Abstract
Melanoma is a rapidly growing and highly metastatic cancer with high mortality rates, for which a resolutive treatment is lacking. Identification of novel therapeutic strategies and biomarkers of tumour stage is thus of particular relevance. We report here on a novel biomarker and possible candidate therapeutic target, the sphingolipid metabolising enzyme acid sphingomyelinase (A-SMase). A-SMase expression correlates inversely with tumour stage in human melanoma biopsies. Studies in a mouse model of melanoma and on cell lines derived from mouse and human melanomas demonstrated that A-SMase levels of expression actually determine the malignant phenotype of melanoma cells in terms of pigmentation, tumour progression, invasiveness and metastatic ability. The action of A-SMase is mediated by the activation of the extracellular signal-regulated kinase, the subsequent proteasomal degradation of the Microphtalmia-associated transcription factor (Mitf) and inhibition of cyclin-dependent kinase 2, Bcl-2 and c-Met, downstream targets of Mitf involved in tumour cell proliferation, survival and metastatisation.
Keywords: melanoma, acid sphingomyelinase, signalling mechanisms, microphtalmia-associated transcription factor
Melanoma is the most aggressive form of skin cancer that hesitates early in the metastatic stage with inauspicious prognosis.1 Significant advances have been made in understanding its pathophysiology and treatment. Conventional therapies, consisting in surgical resection of lesions and chemotherapy, are thus being complemented by new approaches based on growth factors signalling inhibitors, agents stimulating antitumour immune responses, or adoptive T-cell therapies.1 The percentage of patients responding to therapy is however still relatively small, with a majority of patients showing short-lived responses.2, 3 The difficulty in treating melanoma lies mainly in its high degree of heterogeneity4, which accounts for the appearance, during melanoma development, of cell populations differing in tumour marker expression, invasive phenotype, drug resistance and immunogenicity.5 Heterogeneity also impacts on the melanoma staging and prognosis. Melanoma stage is still defined by clinical criteria.6 However, significant recent developments are leading to a classification by molecular criteria.7 A better definition of the melanoma molecular signature will be instrumental to both early diagnosis and efficient and tailored treatments.
We identify an unexpected role for the sphingolipid-metabolising enzyme acid sphingomyelinase (A-SMase) that opens new perspectives in terms of both its use as a melanoma biomarker and as a therapeutic target. A-SMase is activated in response to various proinflammatory and proapoptotic stimuli;8, 9, 10, 11, 12 it contributes to apoptotic death of tumour cells induced by different antineoplastic treatments13 and has a role in immunity and inflammation, regulating macrophages and dendritic cells differentiation/function or cytokines secretion.14, 15, 16 We now report that A-SMase is also a key determinant of melanoma cell behaviour. A-SMase levels of expression correlate with melanoma grade in human biopsies. Studies in vivo in a mouse model of melanoma and in vitro on human and mouse melanoma cells demonstrated that such correlation is not coincidental. A-SMase levels of expression actually determine the malignant phenotype of melanoma cells in terms of pigmentation, tumour progression and metastatic capacity. We have characterised the molecular pathway responsible for A-SMase effect and found a significant role being played by the Microphtalmia-associated transcription factor (Mitf), a gene associated with familial and sporadic melanoma.17, 18, 19 We found that Mitf regulation by A-SMase is mediated by the activation of the extracellular signal-regulated kinase (ERK) responsible for Mitf degradation by proteasome. The loss of A-SMase during melanoma progression accounts for the upregulation of Mitf and its downstream targets cyclin-dependent kinase 2 (CDK2), Bcl-2 and c-Met.
Results
A-SMase expression correlates with melanoma progression
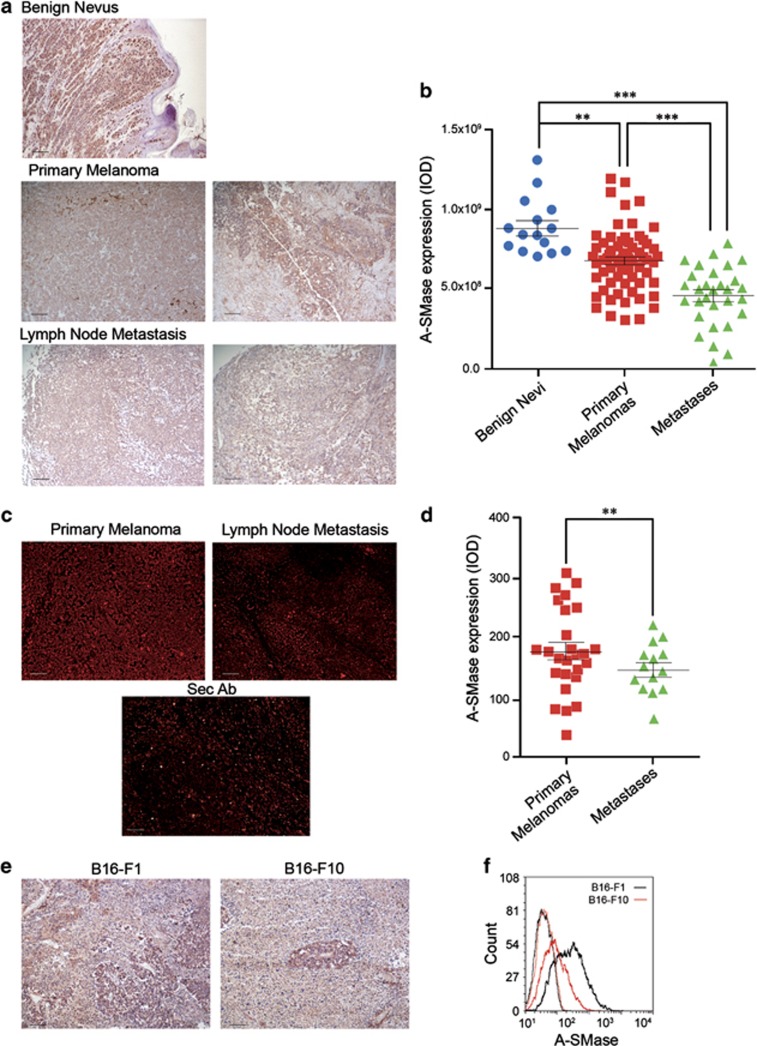
A-SMase expression was evaluated by immunohistochemistry in sections from human bioptic specimens of melanomas at various stages (Supplementary Table S1). Enzyme expression was higher in benign nevi than in primary melanomas, and further reduced in the lymph-node metastases (Figure 1a). Quantitative analyses of results from several samples carried out using two analysis softwares, ImageJ (Figure 1b) and AxioVision 4 (Supplementary Figures S1A and B),20, 21 indicate that the differences in A-SMase expression among the groups of specimens are significant. The different expression of A-SMase in primary melanomas and metastases was confirmed further by immunofluorescence analyses (Figures 1c and d). Similar results were observed in an in vivo model of mouse melanoma in which we injected sub-cutaneously (s.c.) in C57BL/6 mice B16-F1 and B16-F10 murine cells; the former was unable to generate metastases, whereas the latter was able to.22 Expression of A-SMase was higher in B16-F1 melanoma, further suggesting a relationship between melanoma propensity to yield metastases and A-SMase expression (Figures 1e and f).
Figure 1.
