Abstract
Background
BK virus associated nephropathy (BKVN) is associated with an increased risk of graft failure.
Methods
Levels of mRNAs encoding proteins implicated in inflammation and fibrosis were measured in urine collected at the time of biopsy diagnosis of BKVN in 29 kidney graft recipients, and analyzed for prognosticating graft failure using logistic regression.
Results
Ten of 29 BKVN patients had graft failure within 36 months of BKVN diagnosis and the remaining 19 patients did not. Serum creatinine level, BKVN biopsy stage and urinary cell levels of mRNA for PAI-1, vimentin, TIMP-1, fibronectin, granzyme B or perforin were associated with graft failure. A combination of PAI-1 mRNA level, BKVN biopsy stage and creatinine level (P=0.0015, by logistic regression) and a combination of PAI-1 mRNA level and creatinine level (P=0.001) were the best fitting models for prognosticating graft failure, and PAI-1 mRNA level was the only independent predictor (OR=2.8; 95% CI: 1.1 to 6.8, P=0.03) by multivariable analysis. The AUC for the combination of PAI-1 mRNA, biopsy and creatinine was 0.92 (95% CI: 0.80 to 1.0, P<0.001) by ROC curve analysis, and the AUC was 0.92 (95% CI: 0.80 to 1.0, P<0.001) for the combination of PAI-mRNA and creatinine. Graft outcome was correctly predicted in 27 of 29 BKVN patients by either model.
Conclusion
Urinary cell level of PAI-1 mRNA, measured at the time of BKVN diagnosis, is an independent prognosticator of graft failure and a prediction model of serum creatinine and PAI mRNA is as accurate as the model that includes the biopsy result.
Keywords: BK virus nephropathy, Messenger RNA, Kidney transplantation, PCR assay
INTRODUCTION
Polyomavirus BK-associated nephropathy (BKVN) is a serious complication following kidney transplantation and is associated with a high rate of graft failure (1). Biopsy features prognostic of BKVN have been elucidated and the presence of intragraft fibrosis and inflammation have been reported to portend graft functional decline and failure (2, 3). BKVN associated histologic changes however may be focal in nature and sampling errors are a cause of concern (3, 4).
The invasive biopsy procedure has become safer over the years but can still be associated with bleeding, graft loss and rarely death (5, 6). Development of noninvasively measurable biomarkers diagnostic and prognostic of BKVN may improve clinical outcome. We have developed and validated that urinary cell level of BKV-VP1 mRNA is accurately diagnostic of BKVN (7, 8). We have also reported that BKV-VP1 mRNA level, measured at the time of BKVN diagnosis, is not predictive of future graft function whereas levels of granzyme B mRNA and its endogenous antagonist proteinase inhibitor (PI)-9 mRNA are predictive of decline in graft function during the 12-months following BKVN diagnosis (8).
In view of data that the presence of fibrosis as well as inflammation in the kidney allograft with BKVN diagnosis have been associated with subsequent graft functional decline and failure (2, 9) and in the light of our findings that urinary cell mRNA profiles are diagnostic of intragraft inflammation and fibrosis (10–12), we designed and developed a 20 member urinary cell mRNA panel comprised of mRNAs encoding proteins implicated in fibrosis and inflammation and investigated the hypothesis that urinary cell mRNA profiles of urine collected at the time of BKVN diagnosis foretell graft failure in patients with BKVN.
RESULTS
BKVN Patients with or without Graft Failure
One thousand four hundred and fifty patients received a kidney transplant at our center during January 1999 to December 2008, and 38 (2.6%) patients were diagnosed as having BKVN following a clinically indicated kidney allograft biopsy. Urine specimens collected at the time of biopsy were available in 29 of 38 BKVN patients, and these 29 patients constitute the study cohort for this investigation (Table 1).
Table 1.
BKVN Study Cohort
| Patient | Year of Transplant | Year of Diagnosis | Time from Transplant to Biopsy Diagnosis of BKVN (Months) | BKVN Stagea | Plasma BKV DNAb (copies/ml) | Biopsy Creatinine (mg/dL) | Post Biopsy Year 1 Creatinine (mg/dL) | Post Biopsy Year 2 Creatinine (mg/dL) | Post Biopsy Year 3 Creatinine (mg/dL) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1999 | 2000 | 20.2 | C | n/ac | 2 | n/a | n/a | 2 |
| 2 | 2000 | 2000 | 2.8 | A | n/a | 1.4 | 1.8 | 1.8 | 1.7 |
| 3 | 2001 | 2004 | 35.9 | C | 3.8 × 105 | 2.2 | 3.1 | HDd | |
| 4 | 2002 | 2003 | 11.5 | A | n/a | 2.6 | 3 | 2.8 | 2.8 |
| 5 | 2002 | 2005 | 33.6 | B | 1.3 × 106 | 2.2 | 2.7 | 3.5 | 4.1 |
| 6 | 2002 | 2004 | 22.2 | B | 2.0 × 105 | 1.2 | HD | ||
| 7 | 2003 | 2005 | 15.2 | B | 3.2 × 105 | 2 | 2 | 1.9 | 1.7 |
| 8 | 2005 | 2006 | 12.4 | B | 7.5 × 105 | 2 | 2 | 1.8 | 1.6 |
| 9 | 2005 | 2007 | 21 | A | 1.8 × 105 | 1.9 | 2.2 | 1.8 | 2 |
