-repressible MET25 promoter. Arrows depict forward and reverse primers matching the A and B regions, respectively. (b) A library of channel genes carrying random mutations (stars) is generated in an error-prone PCR using the plasmid and primers shown in (a). (c) Diagram depicting heterologous recombination of one channel gene with the linearized vector following preparation by treatment with restr1 and restr2 endo-nucleases. (d) Exemplar images of yeast expressing the Trk1, Kir2.1 or the nonfunctional Kir2.1* mutant on medium supplemented with different amounts of KCl or BaCl2. Trk1p and Kir2.1 but not the inactive Kir2.1* mutant enable yeast growth on medium supplemented with 1.0 and 0.5 mM KCl (-Ura/-Met 1 K and -Ura/-Met 0.5 K media in the protocol). Ba2+ inhibition of Kir2.1, but not the Ba2+-resistant Trk1p channel correlates with yeast growth on the potassium-depleted test medium
Clone the channel of interest into a yeast expression vector using the following scheme: A-restr1-K+CHANNEL-restr2-B, where A and B are regions on the vector flanking the gene of interest (K+CHANNEL), and restr1 and restr2 are sites for endonucleases (in case of the modified pYes2 vector, restr1 and restr2 are HindIII and XhoI, respectively).
To prepare linearized empty vector, cut 10 μg of the construct with resrt1 and restr2 endonucleases, resolve by electrophoresis in agarose, isolate the empty vector from the gel in 50 μl Milli-Q water using the QIAquick gel extraction kit (or similar), and measure DNA concentration by measuring optical of 1.0 = 50.0 μg DNA). Typical condensity at 260 nm (A260 centration of the linearized vector DNA is ~100 μg/μl. It is not necessary to dephosphorylate the vector.
-
Generate mutagenic library in at least four separate identical PCRs (50 μl each). For each reaction, mix the following components in a thin-walled PCR tube:
Template (uncut plasmid carrying the cassette)—20–25 ng.
10× Error-prone buffer—5 μl.
Error-prone dNTP stock solution—4 μl.
Forward primer (A) and reverse primer (B)—2.5 μl of a 20 pmol/μl stock solution (in water) of each primer.
Taq polymerase—5 units.
Water up to 100 μl.
-
Perform PCR cycling using a standard Taq protocol. Example
2 min at 95°C (initial denaturation).
30 s at 95°C (denaturation).
30 s at 62°C (annealing).
[1 min × 1 kb of target DNA length] at 72°C (extension).
Return to step b, 29 times.
5 min at 72°C (final extension).
Analyze the PCR by electrophoresis in DNA-grade agarose.
Isolate the PCR products from the gel (e.g., using QIAquick gel extraction kit from Qiagen) in 30 μl Milli-Q.
Combine the PCR products from the four different reactions in one tube and measure DNA concentration. Typical concentration of the target DNA is ~100 μg/μl. The library is now ready to be mixed with the linearized vector and used for yeast transformation.
3.3 Library Transformation
Streak out yeast from a −80°C stock onto a YPAD/100 mM KCl plate, and grow 3–4 days at 30°C.
Pick 3–4 medium-sized colonies, inoculate each in 5 ml liquid YPAD/100 mM KCl medium, and grow overnight (14–16 h) in a 50 ml conical tube at 30°C at constant shaking at 225 rpm. To provide adequate ventilation, place the tube cap on and secure it with a tape instead of screwing it on.
Add 1.25 ml of the overnight culture to a 250 ml autoclaved flask with 50 ml YPAD/100 mM KCl.
Grow at 30°C at constant shaking at 225 rpm until the culture reaches OD600 0.5–0.8 (usually takes 4.5–5 h).
Collect the cells by centrifugation in a 50 ml conical tube for 10 min at 4,000 × g, and discard supernatant.
Resuspend the cells in 30 ml Li–TE and incubate at 30°C for 1 h without shaking.
Collect the cells by centrifugation and resuspend in 850 μl Li–TE (final volume).
-
For transformation with a plasmid: transfer 100 μl of the cellular suspension into an Eppendorf tube containing 1–2 μg plasmid DNA and 100 μg of single-stranded carrier DNA preheated for 5 min at 95°C (e.g., herring testes carrier DNA from Clontech, cat#S0277).