A-SMase downregulation correlates with increasing melanoma malignancy. (a) Immunohistochemistry of human tissues (nevi and melanomas at different stages) for the evaluation of A-SMase levels. A-SMase is visualised using DAB (brown) and nuclei by haematoxylin (blue) (scale bar, 110 μm). (b) A-SMase quantification in A-SMase/DAB stained human samples (benign nevi, n=15; primary melanomas, n=70; lymph node metastases, n=30) using ImageJ colour deconvolution plug-in as described in Supplementary Figure S1. Error bars: S.E.M. (c) Immunofluorescence staining of human tissues (melanomas at different stages) with A-SMase antibody (scale bar, 110 μm). (d) Quantitative assessment of immunofluorescence staining of A-SMase on human samples (primary melanoma, n=27; lymph node metastases, n=14). (e) Immunohistochemistry of A-SMase expression on tumours derived from B16-F1 and B16-F10 injected s.c. (scale bar, 110 μm). Images shown are representative of one out of three reproducible experiments. (f) A-SMase expression versus secondary antibody staining in B16-F1 (black trace) and B16 F10 (red trace) revealed by flow cytometry. Image shown is representative of one out of three reproducible experiments. Error bars: S.E.M. **P<0.01; ***P<0.001
A-SMase expression by melanoma cells accounts for melanin content
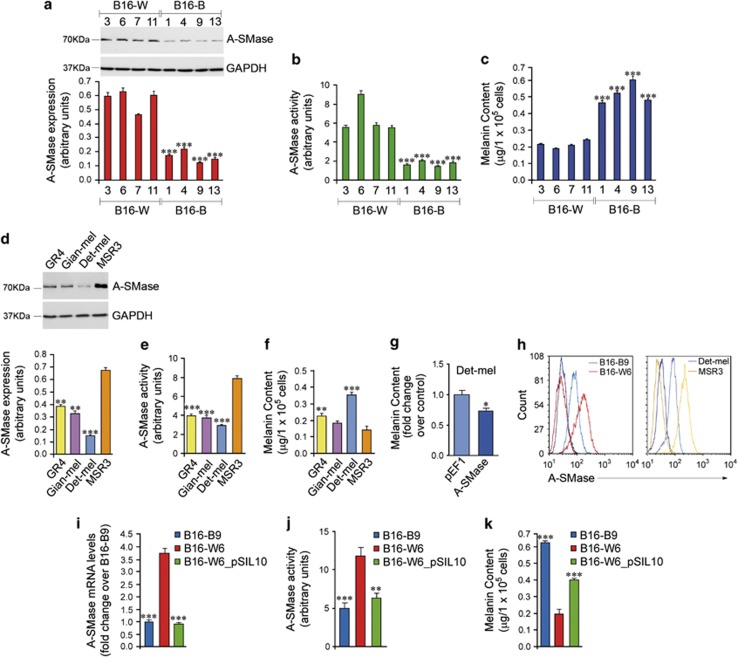
We investigated whether a causal relationship exists between A-SMase expression and tumour behaviour. To this end, taking advantage of the heterogenous A-SMase expression in B16 cells (Figure 1f), we isolated 45 clones from in vivo growing B16-F1 cells; among them, 31 clones expressed the melanoma marker Melan-A (Mel-A), used to identify melanoma cells versus the non-tumour cells in the tumour microenvironment (data not shown). Then, eight representative clones expressing Mel-A (Supplementary Figure S2) were analysed for A-SMase expression and activity (Figures 2a and b). In parallel, we characterised also four human melanoma cell lines GR4, Gian-mel, Det-mel and MSR3,23 which significantly differ in terms of expression/activity of A-SMase (Figures 2d and e).
Figure 2.
A-SMase expression and activity regulates melanoma cell pigmentation. (a–c) A-SMase expression (a) and activity (b) and melanin content (c) measured in four Black and four White representative B16 clones. (a) Western blotting analysis of A-SMase on cell lysates. The graph shows the densitometric values normalised on GAPDH±S.E.M. (n=3). (b) A-SMase activity on cell lysates measured as sphingomyelin hydrolysis to phosphorylcholine at pH 5.5. Values are expressed as arbitrary unit±S.E.M. (n=3). (c) Melanin absorbance measured at 410 nm. Each value (expressed as μg/105 cells) represents the mean±S.E.M. (n=3). (d–g) A-SMase expression (d) and activity (e), and melanin content (f and g) measured in the human melanoma cell lines as in (a–c). (g) Melanin content measured in Det-mel cells transfected with the empty vector pEF1 and with the same vector containing the cDNA of A-SMase here referred as A-SMase. (h) A-SMase expression analysed in B16-B9 and B16-W6 clones, and on the human melanoma cell lines Det-mel and MSR3 by flow cytometry. Images shown are representative of three reproducible experiments. (i–k) A-SMase expression (i), activity (j) and melanin content (k) measured in B16-B9, B16-W6 and B16-W6_pSIL10 cells. (i) qPCR of A-SMase. Values are expressed as mean±S.E.M. (n=6) of the fold increase over B16-B9. (j and k) A-SMase activity and Melanin content (μg/105 cells) performed as in (c). Each value represents the mean±S.E.M. (n=4). Asterisks in a, b, c, i, j and k indicate statistical significance versus the clone B16-W6, which showed the highest A-SMase expression. Asterisks in d, e and f indicate statistical significance versus the human cell line MSR3, which showed the highest A-SMase expression. Asterisks in g indicate the statistical significance versus Det-mel cells transfected with the empty vector. *P<0.05; **P<0.01; ***P<0.001
As the relationship between melanoma pigmentation and progression has long been of research interest, we first aimed at examining the effect of A-SMase on melanin content. We found an inverse correlation between A-SMase expression/activity and melanin content (Figures 2a–c) in the murine clones, with hyper-pigmented clones (B16-Black (B16-B) clones) showing a lower expression/activity of A-SMase with respect to the hypo-pigmented clones (B16-White (B16-W) clones). Such inverse correlation was present also, at least in part, in the human cell lines (Figures 2d–f).
We then investigated the differences between B16-B9 clone and human Det-mel cells, characterised by low expression/activity of A-SMase versus B16-W6 clone and human MRS3 cells, characterised by high expression/activity of A-SMase (Figure 2h). To assess the cause–effect relationship between A-SMase and the observed phenotypes we generated from the A-SMase highly expressing clone B16-W6, a clone (B16-W6_pSIL10) in which A-SMase was stably silenced (Supplementary Figures S3A–C and F; Figure 2i). Silencing of the enzyme and the reduction of its activity (Figure 2j) led to an increase of 50% of the melanin content by B16-W6_pSIL10 with respect to the parental clone (Figure 2k). Expression of tyrosinase (Tyr), a key enzyme of melanogenesis, in B16-B9 and B16-W6_pSIL10 cells was higher than in B16-W6 (Supplementary Figure 4A), whereas no differences were found in Mel-A expression among clones with different A-SMase levels (Supplementary Figure S2), suggesting that A-SMase does not regulate Mel-A. Overexpression of A-SMase in Det-mel cells decreased significantly their melanin content (Figure 2g), thus supporting the role of A-SMase in the modulation of melanoma cell phenotype and confirming previous studies showing that melanin in melanocytes is significantly decreased by ceramide, the main product of A-SMase.24 The tight relationship between ceramide metabolism and melanogenesis is demonstrated further by the similarity in the pattern of ceramide content in B16-B9 and B16-W6_pSIL10 cells, while differing significantly in B16-W6 (Supplementary Figure S4B).
A-SMase expression by melanoma cells inhibits tumour formation and growth
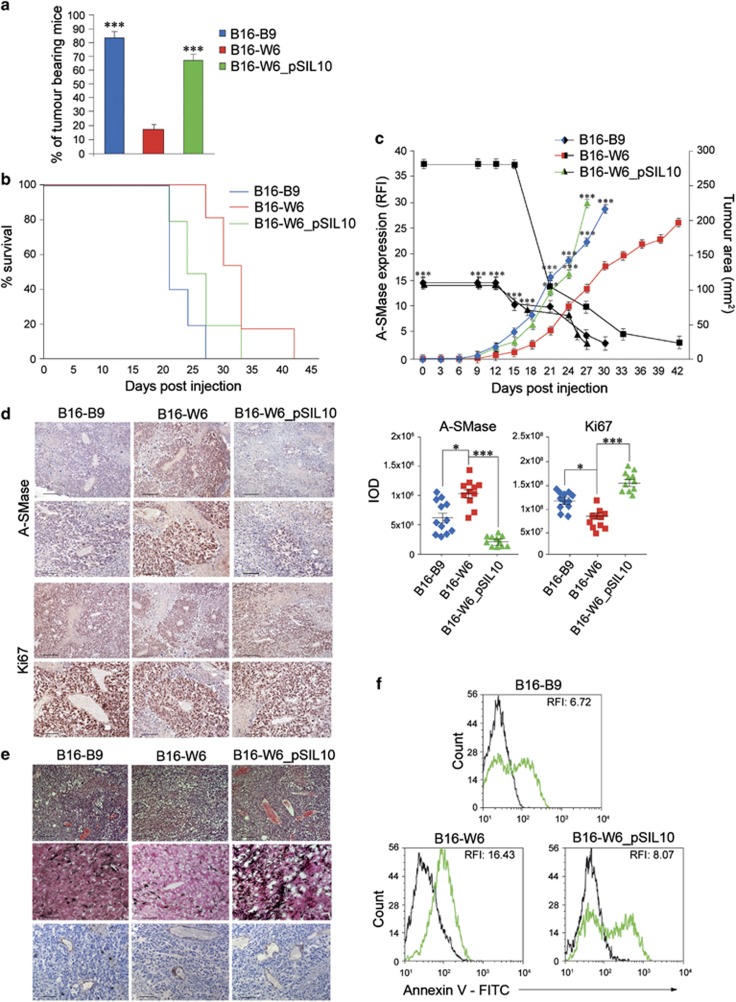
We evaluated the tumourigenic capacity of the B16 clones in vivo following single-cell s.c. injection into C57BL/6 mice. A high percentage of mice injected with B16-B9 and B16-W6_pSIL10 developed detectable tumours (B16-B9=83±3% B16-W6_pSIL10=70±4%) within 60 days (Figure 3a). In contrast, tumours grew only in 18±2% of mice injected with B16–W6 cells. Consistently, B16-B9 and B16-W6_pSIL10 generated a significantly higher number of large colonies than that generated by B16-W6 cells when tested in vitro for their anchorage-independent growth capacity (Supplementary Figure S5A and Supplementary Table S2).