| 10 | 2005 | 2008 | 30.5 | A | 5.8 × 104 | 2.1 | 1.8 | 2.3 | 2.3 |
| 11 | 2005 | 2009 | 43 | A | 2.4 × 105 | 1.8 | 1.9 | 2 | 2.2 |
| 12 | 2005 | 2008 | 33.8 | C | 1.2 × 106 | 2.6 | 3.9 | HD | |
| 13 | 2006 | 2006 | 6.5 | A | 1.7 × 105 | 1.6 | 1.7 | 1.3 | 1.5 |
| 14 | 2006 | 2007 | 13 | A | 1.8 × 106 | 1.4 | 1.4 | 1 | 1 |
| 15 | 2006 | 2009 | 36.7 | B | 1.1 × 107 | 2.2 | 2.9 | 3.2 | 3 |
| 16 | 2006 | 2007 | 8.7 | A | 4.5 × 106 | 2.9 | 4 | HD | |
| 17 | 2006 | 2007 | 19.4 | B | 8.2 × 106 | 1.9 | HD | ||
| 18 | 2006 | 2007 | 12.5 | B | 5.6 × 106 | 3 | HD | ||
| 19 | 2006 | 2006 | 3.2 | B | 5.9 × 105 | 2.6 | HD | ||
| 20 | 2006 | 2006 | 6 | B | 1.6 × 104 | 6.1 | HD | ||
| 21 | 2007 | 2008 | 9.3 | B | 1.7 × 105 | 2.1 | 1.9 | 2 | 1.8 |
| 22 | 2008 | 2008 | 10.7 | B | 1.2 × 105 | 4.7 | 3.2 | 2.9 | 3.4 |
| 23 | 2008 | 2009 | 10.5 | A | 9.0 × 105 | 2.1 | 1.5 | 1.9 | 1.8 |
| 24 | 2008 | 2008 | 5.6 | A | 9.5 × 104 | 2.2 | 2.3 | 2.6 | 2.4 |
| 25 | 2008 | 2008 | 7.3 | A | 2.7 × 105 | 2.1 | 2.3 | 2.8 | 2.8 |
| 26 | 2008 | 2009 | 9.2 | B | 9.0 × 105 | 1.2 | 1.1 | 1.2 | 1 |
| 27 | 2008 | 2008 | 5.5 | A | 7.1 × 105 | 1.9 | 1.8 | 1.9 | 2.4 |
| 28 | 2008 | 2009 | 2.8 | B | 5.1 × 105 | 3.3 | 3.7 | HDc | |
| 29 | 2008 | 2009 | 12.2 | B | 2.5 × 105 | 3.5 | HD |
BKVN stage was determined using the Banff 2009 working proposal (14);
Plasma DNA copies were measured using commercially available assays (http://www.viracor.com)(24)
n/a=not available;
HD = return to hemodialysis.
Ten of the 29 BKVN patients had graft failure requiring chronic hemodialysis therapy (Graft Failure group) within 24 months of biopsy diagnosis of BKVN and the remaining 19 did not experience graft failure for a minimum of 36-months following the biopsy (No Graft Failure group). Five patients were diagnosed with BKVN prior to the 2005 publication of the Interdisciplinary Analyses and Recommendations for monitoring, diagnosis and management of BKVN (13). Two of the five were in the Graft Failure group and three were in the No Graft Failure group (20% vs. 11%; P=1.0). The mean (±SD) graft survival time in the graft failure group was 11±10 months and the mean duration of follow-up in the group without graft failure was 56±21 months, as of February 1, 2013.
The clinical characteristics and kidney graft biopsy findings of the study cohort are summarized in Table 2. Serum creatinine, measured at the time of biopsy diagnosis BKV, was significantly higher in the graft failure group. BKVN in renal allograft biopsies were classified using the Banff 2009 Meeting working proposal (14, 15) and BKVN stage A was more frequent in the group without graft failure and stages B and C in the graft failure group. All patients were managed with a decrease in their immunosuppressive therapy and the clinical management was not significantly different (P>0.05) between the two groups (Table 2).
Table 2.
Clinical Characteristics and Renal Allograft Biopsy Findings in BKVN Patients.
| Baseline parameters | Graft Failure (N=10) | No Graft Failure (N=19) | P Valuea |
|---|---|---|---|
| Age (mean ± SD) | 46±19 | 55±14 | 0.19 |
| Gender, n (% female) | 1 (10%) | 6 (32%) | 0.20 |
| Ethnicity (White/AA /other) | 7/2/1 | 9/7/3 | 0.5 |
| Deceased donor grafts, n (%) | 7 (70%) | 11 (58%) | 0.5 |
| ATG induction, n (%) | 8 (80%) | 15 (79%) | 0.95 |
| Tacrolimus/MMF maintenance | 10 (100%) | 19 (100%) | 1.0 |
| Steroid maintenance, n (%) | 7 (70%) | 8 (42%) | 0.15 |
| Acute Rejection therapy, n (%) | 3 (30%) | 1 (5%) | 0.07 |
| Months from transplantation to biopsy (mean ± SD) | 16±12 | 16± 12 | 0.91 |
| Creatinine (mg/dL) at time of BKVN diagnosis (mean ± SD) | 2.9±1.3 | 2.1±0.7 | 0.02 |
| Plasma BKV DNA (copies/ml) (mean ± SD) | 2.1×106 ± 2.9×106 | 1.2×106 ± 2.7×106 | 0.34 |
| Renal Allograft Biopsy Result | |||
| BKVN Stageb | A - 1 (10%) | A - 11 (58%) | |
| B - 7 (70%) | B - 7 (37%) | ||
| C - 2 (20%) | C - 1 (5%) | 0.02 | |
| Histologic Scoresc (mean ± SD) | |||
| Cytopathic score | 1.9±0.7 | 1.7±0.7 | 0.43 |
| Tubulitis score | 1.3±1.1 | 1.1±0.9 | 0.48 |
| Infiltrate score | 1.4±1.0 | 1.2±0.9 | 0.44 |
| Tubular atrophy score | 1.4±0.8 | 0.8±0.8 | 0.07 |
| Interstitial fibrosis score | 1.1±0.7 | 0.8±0.9 | 0.24 |
| Management of BKVN, n (%) | |||
| Discontinuation of MMF and Reduction of Tacrolimus | 10 (100%) | 19 (100%) | 1.0 |
| Prednisone Maintenance | 8 (80%) | 9 (47%) | 0.12 |
| IVIG therapy | 8 (80%) | 17 (89%) | 0.48 |
| Leflunomide therapy | 9 (90%) | 16 (84%) | 0.67 |
| Acute Rejection Post BKVN | 1 (10%) | 1 (5%) | 1.0 |
Continuous variables were compared using Mann Whitney U test. Categorical variables were compared using Chi-square test or Fisher’s exact test;
BKVN stages are based on the Banff 2009 Meeting Working Proposal for Polyomavirus Nephropathy (14); The proportion of BKVN stage A vs. stages and B and C in those with graft failure were compared to those without graft failure using Fisher’s exact test.