Add 200 μl PEG–TE, mix by gently pipetting the solution up and down 4–5 times, and incubate at room temperature for 1 h.
Heat-shock for 10 min at 42°C in a heat block.
Incubate at room temperature for 5 min.
Plate 100 μl of each reaction onto one -Ura/-Met 100 K plate, and incubate upside down at 30°C for 3–5 days.
-
For transformation with a library. Prepare library mix by combining 3 μg of non-mutagenized vector with 3 μg of the mutagenized gene, and 300 μg of preheated carrier DNA (see above). Combine the DNA mix with 300 μl of yeast cells.
Add two volumes of PEG–TE to one volume of yeast/DNA suspension, and incubate 20 min at 30°C.
Heat-shock for 12 min at 42°C.
Plate the whole suspension onto -Ura/-Met 100 K plates (100–150 μl per plate), and grow at 30°C for 3–5 days. If the number of transformants on -Ura/-Met 100 K is low (i.e., less than 50 colonies), increase the amount of cells and/or library DNA. Alternatively, increase the ratio of mutagenized gene to linearized vector.
3.4 Functional Selection for Yeast Expressing Active Potassium Channels
Allow 3–5 days for colony formation on -Ura/-Met 100 K medium (see Note 3). Only yeast containing a plasmid expressing the URA3 gene will grow. Although the absence of methionine in the medium will drive the expression of the potassium channel from the plasmid, the high concentration of potassium in the medium will enable growth regardless of the activity of the channel.
Transfer the colonies from -Ura/-Met 100 K onto -Ura/-Met 1 K medium by replica plating, and incubate at 30°C for 2–4 days. Low potassium concentration in the medium will select for cells that express an active potassium channel, such as the yeast Trk1p or mammalian Kir2.1. However, cells expressing poorly functional or nonfunctional potassium channels may grow on this medium too. This may happen due to the presence of residual potassium carried over from the potassium-rich -Ura/-Met 100 K medium. Colonies displaying weak growth due to potassium carryover have a different appearance than those relying on the expressed channel for survival. Comparison with the controls can show some differences. Doubts can be removed by a second replica-plating step onto either -Ura/-Met 1 K or -Ura/-Met 0.5 K.
Transfer colonies from -Ura/-Met 1 K onto -Ura/-Met 0.5 K medium by replica plating, and incubate for 3–7 days. Only colonies expressing active potassium channels will grow on this medium.
Using a sterile toothpick, scoop up as many colonies as possible from the -Ura/-Met 0.5 K plate and inoculate each into 5 ml of liquid -Ura/-Met 100 K medium (see Note 4) in a 50 ml conical tube with a loose cap as described above. Pick and inoculate as many colonies as possible, and grow the cultures for 48 h at 30°C at constant shaking at ~225 rpm.
3.5 Plasmid Isolation Using the QIAquick Miniprep Kit, and Retransformation
Pellet the yeast by centrifugation for 10 min at 5,000 × g and discard supernatant. Wash the cells by resuspending in water and pelleting. Resuspend the cells in 250 μl of RNAse-containing resuspension buffer P1.
Transfer the suspension in an Eppendorf tube containing 100 μl glass beads.
Vortex the tube for 4 min at room temperature.
Add 350 μl of the lysis buffer P2, and incubate 10 min at room temperature.
Add 500 μl of N3, mix well, and centrifuge for 10 min at 15,000×g at 4°C.
Isolate plasmid DNA from the supernatant using the QIAprep columns and the protocol as for a regular DNA miniprep. Briefly, bind DNA to a QIAprep spin column, wash with 750 μl of ethanol-containing wash buffer PE, dry the membrane by additional centrifugation at high speed, and elute DNA in 30 μl water.
Use 10–15 μl of the yeast plasmid miniprep to transform E. coli, using chemical or electroporation approaches. Regardless of the choice of the transformation method, it is advisable to use bacteria with highest competence rate possible.
Prepare minipreps from the bacterial colonies, and transform back into yeast as described above.