Figure 3.
A-SMase down-expression results in a more aggressive behaviour in vivo. (a) Tumour incidence after s.c. single-cell injection of B16-B9, B16-W6 and B16-W6_pSIL10 cells (five animals per group). Tumour occurrence was monitored daily for 60 days. Values are expressed as percentage of tumour-bearing mice±S.E.M. (n=3). (b–c) C57BL/6 mice (five animals per group) were injected in the right flank with B16-B9, B16-W6 and B16-W6_pSIL10 (2.5 × 104 cells). (b) Kaplan-Meier survival curve of animals (in one out of four reproducible experiments). (c) Tumour growth monitored by measuring tumour area (mm2) every 3 days (n=4). In parallel A-SMase expression in tumours was evaluated at the indicated times (black traces) by flow cytometry (relative fluorescence intensity, RFI) (n=4). Values are expressed as mean±S.E.M. (d) A-SMase and Ki67 staining and quantification in B16-B9, B16-W6 and B16-W6_pSIL10-derived tumours (three animals per group). Images shown are representative of 1 out of 12 tumours per group (top, scale bar, 110 μm; bottom scale bar, 55 μm). Graphs show the quantification (expressed as IOD). Error bars: S.E.M.; n=4. (e) Images showing haematoxylin–eosin, Masson Fontana staining and CD31 staining of B16-B9, B16-W6 and B16-W6_pSIL10-derived tumours (n=6 animals per group) (haematoxylin–eosin, scale bar, 110 μm; Masson Fontana staining and CD31 scale bar 55 μm). (f) Determination of apoptotic cells in vivo. C57BL/6 mice (three animals per group) were injected as in B and tumours were dissected and dispersed when they reached 4 mm for each diameter. Apoptosis was assessed by flow cytometry measuring phosphatidylserine exposure onto the plasma membrane by the staining with Annexin V-FITC. Graphs are representative of one out of three reproducible experiments. Asterisks in a–c indicate statistical significance versus B16-W6 derived tumours. *P<0.05; **P<0.01; ***P<0.001
We next evaluated the effect of A-SMase on melanoma cell proliferation. In vitro, proliferation of B16-B9, B16–W6 and B16–W6_pSIL10 cells correlated inversely with A-SMase expression both in normal serum conditions and after serum deprivation (Supplementary Figures S5B and C). In vivo, the growth of B16-W6 tumours were slower than that of B16-B9 and B16-W6_pSIL10, resulting in an increase of the median survival, particularly in the first 21 days after injection (median of survival: B16-W6=33 days; B16-B9=21; B16-W6_pSIL10=25.5 days; B16-W6 versus B16-B9, P<0.0001; B16-W6 versus B16-W6_pSIL10, P<0.01) (Figure 3b). Interestingly, the expression of A-SMase by the B16 tumours decreased with time regardless of the clone (Figures 3c and d) and this was associated to an increased proliferation rate. At later stages, when A-SMase expression was very low in all clones, their proliferation rates became super-imposable.
B16-B9 and B16-W6_pSIL10 tumours expressed high levels of Ki67, a marker of proliferative cells (Figure 3d), and showed morphological aspects typical of rapid growth phase tumours (Figure 3e) with proliferating cells highly represented even in the inner mass, indicating that A-SMase-deficient cells are able to proliferate even in conditions of nutrition deprivation and low oxygen. By contrast, in the slow-growing B16-W6 tumour proliferating cells resided only around blood vessels, identified by the expression of CD31 (Figure 3e).
We finally examined the effect of A-SMase expression on melanoma apoptosis in vivo, using flow cytometry after staining of tumours with Annexin V. The higher expression of A-SMase in B16-W6 tumour correlated with an increased number of apoptotic cells when compared with B16-B9 and B16-W6_pSIL10 (Figure 3f), according to the apoptogenic role of A-SMase.
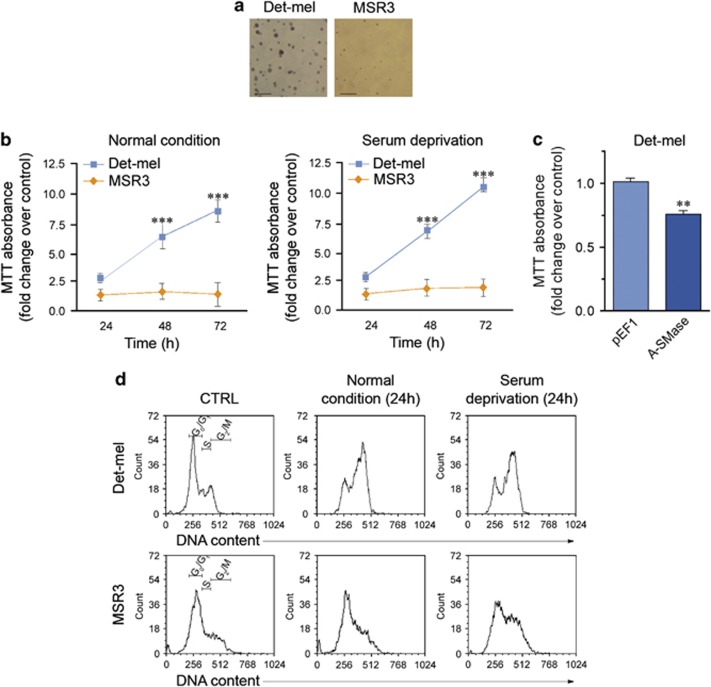
The results obtained in human Det-mel and MSR3 cells were in line with those of mouse models. The reduced expression of A-SMase in Det-mel correlated with an increased anchorage-independent growth capacity (Figure 4a and Supplementary Table S2) and proliferation rate with respect to A-SMase highly expressing MSR3 (Figures 4b and d); consistent with a role of A-SMase in melanoma cell behaviour the overexpression of A-SMase in Det-mel cells reduced significantly their proliferation (Figure 4c).
Figure 4.