Histologic scores were assigned using the Banff ‘97 Meeting Report (22)
Urinary Cell mRNA Levels in BKVN Patients
Figure 1, box and whisker plots, shows the levels of mRNAs in urine collected at the time of biopsy diagnosis of BKVN. The log-transformed mean (±SD) ratio of plasminogen activator inhibitor-1 (PAI-1) mRNA copies to 18S-rRNA copies in urinary cells was higher in urine from BKVN patients with graft failure compared to patients without graft failure (Fig. 1A, 8.1±0.9 vs. 6.3±1.3; P=0.001, Mann Whitney U test). Levels of mRNA for vimentin (Fig. 1B, 11±0.8 vs.10±1.2; P=0.003), tissue inhibitor of metalloproteinase-1 (TIMP-1) (Fig. 1C, 12±1.3 vs. 11±1.4; P=0.007), fibronectin (Fig. 1D, 8.8±1.3 vs. 7.3±1.5; P=0.008), granzyme B (Fig. 1E, 7.6±1.1 vs. 6.1±2; P=0.02), CD25 (P=0.02, not shown in Fig. 1) and perforin (Fig 1F, 6.9±1.4 vs. 5.4±2; P=0.04) were also higher in those with graft failure compared to those without graft failure. In contrast, urinary cell levels of BKV VP1 mRNA (Fig. 1G, 22±2.9 vs. 22±1.7; P=0.77) and 18S rRNA (Fig. 1H, 8.2±1.2 vs. 8.9±1.1; P=0.1) were not significantly different between the two groups. Also, urinary cell levels of mRNA for CD103, E-cadherin, fibroblast growth factor-2, hepatocyte growth factor, cytotoxic T lymphocyte associated antigen-4, transforming growth factor-beta1, fibroblast specific protein1, collagen 1A1, alpha-smooth muscle actin, sodium-potassium chloride cotransporter-2, Forkhead box transciption factor -3, bone morphogenic protein-7, or uterine sensitization associated antigen-1 were not different between the graft failure group and no graft failure group (Supplemental Digital Content [SDC], Figure S1).
Figure 1. Urinary Cell mRNA Levels in BKVN Patients with or without Graft Failure.
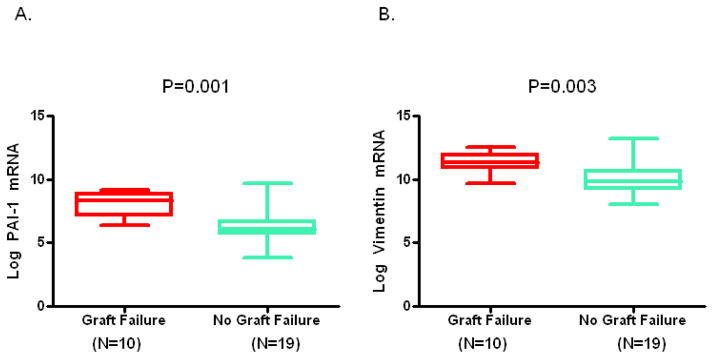
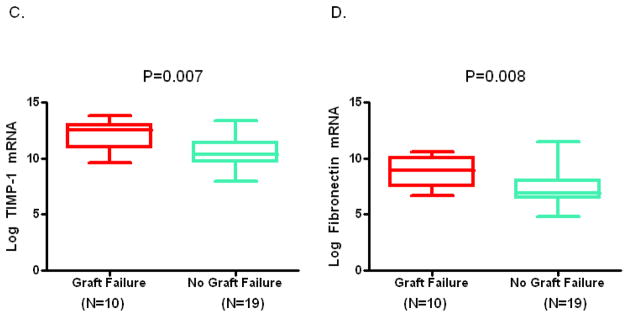
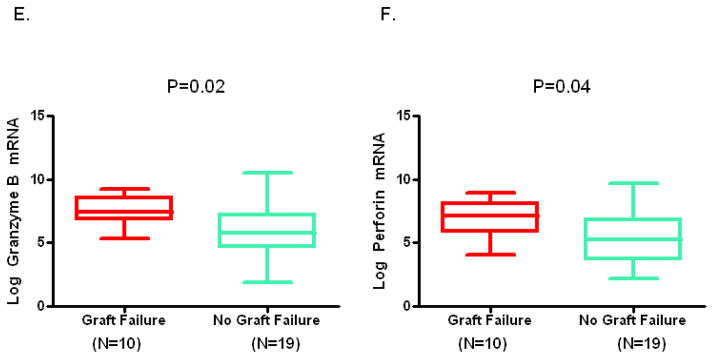
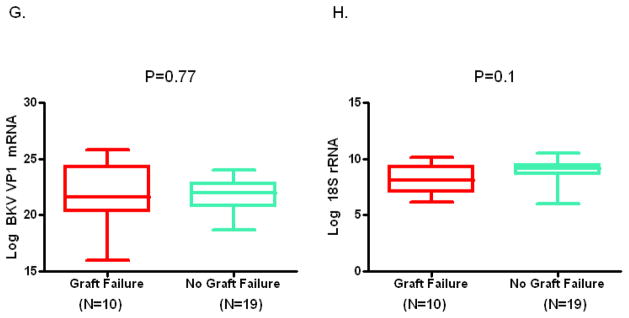
Box and whisker plots show the 10th, 25th, 50th (median), 75th and 90th percentile values of the log-transformed ratios of mRNA copies to 18S rRNA copies for PAI-1, vimentin, TIMP-1, fibronectin, granzyme B, perforin, and BKV VP1 in urine collected at the time of biopsy diagnosis BKVN from 10 kidney graft recipients who experienced graft failure (Graft Failure) and 19 recipients who did not experience graft failure (No Graft Failure) during the three years following BKVN diagnosis. Levels of mRNA for PAI-1, vimentin, TIMP-1, fibronectin, granzyme B, perforin but not the levels of BKV VP1 mRNA and 18S rRNA were higher in the patients with graft failure. Two-tailed P values are based on the Mann Whitney U test.