Pick 1–2 independent yeast colonies for each of the recovered plasmids and patch (~0.5 × 0.5 mm) onto -Ura/-Met 100 K medium. Pass the patch consecutively onto -Ura/-Met 1 K and -Ura/-Met 0.5 K media to confirm the phenotype exhibited by the plasmid in the initial screen (Fig. 1d).
3.6 Yeast Cultivation in the Presence of Chemical Compounds
Sometimes it is desirable to assess activity of a chemical compound against a potassium channel expressed in yeast. In this case, a blocker or an activator of the channel may be directly added to the -Ura/-Met 0.5 K test medium. An activator is expected to facilitate growth, whereas an inhibitor to suppress yeast growth on this medium. Although the compound may be added directly to the hot medium before pouring the plates, we find it more practical to use one of the following two approaches as they avoid potential inactivation of the compound by the hot media. A liquid compound may be applied to a sterile Whatman paper disc (4–5 mm in diameter), and placed onto solidified plates following replica plating of an evenly spread layer of yeast from the -Ura/-Met 100 K or -Ura/-Met 1 K media. In this format, it is possible to test several concentrations of a compound on a single plate, by placing the discs in different sectors of the plate. The halo of yeast growth (or lack thereof) around the disc will be indicative of the effect of the compound. For informative illustrations, see refs. 5, 18. Alternatively, solutions containing the modifier can be applied directly to the plate (e.g., 200 μl of the modifier solution per 10 cm plate with 20–25 ml of solid medium) and spread evenly using a sterile spreader, followed by yeast transfer onto the plate by replica plating. If the plate is prepared properly, the agar will readily imbibe the 200 μl of the modifier solution, and the plate will be ready for replica plating. If the plate stays wet, incubate the plate at room temperature until the modifier solution completely disappears from the surface of the plate.
3.7. Screening for Modulator-Resistant Clones or Modulator-Sensitive Mutations in Potassium Channels
The SGY1528 yeast offers a unique opportunity to pinpoint key structural elements of a potassium channel that mediate its interaction with pharmacological agents. This can be done by utilizing a combination of random or targeted mutagenesis of the potassium channel gene followed by functional selection. For example, it may be desirable to find point mutations in a potassium channel that confer resistance to an inhibitor. In this case, yeast transformed with a library of mutagenized potassium channel are selected on -Ura/-Met 0.5 K medium in the presence of the compound. Yeast cells that bear a mutation in the channel that confers resistance to the inhibitor without compromising channel function will grow on this medium. Plasmids bearing the mutated channels are isolated and retested as described above. To perform functional selection on the -Ura/-Met 0.5 K medium, it is most convenient to apply the compound directly on the plate and spread it evenly using a sterile spreader (see notes in the previous section), and then transfer the yeast onto the plate by replica plating from the -Ura/-Met 100 K or -Ura/-Met 1 K media. Alternatively, the compound may be added onto a Whatman filter and placed in the middle of the -Ura/-Met 0.5 K plate following yeast transfer. For example, see refs. 5, 18.
Acknowledgments
This work was supported by grants to D.L.M. from NIH (NS065448, MH093603) and the American Heart Association (0740019 N), and to S.N.B. from the Life Sciences Research Foundation. D.L.M. is an AHA established investigator. S.N.B. is a Genentech fellow of the Life Sciences Research Foundation.
Footnotes
While we routinely use YPAD/100 K medium for nonselective cultivation of the SGY1528 strain, a simpler YPD medium, which is widely used for cultivation of the standard laboratory yeast strains (e.g., W303, 74D), can be used for this purpose. YPD has the same recipe as YPAD/100 K but lacks the additional adenine hemisulfate and KCl. The absence of adenine in the YPD medium will cause the cells to develop a red-colored pigment, but will not impact viability.
The strain is extremely sensitive to temperature changes. While S. cerevisiae normally tolerates prolonged incubation at 4°C, and is able to grow for several days at 37°C, the SGY1528 strain quickly looses fitness even after an overnight incubation at 4°C, while growth at 37°C is significantly impaired.
At this point, it is advisable to pick 50–100 colonies, isolate the plasmids, and sequence in order to determine the rate of mutations in the library.