A-SMase downregulation triggers proliferation of human melanoma cells in vitro. (a) Soft agar assay for measurement of clonogenic capacity of Det-mel and MSR3. Images are representative of three reproducible experiments (scale bar, 55 μm). (b) MTT analysis of Det-mel and MSR3 cell viability following 24, 48 and 72 h of culture in normal condition (medium added with 10% FBS, left panels) or in serum deprivation (medium added with 2% FBS, right panels). Values are expressed as fold increase over control (time 0 for each cell line)±S.E.M. (n=6). (c) MTT analysis of Det-mel cells transfected with the empty vector pEF1 and with the pEF1-A-SMase vector (A-SMase). (d) Cell cycle analysis of Det-mel and MSR3 cultured for 24 h in medium added with 10% FBS or with 2% FBS, stained with propidium iodide and analysed by flow cytometry. Images shown are representative of four different reproducible experiments. Asterisks in b indicate statistical significance versus MSR3 cells ***P<0.001
A-SMase expression by melanoma cells inhibits their migratory, invasive and metastatic ability
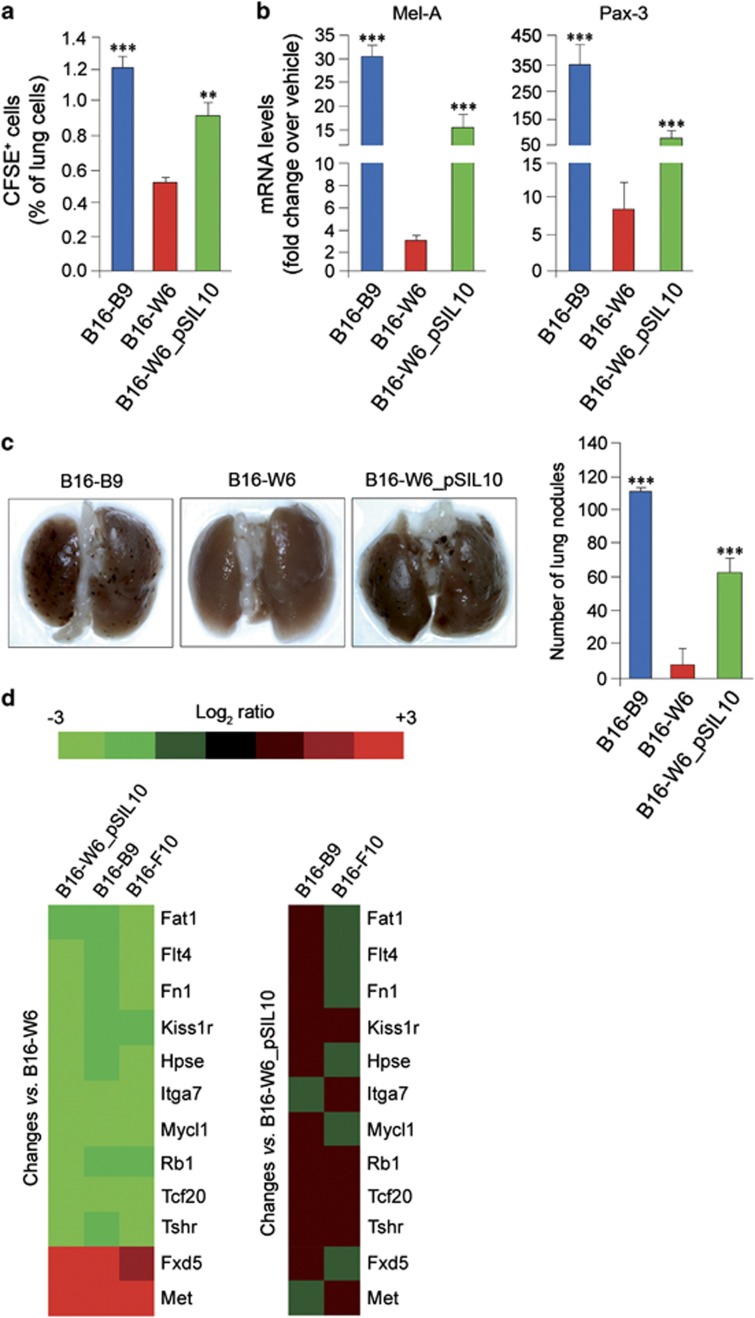
The migratory and invasive properties of B16-B9, B16-W6 and B16-W6_pSIL10 clones were then examined. In vitro, migration, invasion and transmigration of B16-W6 cells were significantly reduced with respect to B16-B9 and B16-W6_pSIL10 cells (Supplementary Figures S6A and B). Thus, A-SMase expression correlates inversely with metastatic potential. In vivo, this correlation was evaluated by injecting B16 clones labelled with the fluorescent dye CFSE into the tail vein of mice. The presence of melanoma cells in lungs was evaluated at 96 h by flow cytometry and quantitative real-time PCR (qPCR). B16-B9 and B16-W6_pSIL10-injected mice displayed a significantly higher number of fluorescent cells at the pulmonary level than B16-W6-injected ones (Figure 5a). Likewise, the specific melanoma markers Mel-A and Pax-3, expressed by all clones at similar degrees (Supplementary Figure S6C), were more abundant in lungs of B16-B9 and B16-W6_pSIL10-injected animals (Figure 5b). After 14 days, we analysed lung metastases by counting the number of nodules. Lung of mice injected with B16-B9 and B16-W6_pSIL10 cells showed a 100- and 60-fold increase of nodules, respectively, compared with those of mice injected with B16-W6, where nodules were virtually undetectable (Figure 5c).
Figure 5.
A-SMase expression affects in vitro and in vivo metastatic potential of B16 clones. (a–c) Evaluation of metastatic potential in vivo. C57BL/6 mice (eight animals per group) were injected into the tail vein with 5 × 105 of CFSE-labelled (a) or native (b and c) B16-B9, B16-W6 and B16-W6_pSIL10 cells. After 96 h (a and b) or 14 days (c) mice were killed and lungs were removed. (a) Flow cytometric analysis of CFSE-positive cells present at pulmonary level (n=4). (b) qPCR evaluation of Mel-A and Pax-3 expression level on mRNA extract from lung samples (n=4). (c) Lung metastasis assay. Lungs were scored for the number of metastatic nodules on the surface. Images shown are representative of one out of four different reproducible experiments. (d) Heat map representing gene expression changes in B16-W6_pSIL10, B16-B9 and B16-F10 versus B16-W6 cells (left panels) and in B16-B9 and B16-F10 versus B16-W6_pSIL10 cells (right panel). Values in the graphs are expressed as mean±S.E.M. Asterisks in a–c indicate statistical significance versus B16-W6 cells; **P<0.01, ***P<0.001
A-SMase downregulation in melanoma cells affects the expression of metastasis-related genes
We evaluated whether A-SMase expression affects the molecular signature of murine melanoma clones using a qPCR array targeting 84 genes known to be involved in tumour growth and metastasis (Supplementary Table S3). B16-F10 murine cells were used as a positive control because they are a model with high metastatic potential.22 These cells displayed A-SMase expression/activity levels and melanin content comparable to B16-B9 and B16-W6_pSIL10 cells (Supplementary Figures S7A–C). qPCR array data are summarised in the correlation and linear regression analysis of Supplementary Figure S6D, where the gene expression ratios (ΔCt) in three experiments are presented. The gene expression levels measured in each array strongly correlated, the Pearson correlation coefficients for each array pair comparison being >0.90 (P<0.001). The paired comparisons of B16-W6_pSIL10, B16-B9 and B16-F10 appear ‘tighter' and closer to the respective linear regression fit line than data resulting from their comparisons with B16-W6. These monotonic relationships between B16-W6_pSIL10, B16-B9 and B16-F10 cells indicate a very high degree of gene expression homology, whereas B16-W6 cells do differ.
The expression of several genes did not change in B16-W6_pSIL10 cells when compared with B16-W6 (less than twofold change) (heat-map in Supplementary Figure S6E). Similar results were obtained with B16-B9 and B16-F10 cells. Several other genes, common to B16-W6_pSIL10, B16-B9 and B16-F10, however, showed at least a twofold change versus B16-W6 (Figure 5d). Among these we found 12 genes whose expression was similar in B16-B9, B16-F10 and B16-W6_pSIL10, suggesting that their regulation is tightly coupled to A-SMase signalling (Figure 5d). Of interest, the proto-oncongene c-Met, highly expressed in melanomas and linked to metastatic potential,25 was upregulated by five folds.