Plasma BK virus DNA copy numbers were measured in 26 of 29 BKVN patients at the time of BKVN diagnosis and the copy number was not different between the graft failure group and no graft failure group (Table 2). Also, there was no significant association between plasma BKV copy number and urinary cell BKV-VP1 mRNA copy number or the copy numbers of any of the mRNAs measured in urinary cells. (SDC, Table S1).
Development of Models Prognostic of Graft Failure in BKVN Patients
We used logistic regression to select variables prognostic of future graft outcome in the patients with BKVN. Levels of mRNA for PAI-1 (P=0.006), vimentin (P=0.02), TIMP-1 (P=0.02), fibronectin (P=0.02) and BKVN stage (P=0.03) were strongly associated (P<0.05) with graft failure by bivariate logistic regression, and the levels of granzyme B (P=0.05), perforin (P=0.06), E-cadherin (P=0.07) and serum creatinine level (P=0.08) were modestly associated (P=0.05 to P<0.1) (SDC, Table S2). SDC Table S3 shows the bivariate relationship between all the variables studied and the decline in kidney graft function during the three-year follow up.
We constructed multivariable models incorporating the variables found associated (P<0.1) with graft failure by bivariate logistic regression. Prior to inclusion of mRNAs in the multivariable model, we examined whether the mRNAs were associated with each other to avoid co-linearity. Spearman’s rank-order correlation analysis showed that the mRNAs are highly related with each other (SDC, Table S4). We therefore added only one mRNA measure to each multivariable model that included the clinical parameters, serum creatinine level and the biopsy stage, and examined the model’s prognostic accuracy. Among the 7 models created with the addition of different mRNAs to serum creatinine and biopsy stage, the combination of creatinine, biopsy stage and PAI-1 mRNA was the best predictor (SDC Table S5) and this model was selected for additional analysis.
The odds ratio and the P value for each variable included in this best fitting model are provided in Table 3A and demonstrate that urinary cell level of PAI-1 mRNA is an independent predictor of graft failure (P=0.03) and that neither serum creatinine level (P=0.33) nor BKVN biopsy stage (P=0.22) is significantly associated with graft failure in this model.
Table 3.
Multivariable Logistic Regression Models Predicting Graft Failure
| A. Three variable model for predicting graft failure in BKVN patientsa.
| |||
|---|---|---|---|
| Variable | Coefficient | Odds Ratio (95% Confidence Intervals) | P Value |
| Constant | −10.59 | ||
| Urinary cell PAI-1 mRNA | 1.01 | 2.8 (1.12 – 6.78) | 0.03 |
| Serum Creatinine | 0.59 | 1.8 (0.55 – 6.02) | 0.33 |
| Biopsy B/C | 1.65 | 5.2 (0.38 – 70.38) | 0.22 |
|
| |||
| Model pseudo-R2 valueb: 0.4142, P=0.0015. | |||
| Composite Score Equation: −10.59 + 1.01Ln(PAI-1/18S) + 0.59(serum creatinine mg/dL) + 1.65(biopsy BKVN stage B/C[1] or A[0]). | |||
| B. Two variable model for predicting graft failure in BKVN patientsc.
| |||
|---|---|---|---|
| Variable | Coefficient | Odds Ratio (95% Confidence Interval) | P value |
| Constant | −10.61 | ||
| Urinary cell PAI-1 mRNA | 1.14 | 3.1 (1.28 – 7.54) | 0.01 |
| Serum Creatinine | 0.72 | 2.1 (0.63 – 6.77) | 0.23 |
|
| |||
| Model pseudo-R2 valueb: 0.3682, P=0.001. | |||
| Composite Score Equation: −10.61 + 1.14Ln(PAI-1/18S) + 0.72(serum creatinine mg/dL). | |||
Multivariable logistic regression models were constructed including variables associated (P<0.1) with graft failure by bivariate analysis (SDC Table S2). Since Spearman’s rank-order correlation analysis showed that the mRNAs are highly related with each other (SDC, Table S4), only one mRNA measure was added to each logistic model that included serum creatinine level and BKVN biopsy stage. Among the 7 models created with the addition of different mRNAs to serum creatinine and biopsy stage (SDC Table S5), the combination of creatinine, biopsy stage and PAI-1mRNA was the best predictor. The coefficient, odds ratio and the P value for each variable included in this optimum model are provided in Table 3A.
R2 value indicates the percent of variation in the dependent variable (graft failure) that can be explained by the predictor variables (e.g., PAI-mRNA level) in the model. It ranges from 0 to 1.0 with a value of 1.0 indicating the perfect predictor. The equivalent terminology for the term R2 used in the linear regression model is pseudo-R2 in the logistic regression model;
Table 3B shows the coefficient, odd ratios and P values for the noninvasive prediction model comprised of serum creatinine level and urinary cell PAI-1 mRNA level and without the biopsy results. None of the other mRNAs included in the prediction models shown in SDC Table S5 was an independent predictor (P<0.05) in the multivariable logistic regression analysis.