Beware of spontaneous mutants that grow in most stringent potassium-depleted conditions. These colonies are usually characterized by robust growth, and it is therefore advisable not to pick them. If the colony is picked for plasmid isolation and retransformation, the phenotype will not be confirmed in the rescreening round.
References
- 1.Ko CH, Gaber RF. TRK1 and TRK2 encode structurally related K+ transporters. Mol Cell Biol. 1991;11:4266–4273. doi: 10.1128/mcb.11.8.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ko CH, Buckley AM, Gaber RF. TRK2 is required for low affinity K+ transport in Saccharomyces cerevisiae. Genetics. 1990;125:305–312. doi: 10.1093/genetics/125.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gazzarrini S, Kang M, Abenavoli A, Romani G, Olivari C, Gaslini D, Ferrara G, van Etten JL, Kreim M, Kast SM, Thiel G, Moroni A. Chlorella virus ATCV-1 encodes a functional potassium channel of 82 amino acids. Biochem J. 2009;420:295–303. doi: 10.1042/BJ20090095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balss J, Papatheodorou P, Mehmel M, Baumeister D, Hertel B, Delaroque N, Chatelain FC, Minor DL, Jr, Van Etten JL, Rassow J, Moroni A, Thiel G. Transmembrane domain length of viral K+ channels is a signal for mitochondria targeting. Proc Natl Acad Sci U S A. 2008;105:12313–12318. doi: 10.1073/pnas.0805709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatelain FC, Gazzarrini S, Fujiwara Y, Arrigoni C, Domigan C, Ferrara G, Pantoja C, Thiel G, Moroni A, Minor DL., Jr Selection of inhibitor-resistant viral potassium channels identifies a selectivity filter site that affects barium and amantadine block. PLoS One. 2009;4:e7496. doi: 10.1371/journal.pone.0007496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sesti F, Rajan S, Gonzalez-Colaso R, Nikolaeva N, Goldstein SA. Hyperpolarization moves S4 sensors inward to open MVP, a methanococcal voltage-gated potassium channel. Nat Neurosci. 2003;6:353–361. doi: 10.1038/nn1028. [DOI] [PubMed] [Google Scholar]
- 7.Paynter JJ, Andres-Enguix I, Fowler PW, Tottey S, Cheng W, Enkvetchakul D, Bavro VN, Kusakabe Y, Sansom MS, Robinson NJ, Nichols CG, Tucker SJ. Functional complementation and genetic deletion studies of KirBac channels: activatory mutations highlight gating-sensitive domains. J Biol Chem. 2010;285:40754–40761. doi: 10.1074/jbc.M110.175687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paynter JJ, Sarkies P, Andres-Enguix I, Tucker SJ. Genetic selection of activatory mutations in KcsA. Channels (Austin) 2008;2:413–418. doi: 10.4161/chan.2.6.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paynter JJ, Shang L, Bollepalli MK, Baukrowitz T, Tucker SJ. Random mutagenesis screening indicates the absence of a separate H(+)-sensor in the pH-sensitive Kir channels. Channels (Austin) 2010;4(5):390–397. doi: 10.4161/chan.4.5.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson JA, Huprikar SS, Kochian LV, Lucas WJ, Gaber RF. Functional expression of a probably Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992;89:3736–3740. doi: 10.1073/pnas.89.9.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sentenac H, Bonneaud N, Minet M, Lacroute F, Salmon J-M, Gaymard F, Grignon C. Cloning and expression in yeast of a plant potassium ion transport system. Science. 1992;256:663–665. doi: 10.1126/science.1585180. [DOI] [PubMed] [Google Scholar]
- 12.Schachtman DP, Schroeder JI, Lucas WJ, Anderson JA, Gaber RF. Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1 cDNA. Science. 1992;258:1654–1658. doi: 10.1126/science.8966547. [DOI] [PubMed] [Google Scholar]