A-SMase modulates Mitf and its downstream signalling targets to regulate cell malignant properties
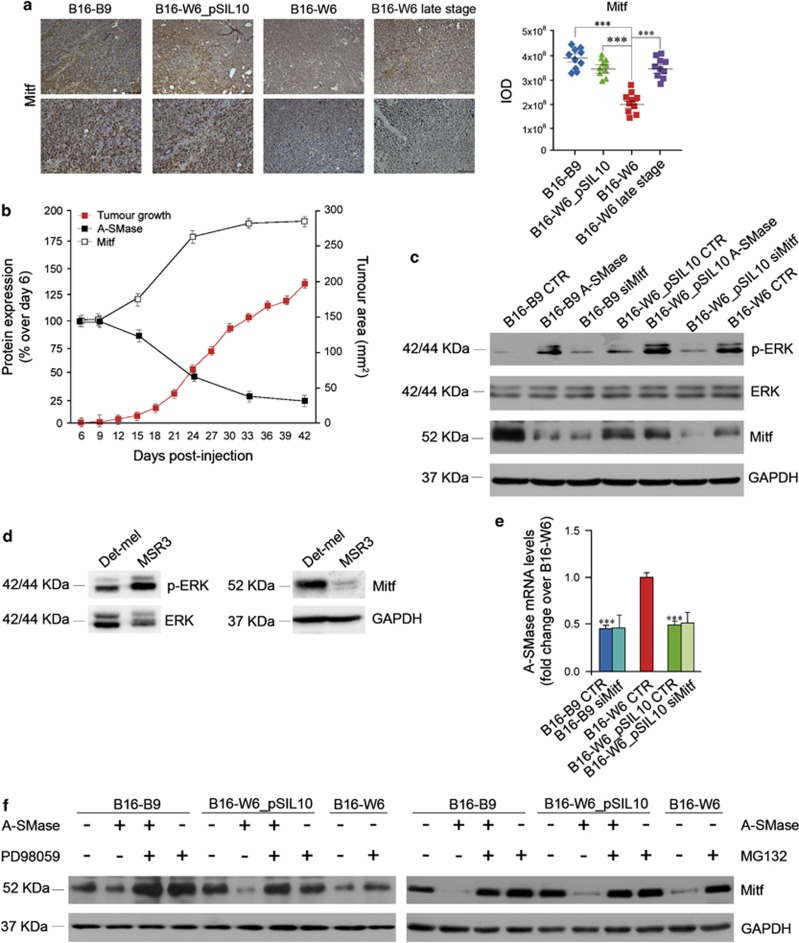
The increased c-Met expression in melanoma cells occurs at transcriptional levels requiring Mitf, a master regulator gene of melanocyte development and melanoma development and progression.26 We thus investigated the relationship between A-SMase and Mitf. As shown by immunohistochemical analysis of tumours, Mitf expression was regulated by A-SMase; in low A-SMase-expressing B16-B9, B16-W6_pSIL10 cells it was upregulated with respect to B16-W6 cells (Figure 6a). Of importance, Mitf expression increased in B16-W6 tumours at late stage of progression (Figure 6a), that is when A-SMase became downregulated, with a clear inverse relationship (Figure 6b), consistent with the role of Mitf on melanoma growth.27, 28 Similar results were obtained in vitro in murine cells including B16-F10, as well as in human cells (Figures 6c and d and Supplementary Figure S7D); notably, Mitf silencing did not affect A-SMase expression (Figure 6e and Supplementary Figure S8A). We then investigated how A-SMase regulates Mitf. Ceramide induces Mitf phosphorylation and its degradation by proteasome through ERK activation.24 Consistently, phosphorylated ERK (pERK) was significantly increased in B16-W6 and B16-B9/B16-W6_pSIL10 transfected with A-SMase when compared with untransfected B16-B9/B16-W6_pSIL10 and B16-F10 cells (Figure 6c and Supplementary Figure S7D). The correlation between pERK and Mitf was also observed in Det-mel and MSR3 cells (Figure 6d). The effect of A-SMase was inhibited by PD98059, an inhibitor of MAP kinases (Figure 6f and Supplementary Figure S8B). pERK was not affected by Mitf silencing confirming that MAPK activity is upstream Mitf (Figure 6c and Supplementary Figure S8A). Moreover, the proteasome inhibitor MG132 blocked the effect of A-SMase, indicating that A-SMase-induced ERK activation coupled to Mitf degradation (Figure 6f).
Figure 6.
A-SMase modulates melanoma progression through the regulation of Mitf. (a) Mitf staining and quantification in B16-B9, B16-W6_pSIL10, B16-W6 and B16-W6 late stage-derived tumours (three animals per group). Images shown are representative of 1 out of 12 tumours per group (top, scale bar, 110 μm; bottom scale bar, 55 μm). Graphs show the quantification (expressed as IOD). Error bars: S.E.M.; n=4. (b) C57BL/6 mice were injected in the right flank with B16-W6 (2.5 × 104 cells); tumour growth (gray trace) was monitored by measuring tumour area (mm2) every 3 days (n=4). Values are expressed as mean±S.E.M. In parallel A-SMase (full squares) and Mitf (open squares) expression in tumours was evaluated at the indicated times by immunohistochemistry quantification (n=4). Values are expressed as the % of protein expression over tumour at day 6 after cell injection±S.E.M. (c) Western blotting of Mitf, phospho-ERK (p-ERK), ERK and GAPDH performed on B16 cells untreated (B16-B9, B16-W6 and B16-W6_pSIL10) here referred to as CTR, and transfected with a pEF1-A-SMase vector or with a specific Mitf siRNA (B16-B9 and B16-W6_pSIL10), here referred as A-SMase and siMitf respectively. Images shown are representative of one out of four reproducible experiments. (d) Western blotting of Mitf, GAPDH, phospho-ERK (p-ERK) and ERK performed on Det-mel and MSR3 cells. Images shown are representative of one out of three reproducible experiments. (e) qPCR of A-SMase on mRNA extract from B16 cells untreated (B16-B9-, B16-W6- and B16-W6_pSIL10-CTR) and transfected with Mitf siRNA (B16-B9- and B16-W6_pSIL10-siMitf). Values are expressed as mean±S.E.M. (n=4). (f) Western blotting of Mitf and GAPDH performed on B16 cells untransfected or transfected with a A-SMase cDNA (A-SMase) in the presence or absence of PD98059 (40 μM) (left panel) or MG132 (1 μM) (right panel). Images shown are representative of one out of three reproducible experiments. Asterisks in a and c indicate statistical significance versus B16-W6; ***P<0.001
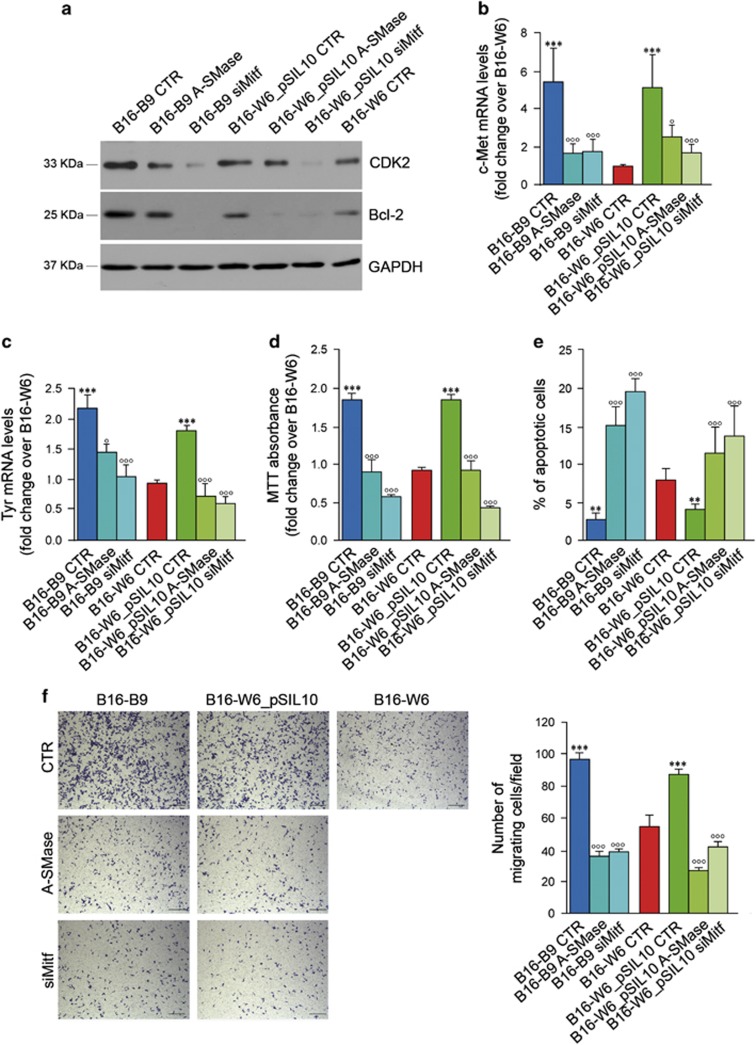
Finally, we investigated whether Mitf regulation by A-SMase led to changes in Mitf signalling. Whenever Mitf was upregulated, that is in the low A-SMase-expressing cells, the proliferation marker CDK2, the apoptosis modulator Bcl-2, the melanin-synthesising enzyme Tyr and the pro-metastatic c-Met were upregulated (Figures 7a–c). Of importance, A-SMase overexpression or Mitf silencing into B16-B9 and B16-W6_pSIL10 cells reduced the expression of all Mitf targets to levels similar to those found in B16-W6 cells (Figures 7a–c) and specifically inhibited proliferation, survival and migratory ability of tumour cells (Figures 7d–f and Supplementary Figures S9A–C).
Figure 7.