Because our goal was to develop a noninvasive prediction model, we investigated whether graft failure can be prognosticated accurately without the inclusion of the biopsy information in our model. This analysis showed that a model comprised of levels of serum creatinine and urinary cell PAI-1 mRNA and without the biopsy result is also a strong predictor of graft outcome (pseudo-R2: 0.37, P=0.001, Table 3B). PAI-mRNA level was again an independent predictor of graft failure (P=0.01) and serum creatinine level (P=0.23) was not.
ROC Curve Analysis of Composite Scores
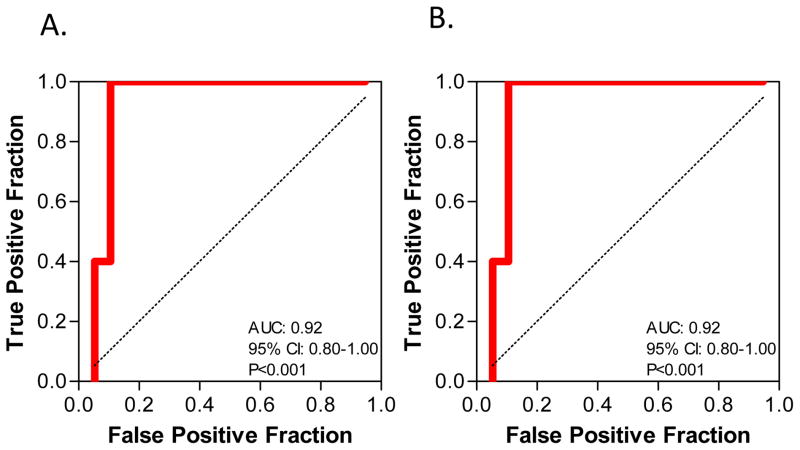
The composite score for the combination of serum creatinine, biopsy stage and PAI-1mRNA, calculated using the equation: −10.59 + 1.01Ln(PAI-1/18S) + 0.59(serum creatinine mg/dL) + 1.65[biopsy BKVN stage B/C(1) or A(0)]) (Table 3A) was examined for prognostic accuracy. Analysis involving ROC curve showed that at a cutoff value of −0.724, the composite score predicted graft failure with a sensitivity of 100.0% and a specificity of 89.5% and the AUC was 0.92 (95% CI: 0.80–1.0, P<0.001) (Fig. 2A).
Figure 2. ROC Curve Analysis of the Composite Score.
Logistic regression was used to derive the composite score for the combination of biopsy stage, serum creatinine level and urinary cell level of 18S rRNA normalized PAI-mRNA. The fraction of true positive results (sensitivity) and false positive results (1-specificity) for the score of this combination are shown in Panel A. At a cutoff value of −0.724, the score predicted graft failure with a sensitivity of 100.0% and a specificity of 89.5% and the AUC was 0.92 (95% CI: 0.80–1.0, P<0.001. In Panel B, ROC curve analysis of the composite score of the combination of serum creatinine levels and 18S rRNA normalized PAI-1 mRNA level is shown. At a cutoff value of −0.858, the score predicted graft failure with a sensitivity of 100.0% and a specificity of 89.5% and the AUC was 0.92 (95% CI: 0.80–1.0, P<0.001).
The composite score for the combination of levels of serum creatinine level and urinary cell PAI-1 mRNA, calculated using the equation: −10.61 + 1.14Ln(PAI-1/18S) + 0.72(serum creatinine mg/dL) (Table 3B) was also examined for prognostic accuracy. Analysis involving ROC curve showed that at a cutoff value of −0.858, the composite score predicted graft failure with a sensitivity of 100.0% and a specificity of 89.5% and the AUC was 0.92 (95% CI: 0.80–1.0, P<0.001) (Fig. 2B).
A comparison of the AUCs showed that the AUC for the prediction model that included biopsy stage is not significantly different from the AUC of the model that did not include biopsy (P=0.33, DeLong test).
DISCUSSION
Our goal was to develop noninvasively measurable biomolecular markers prognostic of kidney allograft failure in patients whose for-cause biopsies displayed BKVN. Currently, prognostication of kidney graft outcome following BKVN diagnosis relies primarily on renal allograft biopsy features. The biopsy procedure has become safer over the years and the reporting of results more standardized; however the invasive procedure can be associated with significant morbidity and adverse outcomes including graft loss. Our investigation demonstrates that a combination of levels of serum creatinine and urinary cell PAI-mRNA foretells graft failure in patients with BKVN with a high degree of accuracy, and that this novel prognostic model comprised of noninvasively ascertained parameters is as accurate as the prediction model that includes results from the invasive biopsy procedure.
Functional attributes such as GFR and serum creatinine level have been associated with kidney graft outcome in a number of clinical settings including in patients with BKVN ((3, 8)). In the current investigation, serum creatinine level was significantly higher in BKVN cohort at risk for graft failure compared to the no graft failure group, and was marginally associated with graft failure by bivariate logistic regression. Our multivariable model however showed that serum creatinine is not an independent predictor of graft failure in the prediction model that included urinary cell PAI-1 mRNA level. The lack of significant association between serum creatinine level and graft outcome may be due to the small number patients and outcomes (graft failure) studied and/or due to serum creatinine level being a functional parameter rather than a mechanistic contributor to graft failure.
Drachenberg and colleagues reported histological parameters of BKVN as predictors of allograft failure (3). BKVN patients with biopsies with minimal fibrosis, classified as Stage A, had good renal allograft outcome whereas patients with biopsies with advanced fibrosis had inferior outcome. Wadei and colleagues have reported that the degree of intragraft interstitial fibrosis is associated with kidney graft dysfunction in BKVN patients (9). In our study, the scores for histological hallmarks of fibrosis - tubular atrophy and interstitial fibrosis -were higher in the graft failure group compared to the group without graft failure but these numerical differences were not statistically significant. Our findings that urinary cell levels of mRNAs encoding proteins implicated in fibrosis are significantly higher in patients with graft failure compared to those without graft failure suggest the hypothesis that molecular parameters are more sensitive compared to histological features in predicting outcome.