- 13.Lai HC, Grabe M, Jan YN, Jan LY. The S4 voltage sensor packs against the pore domain in the KAT1 voltage-gated potassium channel. Neuron. 2005;47:395–406. doi: 10.1016/j.neuron.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura RL, Anderson JA, Gaber RF. Determination of key structural requirements of a K+ channel pore. J Biol Chem. 1997;272:1011–1018. doi: 10.1074/jbc.272.2.1011. [DOI] [PubMed] [Google Scholar]
- 15.Rubio F, Schwarz M, Gassmann W, Schroeder JI. Genetic selection of mutations in the high affinity K+ transporter HKT1 that define functions of a loop site for reduced Na+ permeability and increased Na+ tolerance. J Biol Chem. 1999;274:6839–6847. doi: 10.1074/jbc.274.11.6839. [DOI] [PubMed] [Google Scholar]
- 16.Rubio F, Gassmann W, Schroeder JI. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science. 1995;270:1660–1663. doi: 10.1126/science.270.5242.1660. [DOI] [PubMed] [Google Scholar]
- 17.Bichet D, Lin YF, Ibarra CA, Huang CS, Yi BA, Jan YN, Jan LY. Evolving potassium channels by means of yeast selection reveals structural elements important for selectivity. Proc Natl Acad Sci U S A. 2004;101:4441–4446. doi: 10.1073/pnas.0401195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatelain FC, Alagem N, Xu Q, Pancaroglu R, Reuveny E, Minor DL., Jr The pore helix dipole has a minor role in inward rectifier channel function. Neuron. 2005;47:833–843. doi: 10.1016/j.neuron.2005.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minor DL, Jr, Masseling SJ, Jan YN, Jan LY. Transmembrane structure of an inwardly rectifying potassium channel. Cell. 1999;96:879–891. doi: 10.1016/s0092-8674(00)80597-8. [DOI] [PubMed] [Google Scholar]
- 20.Sadja R, Smadja K, Alagem N, Reuveny E. Coupling Gbetagamma-dependent activation to channel opening via pore elements in inwardly rectifying potassium channels. Neuron. 2001;29:669–680. doi: 10.1016/s0896-6273(01)00242-2. [DOI] [PubMed] [Google Scholar]
- 21.Tang W, Ruknudin A, Yang W, Shaw S, Knickerbocker A, Kurtz S. Functional expression of a vertebrate inwardly rectifying K+ channel in yeast. Mol Biol Cell. 1995;6:1231–1240. doi: 10.1091/mbc.6.9.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi BA, Lin YF, Jan YN, Jan LY. Yeast screen for constitutively active mutant G protein-activated potassium channels. Neuron. 2001;29:657–667. doi: 10.1016/s0896-6273(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 23.Zaks-Makhina E, Kim Y, Aizenman E, Levitan ES. Novel neuroprotective K+ channel inhibitor identified by high-throughput screening in yeast. Mol Pharmacol. 2004;65:214–219. doi: 10.1124/mol.65.1.214. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez-Navarro A. Potassium transport in fungi and plants. Biochim Biophys Acta. 2000;1469:1–30. doi: 10.1016/s0304-4157(99)00013-1. [DOI] [PubMed] [Google Scholar]
- 25.Madrid R, Gomez MJ, Ramos J, Rodriguez-Navarro A. Ectopic potassium uptake in trk1 trk2 mutants of Saccharomyces cerevisiae correlates with a highly hyperpolarized membrane potential. J Biol Chem. 1998;273:14838–14844. doi: 10.1074/jbc.273.24.14838. [DOI] [PubMed] [Google Scholar]
- 26.Minor DL., Jr Searching for interesting channels: pairing selection and molecular evolution methods to study ion channel structure and function. Mol Biosyst. 2009;5:802–810. doi: 10.1039/b901708a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 28.Kerjan P, Cherest H, Surdin-Kerjan Y. Nucleotide sequence of the Saccharomyces cerevisiae MET25 gene. Nucleic Acids Res. 1986;14:7861–7871. doi: 10.1093/nar/14.20.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnold FH, Georgiou G. Directed evolution library creations—Methods and protocols. Humana; Totowa, NJ: 2003. [Google Scholar]
- 30.Bagriantsev S, Clark KA, Peyronnet R, Honoré E, Minor DL., Jr Multiple modalities act through a common gate to control K2P channel function. The EMBO Journal. 2011;30:3594–3606. doi: 10.1038/emboj.2011.230. [DOI] [PMC free article] [PubMed] [Google Scholar]