A-SMase regulates melanoma cells behaviour and phenotype through the modulation of Mitf downstream targets. Proliferation (a and d), apoptosis (a and e), migration (b and f) and pigmentation (c) rate evaluation on B16 cells untreated (B16-B9, B16-W6 and B16-W6_pSIL10) here referred to as CTR, and transfected with a A-SMase cDNA or with Mitf siRNA, here referred to as B16-B9- and B16-W6_pSIL10-A-SMase and B16-B9- and B16-W6_pSIL10-siMitf respectively. (a) Western blotting of CDK2, Bcl-2 and GAPDH. Images shown are representative of one out of four reproducible experiments. (b–c) qPCR of c-Met and Tyr on mRNA extract. Values are expressed as mean±S.E.M. (n=4). (d) MTT analysis of cell viability following 48 h of culture in normal condition (medium added with 10% FBS). Values are expressed as fold increase over control (B16-W6 cells)±S.E.M. (n=6). (e) Apoptosis was assessed by flow cytometry measuring phosphatidylserine exposure onto the plasma membrane by the staining with Annexin V-FITC. Values are expressed as % of apoptotic cells±S.E.M. (n=4). (f) Migration assay. Cells were plated onto a polycarbonate membrane using 3T3 fibroblast-conditioned medium as source of chemoattractants. After 6 h of incubation, the cells migrated onto the lower surface of the membrane were stained with crystal violet and visually counted in 10 random fields. Images are representative of three different reproducible experiments (scale bar, 110 μm). Values are expressed as mean±S.E.M. (n=3). Circles and asterisks in b–f indicate statistical significance versus the untransfected parental cells and B16-W6, respectively; °,*P<0.05; °°,**P<0.01; °°°,***P<0.001
Discussion
In this study we have identified and characterised a key pathophysiological role of A-SMase. To date knowledge on the role of this enzyme in tumours was limited to its ability of mediating apoptotic cell death in response to chemotherapeutic drugs and radiation.13 Our results expand significantly the role of A-SMase in melanoma showing that it controls tumour ability to grow, invade and metastasise.
Malignant melanomas are composed of subpopulations of cells with a wide range of phenotypic variations, especially in terms of pigment synthesis.29 In melanomas it is possible to identify cells with different grade of pigmentation and also cells unable to generate melanin (amelanotic melanoma).29 Multiple mechanisms to explain amelanosis have been suggested but appropriate studies to compare outcomes of amelanotic versus pigmented melanomas have not been performed.30 On the other hand, it was observed that more pigmented melanoma cells had an enhanced ability to metastasise and that pigmented cell lines are more tumourigenic, invasive and metastatic than the non-pigmented ones.31, 32, 33 Recently, the melanocytic differentiation programme has been suggested to predispose melanomas to metastasis after oncogenic transformation.34 Although the correlation between pigmentation and malignancy is not clearly established, our finding that melanoma clones showed an inverse correlation between A-SMase and melanin content indicates a key role of A-SMase in melanogenesis and possibly melanoma development. In this respect, the central role of the enzyme in determining melanoma cell fate is manifested by what we found in human specimens: A-SMase expression corresponds inversely with tumour malignancy, being lower in metastases with respect to benign lesions. We investigated this aspect in vivo and found that A-SMase expression in melanoma cells decreased progressively with time, an event associated with an increased tumour growth and decreased animal survival. A possible explanation of this event is that microenvironmental factors act on melanoma cells, thus modulating A-SMase during tumour growth. Indeed, it has been shown that tumour microenvironment can modulate the expression patterns of cancer cells involved in proliferation, apoptosis and cell motility.35, 36 Although it is tempting to speculate a crosstalk in tumour microenvironment inducing changes on A-SMase, further studies are needed to understand fully what may have occurred in our system.
The correlation between A-SMase expression and tumour regression we now describe prompted us to propose A-SMase as a key factor in melanoma. The involvement of A-SMase in several critical phases of melanoma cell behaviour from proliferation to migration and ability to metastasise is intriguing and the possibility that the enzyme may be a new useful molecular biomarker of disease progression deserves to be investigated.
The broad role of A-SMase in tumour pathogenesis, identified by us, indicates also that the enzyme is at the crossroad of key pathways in tumourigenesis. This aspect has clear potential in therapeutic perspective and we have characterised it further.
To date analyses of signalling pathways dependent on A-SMase have concentrated mainly on pro-apoptotic effects or in the regulation of the immune system.10, 11, 15, 37, 38, 39 None of its already identified targets can conceivably explain the role of A-SMase in melanoma.
We thus carried out an extensive study of potential effectors of A-SMase in melanoma. The gene expression profile of B16 clones revealed that several factors were differentially expressed depending on the levels of A-SMase. Of particular interest is the inhibition of Mitf expression by A-SMase in tumours, which leads to inhibition of tumour growth, cell proliferation/differentiation and metastatic ability.
In line with previous reports on the modulation of Mitf by ceramide,24 we demonstrated that A-SMase induces the proteasomal degradation of Mitf through a sustained activation of the MAPK ERK. The role of Mitf in melanocytes/melanomas has been object of debate. This transcription factor is expressed by nearly all melanomas, although with a high variability across specimens.40, 41 Mitf is associated with key cellular pathways critical for tumourigenesis.42 In particular, in agreement with our results, many published data are consistent with a role of Mitf in promoting proliferation/progression/invasiveness/resistance to chemotherapy, in suppressing senescence and in forming metastases.28, 32, 40, 43, 44, 45, 46, 47, 48, 49, 50, 51 In addition, Mitf expression is enhanced in ∼20% of human metastatic melanomas and in a genetic mouse model of melanoma.40, 41 Taken together, these observations provide compelling support for a key role of Mitf in favouring melanoma development. However, different reports suggest for Mitf an anti-melanoma activity.28, 49, 52 A model proposes Mitf as a rheostat to control melanoma cell phenotype, allowing a switch from poorly differentiated and highly metastatic cells to fast-growing progeny.49 It has been recently shown that the transient loss of Mitf decreases melanoma cell growth in culture, but favours the development of melanomas in mice.52 These in vivo data are in apparent contrast with our results. However, it should be noted that cells in which Mitf was constitutively silenced did not form tumours in vivo, indicating that Mitf re-expression is required to melanoma growth.52 It has been thus hypothesised that Mitf-depleted cells are able to re-express Mitf giving rise to fast-growing cells and that melanomas contain spontaneously a small population of Mitf-negative cells, which can readily generate a Mitf-expressing progeny with fast-growing properties.28, 52 In agreement with this model, we found that Mitf expression increases during tumour growth. In addition, the inverse correlation between Mitf and A-SMase observed in tumours prompted us to suggest the inhibition of A-SMase as a signal allowing the expression of Mitf and the subsequent increase of tumour growth.
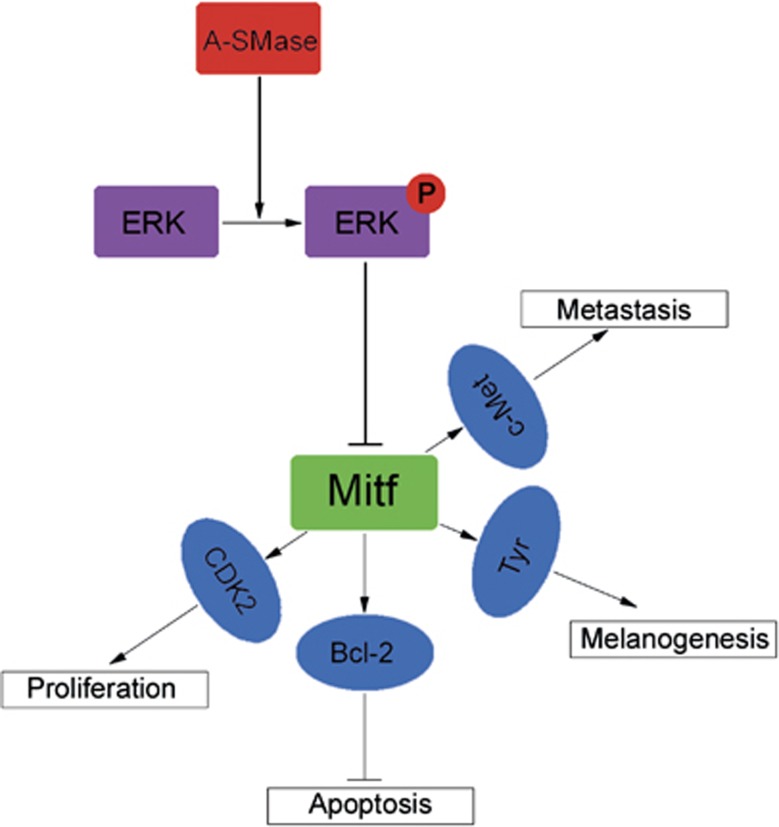
Among the factors we found regulated by A-SMase are CDK2, Bcl2 and c-Met, which are involved in tumoural cell invasion, apoptosis and proliferation, respectively.26, 42, 50 They are downstream targets of A-SMase–Mitf axis, likely modulating cell growth/function. Thus, the A-SMase-induced inhibition of Mitf and its signalling may well explain why this enzyme is implicated in so many diverse phases of melanoma progression (Figure 8). The molecular signalling we describe here for A-SMase has therapeutic implications. Inhibition of CDK2, Bcl2 and c-Met have all been suggested as therapeutic approaches for melanoma treatment.53, 54
Figure 8.