BKVN has recently been classified as stage A, B or C with stage A differing from B primarily by the degree viral induced epithelial cell lysis and stage C differing from A and B by the degree of chronic tubulointerstitial injury. In this study, BKVN stage A was more frequent in those without graft failure compared to those with graft failure and stage B was more frequent in the graft failure group (Table 2). However, and as observed with serum creatinine level, biopsy stage was not an independent predictor of graft failure in the multivariable analysis.
Masutani et al. reported that that the presence of inflammation in renal allograft biopsies of BKVN patients is associated with higher serum creatinine level (2). In our study, inflammation, as quantified using the scores for cytopathic changes, tubulitis and infiltration were numerically higher in the graft failure group compared to the group without graft failure but the differences were not statistically significant. Our previous publications (8, 16) and our current observation that urinary cell levels of mRNAs encoding cytotoxic attack molecules perforin and granzyme B are higher in patients with graft failure compared to those without graft failure (Fig. 1) suggest the hypothesis that molecular parameters of inflammation are of value in predicting critical outcomes such as graft failure.
The central finding from this investigation is that urinary cell level of PAI-1 mRNA level, measured at the time of (for-cause) biopsy diagnosis of BKVN, is an independent predictor of graft failure. In addition to PAI-1 mRNA, we also found that urinary cell levels of mRNA for TIMP-1, fibronectin, vimentin, perforin and granzyme B are all associated with graft failure. These additional mRNAs however were not significantly associated with graft failure in a stepwise backward elimination process once PAI-1 mRNA entered the multivariable logistic model.
A biologically significant question is whether the urinary cell mRNA expression pattern elucidated in this study is a BKVN-specific profile or could be observed with other diagnosis. Our earlier demonstration that the levels of mRNA for perforin and granzyme B are both higher in urine from patients with acute rejection compared to patients with normal biopsy results (10) and that the levels of mRNA for PAI-1, TIMP-1, fibronectin, and vimentin are all higher in urine from patients with tubulointerstitial fibrosis in the kidney allograft unrelated to BKVN (12) suggests that the mRNA expression pattern characterized in this study reflects intragraft inflammation and repair rather than a specific response to the BK virus. We suggest that our strategy of simultaneously profiling urinary cells for BKV VP-1 mRNA as well as mRNAs encoding proteins implicated in inflammation and fibrosis should help resolve whether the mRNA expression pattern is related to BKVN or due to causes unrelated to BKVN.
An additional issue is whether the urinary cell mRNA profile prognostic of BKVN is also prognostic of outcome following an episode of acute rejection. In a small study comprised of 14 kidney graft recipients with acute rejection, we observed that urinary cell level of PAI-1 mRNA, measured at the time of biopsy diagnosis of acute rejection, is associated with graft loss by logistic regression (odds ratio: 6.7, P=0.02). Despite this encouraging result, since acute rejection is treated with an increase in immunosuppressive therapy and BKVN by a decrease, the prediction models developed in this investigation may not function similarly across diagnoses.
How may a transplant clinician apply the results of this study to an individual patient? The box and whisker plots illustrated in Figure 1, while demonstrating that the urinary cell levels of mRNAs distinguish the graft failure group from the group without graft failure, are not helpful in prognosticating graft outcome in an individual patient. The composite score for the combination of levels of serum creatinine and urinary cell PAI-1 mRNA, on the other hand, may be helpful in prognosticating outcome in a single patient. For example, a patient with a composite score of less than −0.8585 for the combination of serum creatinine and urinary cell PAI-mRNA is unlikely to experience graft failure in the 3 years following diagnosis of BKVN. That said the prediction model developed in the current study must be interpreted with caution in view of the relatively small number of BKVN patients studied and because our prediction models have not been validated using an independent group of BKVN patients.
From a mechanistic perspective, our finding that PAI-1 mRNA level is an independent predictor of graft failure following BKVN diagnosis suggests that this molecule plays an important role in the dysregulation of extracellular matrix deposition in individuals with BKVN. Fibrosis of renal allografts is associated with persistent fibrin deposition which results from an imbalance between factors that promote fibrin deposition and factors that promote fibrin degradation such as plasmin (17). The activity of plasmin is controlled both by plasminogen activators and by plasminogen activator inhibitors such as PAI-1. Inhibition of plasmin by PAI-1 has been implicated in an increase in extracellular matrix accumulation in animal models (18). In fact, renal allograft biopsies with chronic allograft nephropathy demonstrate increased expression of PAI-1 within the allograft compared to those with normal allograft histology (19) ((20). Increased levels of PAI-1 mRNA have also been associated with inflammation. Microdissected glomeruli from acute rejection biopsies show increased expression of PAI-1 mRNA compared to no rejection biopsies; the investigators also noted that within the study cohort, PAI-1 mRNA levels in the initial biopsy (rejecting or non-rejecting) correlated with long-term renal allograft dysfunction (21).
In sum, the transplant clinician currently utilizes clinical parameters such as serum creatinine level as a predictor of graft outcome and complements the functional information with biopsy results to improve prediction. Our findings that the composite score of a combination of levels of serum creatinine and urinary cell PAI-1 mRNA predicts graft outcome as accurately as the score from a combination of levels of serum creatinine and urinary cell PAI-1 and biopsy stage are important not only from the perspective of developing noninvasive prognostic biomarkers of BKVN but also for reducing the need for the invasive biopsy procedure and associated morbidity. Also, biomarkers such as PAI-1 mRNA have the potential to suggest mechanistic basis for disease progression and inform the design of targeted therapies.