Schematic representation of A-SMase-driven Mitf pathway
In view of its centrality to these and other tumour-related pathways A-SMase might become an interesting therapeutic target exploiting the synergism of multiple significant pathways. The prognostic and therapeutic relevance of A-SMase is consistent with preclinical studies showing a role of ceramide in delaying tumour growth.55, 56 Recently, the use of adenoviral-mediated gene transfer of A-SMase has been proposed as a candidate therapy for Niemann Pick disease.57 In view of the reduced expression of A-SMase in metastatic melanoma it would be important to address whether such gene transfer is applicable also to melanoma, in order to set up combinatorial therapeutic strategies with classical chemotherapy or the novel monoclonal-based therapies.1
Materials and Methods
Animals and cells
Female C57BL/6 mice (6–8 weeks old) were purchase from Charles River Laboratories (Italy) and treated in accordance with the European Communities Council Directive EEC/609/86 and with the approval of the Institutional Ethical Committee. Murine melanoma cell lines B16-F10 and B16-F1, the derived clones and the human melanoma cell lines Det-mel, GR4, Gian-mel and MSR3, derived from metastases of malignant melanomas, were used.23, 58
B16 cell clones isolation
A C57BL/6 mouse was injected with 1 × 105 B16-F1 cells in the lower right flank; when the tumour reached the proper dimension, it was removed and cells were dispersed and plated at a density allowing the formation of single-cell clones (clonal density). The clones were isolated and identified as cancer cells by immunofluorescence with specific melanoma cell marker Mel-A (Supplementary Figure S2).
B16-W6_pSIL10 cell line generation
A stable transfected clone expressing a short hairpin RNA (shRNA) sequence able to silence ASMase expression was generated from B16-W6 clone. To this purpose three independent specific siRNA sequences targeting A-SMase mRNA were tested (smpd1 siRNA1: sense 5′-GGCUACCGAGUUUACCAAAtt-3′/antisense 5′-UUUGGUAAACUCGGUAGCCag-3′ smpd1 siRNA2: sense 5′-CUCUCUCAAUAUGAAUUUUtt-3′/antisense 5′-AAAAUUCAUAUUGAGAGAGat-3′ smpd1 siRNA3: sense 5′-GAGCUGGAAUUAUUACAAAtt-3′/antisense 5′-UUUGUAAUAAUUCCAGCUCca-3′ smpd1 scrambled: sense 5′- GCCTATGCTAATCGCAAATGTtt-3′/antisense 5′-ACAUUUGCGAUUAGCAUAGGCag-3′). As the smpd1 siRNA1 and siRNA2 silenced A-SMase (Supplementary Figure S2B), their specificity was tested by the qPCR analysis of off-target gene expression (Supplementary Figures S2C and D). The smpd1 siRNA1 sequence, which gave the best silencing (Supplementary Figure S2B), was used to create a short hairpin sequence (shRNA) to be cloned into a pSilencer 4.1-CMV neo vector. Transfection of the shRNA vector was performed in B16-W6 cells using Fugene transfection reagent according to the manufacturer's protocol. Approximately 30 colonies resistant to the antibiotic G418 (500 μg/ml) were tested for A-SMase silencing through FACS analysis. The selected clone (here referred to as B16-W6_pSIL10) showed an 80% decrease in A-SMase expression when compared with B16-W6 clone (Supplementary Figure S2E). B16-W6 cells transfected with the pSilencer 4.1-CMV neo Negative Control plasmid supplied with the kit were also generated. As shown in Supplementary Figures S3D and E, similar levels of A-SMase expression and cell proliferation were measured in untransfected B16-W6 cells when compared with B16-W6 cells transfected with the pSilencer 4.1-CMV neo Negative Control plasmid. These results suggest that the transfection of cells did not modify unspecifically the cell content of A-SMase and the cell behaviour.
Tumour growth
C57BL/6 mice (five animals per group) received 2.5 × 104 B16 cells s.c. in the lower-right flank on day 0. Tumour growth was monitored three times a week. Mice were killed when their tumours reached 15 mm on either diameter.16
For in vivo tumourigenicity experiments, single-cell suspension was obtained by serial dilution and injected s.c. in the lower-right flank on day 0. Tumour occurrence was checked every day for 60 days.
Immunohistochemistry and immunofluorescence
For immunohistochemistry and immunofluorescence of formalin-fixed, paraffin-embedded sections the slides were first dewaxed in xylene and hydrated with 2% bovine serum albumin (BSA)-PBS and then incubated overnight at 4 °C with primary antibody (Ab) for A-SMase.9, 11 For immunohistochemistry, slides were incubated with the biotinylated secondary Ab followed by incubation with steptavidin horseradish peroxidase-conjugated, after washing. Reactions were developed using 3,3-diaminobenzidine (DAB; Sigma-Aldrich) and sections counterstained with haematoxylin, dehydrated with a graded series of ethanol, cleaned with xylene, mounted and visualised by light microscopy. For immunofluorescence, after washing, slides were incubated with a secondary Ab conjugated with Alexa Fluor 546. Images were acquired with a Leica DMI4000B microscope.
For histochemistry and immunohistochemistry of murine melanomas, tumours were induced by s.c. injection of B16 cells, removed when they reached 7 mm on either diameter, embedded in OCT and sectioned. Sections of 5–6 μm of thickness were previously air-dried and first stained with haematoxylin–eosin for the initial morphological evaluation.
For immunohistochemistry the air-dried sections were treated in 0.3% H2O2 in methanol to quench endogenous peroxidase activity, and incubated overnight at 4 °C with the specific primary Abs against A-SMase, Ki67 and CD31. For secondary Ab staining and image visualisation the procedure followed was as above.
For cells immunofluorescence studies, cells were seeded on glass coverslips coated with poly-L-lysine. After their adhesion, cells were fixed for 10 min with 4% paraformaldehyde at room temperature, incubated for 5 min with 1% glycine-PBS, and blocked for 30 min with 10% goat serum, 1% BSA, 0.1% saponin in PBS at room temperature. Cells were then incubated with specific primary Abs in 1% BSA, 0.1% saponin in PBS for 1 h at room temperature. For fluorescent detection appropriate secondary Ab conjugated with Alexa Fluor 546 was used. Images were acquired with a Leica DMI4000B microscope.9 In all experiments, the secondary Ab alone was assessed as a negative control.
In vivo metastasis assay
C57BL/6 mice (eight animals per group) were injected in the tail vein with 1 × 105 cells loaded with 10 μM carboxyfluorescein succinimidyl ester (CFSE). After 96 h, mice were killed and lungs were removed and dispersed to cells. The presence of melanoma fluorescent cells was detected by flow cytometry using a Fluorescence-activated Cell Sorter (FC500 Dual Laser system; Beckman Coulter), by comparison with lung cells obtained from mice injected with vehicle and also performing qPCR for the specific markers of melanoma cells Mel-A and Pax-3.59 For the evaluation of macrometastasis nodules, mice were injected as above and killed after 14 days. Lungs were scored under a dissecting microscope for the number of metastatic foci on the surface.