METHODS
Study Cohort and Design
Between January 1999 and December 2008, 38 of 1450 patients who received their kidney transplants at our center were diagnosed as having BKVN. Inclusion criteria for the current study were (1) presence of a positive SV40 immunostaining on allograft biopsy (N=38), (2) enrollment in the IRB approved protocol entitled “Use of PCR to Evaluate Renal Allograft Status”(N=37), (3) availability of cDNA from urine specimens that were collected at the time of biopsy (N=31). Exclusion criteria were (1) inadequate yield of urinary cell mRNA, defined as 18S rRNA copies less than 5×108 copies/microgram of total RNA and (2) less than three years of follow-up following biopsy diagnosis of BKVN in those without graft failure (N=29). The institutional review board at Weill Cornell Medical College approved the study, and each patient gave written informed consent.
Renal Allograft Biopsy Evaluation
Percutaneous core needle biopsies of renal allografts were classified by a single renal pathologist (S.V.S) blinded to the results of urinary cell mRNA levels. Biopsy specimens were processed, stained and evaluated using standard procedures described previously (8). The mean (±SD) number of glomeruli in the biopsy specimens was 9±4.9 and the renal allograft biopsy specimens were classified using the Banff 1997 criteria for active and chronic tubulointerstitial parameters (22). BKVN staging (A, B, C) was based on the criteria developed by Banff 2009 working group (14).
Pre-amplification Enhanced Real -Time Quantitative PCR Assay for the Measurement of mRNA Levels
Urine, collected at the time of biopsy diagnosis of BKVN, were centrifuged and total RNA was extracted from the urine cell pellet using RNeasy Minikit® (Qiagen), and reverse transcribed to cDNA as described (7, 10). BKV VP1 mRNA copy number was quantified using a standard curve method (7). We utilized pre-amplification enhanced real-time quantitative PCR assays (11) to measure levels of mRNAs. The sequence and location of the gene specific primers and TaqMan® probes used in the PCR assays are shown in SDC Table S6. Natural log transformed ratio of mRNA copy number to 18S rRNA (reference gene) copy number was used in data analysis. For gene transcript level of zero value, the zero was substituted with half of the lowest ratio (mRNA/18S rRNA) obtained for that mRNA within the entire study cohort, irrespective of graft outcome.
Renal Allograft Function and Patient Survival
Graft failure was defined as the need to initiate dialysis therapy. Ten renal allograft recipients experienced graft failures at median of 9 months (mean±SD: 13±13) following BKVN diagnosis and 19 patients did not experience graft failure for a minimum of 36 months following biopsy diagnosis of BKVN.
Statistical Analysis
The prognostic value of clinical variables and urinary cell gene expression profiles were analyzed using the statistical software packages, STATA version 12 and GraphPad Prism version 4.0. Categorical variables were compared using Fisher’s exact test, and continuous variables were compared using the Mann Whitney U test. The prognostic value of each mRNA levels for graft failure was summarized in terms of the odds ratio and 95% confidence interval using bivariate logistic regression models. All mRNAs associated at P<0.10 in the bivariate analysis were entered into separate multivariable logistic regression models with clinical variables to identify independent predictors of graft failure. Based on the logistic regression equation, a composite BKVN score was derived using the combination of PAI-1 mRNA level and serum creatinine level and with and without biopsy stage. The composite score was used to compute the AUC for prediction of graft failure by ROC curve analysis (23). All p-values are two-sided with statistical significance evaluated at the 0.05 alpha level.
Supplementary Material
Acknowledgments
This work was supported by the awards 2R37AI051652 and 5RO1 AI060706 from the National Institute of Allergy and Infectious Disease, National Institutes of Health (MS) and by a grant from UL1 TR000457-06 of the Clinical and Translational Science Center at Weill Cornell Medical College and an award NPRP 08-503-3-111 from the Qatar National Research Foundation.
Footnotes
Author Contributions
Darshana Dadhania - Participated in research design, performance of the research, data analysis, and in the writing of the paper. No conflict of interest.
Catherine Snopkowski - Participated in the performance of the research and data analysis. No conflict of interest.
Thangamani Muthukumar - Participated in the performance of the research and data analysis. No conflict of interest.
John Lee - Participated in the performance of the research and in the review of the paper. No conflict of interest.
Ruchuang Ding - Participated in the performance of data analysis and contributed new reagents or analytic tools. No conflict of interest.
Paul Christos - Participated in data analysis and in the writing of the paper. No conflict of interest. Paul Christos is supported by the Clinical Translational Science Center (CTSC) (2UL1TR000457-06).
Vijay K. Sharma - Participated in data analysis and in the review of the paper. No conflict of interest.
Heejung Bang - Participated in data analysis and in the writing of the paper. No conflict of interest.
Surya V. Seshan - Participated in the performance of the research, data analysis, and in the writing of the paper. No conflict of interest.
Sandip Kapur - Participated performance of research and in the review of the paper. No conflict of interest.
Manikkam Suthanthiran - Participated in research design, performance of data analysis, and in the writing of the paper. No conflict of interest.
References
- 1.Dharnidharka VR, Cherikh WS, Abbott KC. An OPTN analysis of national registry data on treatment of BK virus allograft nephropathy in the United States. Transplantation. 2009;87 (7):1019. doi: 10.1097/TP.0b013e31819cc383. [DOI] [PubMed] [Google Scholar]
- 2.Masutani K, Shapiro R, Basu A, Tan H, Wijkstrom M, Randhawa P. The Banff 2009 Working Proposal for polyomavirus nephropathy: a critical evaluation of its utility as a determinant of clinical outcome. Am J Transplant. 2012;12 (4):907. doi: 10.1111/j.1600-6143.2012.03993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drachenberg CB, Papadimitriou JC, Hirsch HH, et al. Histological patterns of polyomavirus nephropathy: correlation with graft outcome and viral load. Am J Transplant. 2004;4 (12):2082. doi: 10.1046/j.1600-6143.2004.00603.x. [DOI] [PubMed] [Google Scholar]
- 4.Drachenberg CB, Hirsch HH, Ramos E, Papadimitriou JC. Polyomavirus disease in renal transplantation: review of pathological findings and diagnostic methods. Hum Pathol. 2005;36 (12):1245. doi: 10.1016/j.humpath.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Kim D, Kim H, Shin G, et al. A randomized, prospective, comparative study of manual and automated renal biopsies. Am J Kidney Dis. 1998;32 (3):426. doi: 10.1053/ajkd.1998.v32.pm9740159. [DOI] [PubMed] [Google Scholar]
- 6.Manno C, Strippoli GF, Arnesano L, et al. Predictors of bleeding complications in percutaneous ultrasound-guided renal biopsy. Kidney Int. 2004;66 (4):1570. doi: 10.1111/j.1523-1755.2004.00922.x. [DOI] [PubMed] [Google Scholar]
- 7.Ding R, Medeiros M, Dadhania D, et al. Noninvasive diagnosis of BK virus nephritis by measurement of messenger RNA for BK virus VP1 in urine. Transplantation. 2002;74 (7):987. doi: 10.1097/00007890-200210150-00016. [DOI] [PubMed] [Google Scholar]
- 8.Dadhania D, Snopkowski C, Ding R, et al. Validation of noninvasive diagnosis of BK virus nephropathy and identification of prognostic biomarkers. Transplantation. 2010;90 (2):189. doi: 10.1097/TP.0b013e3181e2a932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wadei HM, Rule AD, Lewin M, et al. Kidney transplant function and histological clearance of virus following diagnosis of polyomavirus-associated nephropathy (PVAN) Am J Transplant. 2006;6 (5 Pt 1):1025. doi: 10.1111/j.1600-6143.2006.01296.x. [DOI] [PubMed] [Google Scholar]
- 10.Li B, Hartono C, Ding R, et al. Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med. 2001;344 (13):947. doi: 10.1056/NEJM200103293441301. [DOI] [PubMed] [Google Scholar]
- 11.Muthukumar T, Dadhania D, Ding R, et al. Messenger RNA for FOXP3 in the urine of renal-allograft recipients. N Engl J Med. 2005;353 (22):2342. doi: 10.1056/NEJMoa051907. [DOI] [PubMed] [Google Scholar]
- 12.Anglicheau D, Muthukumar T, Hummel A, et al. Discovery and validation of a molecular signature for the noninvasive diagnosis of human renal allograft fibrosis. Transplantation. 2012;93 (11):1136. doi: 10.1097/TP.0b013e31824ef181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirsch HH, Brennan DC, Drachenberg CB, et al. Polyomavirus-associated nephropathy in renal transplantation: interdisciplinary analyses and recommendations. Transplantation. 2005;79 (10):1277. doi: 10.1097/01.tp.0000156165.83160.09. [DOI] [PubMed] [Google Scholar]
- 14.Sar A, Worawichawong S, Benediktsson H, Zhang J, Yilmaz S, Trpkov K. Interobserver agreement for Polyomavirus nephropathy grading in renal allografts using the working proposal from the 10th Banff Conference on Allograft Pathology. Hum Pathol. 2011;42 (12):2018. doi: 10.1016/j.humpath.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Sis B, Mengel M, Haas M, et al. Banff ‘09 meeting report: antibody mediated graft deterioration and implementation of Banff working groups. Am J Transplant. 2010;10 (3):464. doi: 10.1111/j.1600-6143.2009.02987.x. [DOI] [PubMed] [Google Scholar]
- 16.Dadhania D, Snopkowski C, Ding R, et al. Epidemiology of BK virus in renal allograft recipients: independent risk factors for BK virus replication. Transplantation. 2008;86 (4):521. doi: 10.1097/TP.0b013e31817c6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeisberg M, Neilson EG. Mechanisms of tubulointerstitial fibrosis. J Am Soc Nephrol. 2010;21 (11):1819. doi: 10.1681/ASN.2010080793. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh AK, Vaughan DE. PAI-1 in tissue fibrosis. J Cell Physiol. 2011;227 (2):493. doi: 10.1002/jcp.22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Revelo MP, Federspiel C, Helderman H, Fogo AB. Chronic allograft nephropathy: expression and localization of PAI-1 and PPAR-gamma. Nephrol Dial Transplant. 2005;20 (12):2812. doi: 10.1093/ndt/gfi172. [DOI] [PubMed] [Google Scholar]
- 20.Grandaliano G, Di Paolo S, Monno R, et al. Protease-activated receptor 1 and plasminogen activator inhibitor 1 expression in chronic allograft nephropathy: the role of coagulation and fibrinolysis in renal graft fibrosis. Transplantation. 2001;72 (8):1437. doi: 10.1097/00007890-200110270-00018. [DOI] [PubMed] [Google Scholar]
- 21.Delarue F, Hertig A, Alberti C, et al. Prognostic value of plasminogen activator inhibitor type 1 mRNA in microdissected glomeruli from transplanted kidneys. Transplantation. 2001;72 (7):1256. doi: 10.1097/00007890-200110150-00013. [DOI] [PubMed] [Google Scholar]
- 22.Racusen LC, Solez K, Colvin RB, et al. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999;55 (2):713. doi: 10.1046/j.1523-1755.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- 23.Seshan VE, Gonen M, Begg CB. Comparing ROC curves derived from regression models. Stat Med. 2012 doi: 10.1002/sim.5648. [DOI] [PMC free article] [PubMed]
- 24.Rennert H, Jenkins SG, Azurin C, Sipley J. Evaluation of a BK virus viral load assay using the QIAGEN Artus BK Virus RG PCR test. J Clin Virol. 2012;54 (3):260. doi: 10.1016/j.jcv.2012.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.