RT2 profiler PCR array
Total RNA was isolated and purified from B16-W6, B16-W6_pSIL10, B16-B9 and B16-F10 cells using SABiosciences RT2 qPCR-Grade RNA Isolation Kit (Qiagen). cDNA was transcribed using a SABiosciences RT2 First Strand Kit following the manufacturer's instructions. For PCR array experiments, the SABiosciences mouse tumour metastasis RT2 Profiler PCR array (catalog number PAM-028) and the SABiosciences RT2 SYBR Green qPCR Mastermix were used to simultaneously examine the mRNA levels of 84 genes closely associated with tumour metastasis and 5 ‘housekeeping genes' (Supplementary Table S3). Arrays were performed on Roche LightCycler 480 Instrument. Ct values for housekeeping genes were monitored for consistency between the arrays and the negative controls ensured a lack of DNA contamination and set the threshold for the absent/present calls. Data were normalised using multiple housekeeping genes and analysed by comparing 2−ΔΔCt of the normalised sample using the software supplied on the SABiosciences website (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php). The resulting values were reported as fold change; only genes showing twofold or greater change were considered. Matrix2png software version 1.2.2 (http://www.chibi.ubc.ca/matrix2png/) was used to create the heat-maps.60
Statistical analysis
Upon verification of normal distribution, statistical significance of raw data between the groups in each experiment was evaluated using unpaired Student's t-test or ANOVA followed by the Bonferroni post-test. Kaplan–Meier data were analysed with the multiple comparison survival curve method using the Log-rank (Mantel-Cox) test. Results are expressed as means±standard error of the mean (S.E.M.).
Acknowledgments
The study was supported by grants from ‘Associazione Italiana Ricerca sul Cancro' (AIRC, IG11365), ‘Regione Lombardia' (NEPENTE Project), an unrestricted grant from Sanofi Aventis Italia, ‘Ministero dell'Istruzione, Università e Ricerca' (MIUR, PRIN2010-2011 projects to EC and DC.), ‘Ministerio de Ciencia e Innovación' (Project SAF2011-22444) and ‘Generalitat de Catalunya' (SGR 2009 1072) and a predoctoral fellowship from ‘Ministerio de Educación y Ciencia'. We thank Biagio Eugenio Leone (Università di Milano-Bicocca, Italy) for the help with pathology analysis. We thank Laura Pozzi and Stefania Russo Scientific Institute IRCCS E Medea, Bosisio Parini, Lecco, Italy, for the technical assistance.
Glossary
- A-SMase
acid sphingomyelinase
- Mitf
Microphtalmia-associated transcription factor
- ERK
extracellular signal-regulated kinase
- pERK
phosphorylated ERK
- CDK2
cyclin-dependent kinase 2
- Mel-A
Melan-A
- B16-B clones
B16-Black clones
- B16-W clones
B16-White clones
- shRNA
short hairpin RNA
- siRNA
small interference RNA
- qPCR
quantitative real-time PCR
- FACS
fluorescence-activated cell sorting
- DAB
3,3-diaminobenzidine
- PBS
phosphate-buffered saline
- BSA
bovine serum albumin
- CFSE
carboxyfluorescein succinimidyl ester
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- Nikolaou VA, Stratigos AJ, Flaherty KT, Tsao H. Melanoma: new insights and new therapies. J Invest Dermatol. 2012;132 (3 Pt 2:854–863. doi: 10.1038/jid.2011.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Hess AR, Seftor RE. Molecular plasticity of human melanoma cells. Oncogene. 2003;22:3070–3075. doi: 10.1038/sj.onc.1206447. [DOI] [PubMed] [Google Scholar]
- Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch CM, Gershenwald JE, Soong SJ, Thompson JF. Update on the melanoma staging system: the importance of sentinel node staging and primary tumor mitotic rate. J Surg Oncol. 2011;104:379–385. doi: 10.1002/jso.21876. [DOI] [PubMed] [Google Scholar]
- Tremante E, Ginebri A, Lo Monaco E, Frascione P, Di Filippo F, Terrenato I, et al. Melanoma molecular classes and prognosis in the postgenomic era. Lancet Oncol. 2012;13:e205–e211. doi: 10.1016/S1470-2045(12)70003-7. [DOI] [PubMed] [Google Scholar]
- Barsacchi R, Perrotta C, Sestili P, Cantoni O, Moncada S, Clementi E. Cyclic GMP-dependent inhibition of acid sphingomyelinase by nitric oxide: an early step in protection against apoptosis. Cell Death Differ. 2002;9:1248–1255. doi: 10.1038/sj.cdd.4401095. [DOI] [PubMed] [Google Scholar]
- Perrotta C, Bizzozero L, Cazzato D, Morlacchi S, Assi E, Simbari F, et al. Syntaxin 4 is required for acid sphingomyelinase activity and apoptotic function. J Biol Chem. 2010;285:40240–40251. doi: 10.1074/jbc.M110.139287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J, Huy C, Brenmoehl J, Obermeier F, Bock J. Matrix metalloproteinase-1 expression induced by IL-1beta requires acid sphingomyelinase. FEBS Lett. 2009;583:915–920. doi: 10.1016/j.febslet.2009.02.008. [DOI] [PubMed] [Google Scholar]
- Edelmann B, Bertsch U, Tchikov V, Winoto-Morbach S, Perrotta C, Jakob M, et al. Caspase-8 and caspase-7 sequentially mediate proteolytic activation of acid sphingomyelinase in TNF-R1 receptosomes. EMBO J. 2011;30:379–394. doi: 10.1038/emboj.2010.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotta C, Clementi E. Biological roles of Acid and neutral sphingomyelinases and their regulation by nitric oxide. Physiology (Bethesda) 2010;25:64–71. doi: 10.1152/physiol.00048.2009. [DOI] [PubMed] [Google Scholar]
- Smith EL, Schuchman EH. The unexpected role of acid sphingomyelinase in cell death and the pathophysiology of common diseases. FASEB J. 2008;22:3419–3431. doi: 10.1096/fj.08-108043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmann T, Buechler C, Ries S, Schaeffler A, Aslanidis C, Schuierer M, et al. Transcription factors Sp1 and AP-2 mediate induction of acid sphingomyelinase during monocytic differentiation. J Lipid Res. 1999;40:870–880. [PubMed] [Google Scholar]
- Sakata A, Ochiai T, Shimeno H, Hikishima S, Yokomatsu T, Shibuya S, et al. Acid sphingomyelinase inhibition suppresses lipopolysaccharide-mediated release of inflammatory cytokines from macrophages and protects against disease pathology in dextran sulphate sodium-induced colitis in mice. Immunology. 2007;122:54–64. doi: 10.1111/j.1365-2567.2007.02612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotta C, Bizzozero L, Falcone S, Rovere-Querini P, Prinetti A, Schuchman EH, et al. Nitric oxide boosts chemoimmunotherapy via inhibition of acid sphingomyelinase in a mouse model of melanoma. Cancer Res. 2007;67:7559–7564. doi: 10.1158/0008-5472.CAN-07-0309. [DOI] [PubMed] [Google Scholar]
- Cronin JC, Wunderlich J, Loftus SK, Prickett TD, Wei X, Ridd K, et al. Frequent mutations in the MITF pathway in melanoma. Pigment Cell Melanoma Res. 2009;22:435–444. doi: 10.1111/j.1755-148X.2009.00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, Bille K, et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480:94–98. doi: 10.1038/nature10539. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Woods SL, Boyle GM, Aoude LG, MacGregor S, Zismann V, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480:99–103. doi: 10.1038/nature10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konsti J, Lundin M, Joensuu H, Lehtimaki T, Sihto H, Holli K, et al. Development and evaluation of a virtual microscopy application for automated assessment of Ki-67 expression in breast cancer. BMC Clin Pathol. 2011;11:3. doi: 10.1186/1472-6890-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashani-Sabet M, Rangel J, Torabian S, Nosrati M, Simko J, Jablons DM, et al. A multi-marker assay to distinguish malignant melanomas from benign nevi. Proc Natl Acad Sci USA. 2009;106:6268–6272. doi: 10.1073/pnas.0901185106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Yoshikawa N, Yamaguchi Y, Kagota S, Shinozuka K, Kunitomo M. Characterization of mouse melanoma cell lines by their mortal malignancy using an experimental metastatic model. Life Sci. 2002;70:791–798. doi: 10.1016/s0024-3205(01)01454-0. [DOI] [PubMed] [Google Scholar]
- Villablanca EJ, Raccosta L, Zhou D, Fontana R, Maggioni D, Negro A, et al. Tumor-mediated liver X receptor-alpha activation inhibits CC chemokine receptor-7 expression on dendritic cells and dampens antitumor responses. Nat Med. 2010;16:98–105. doi: 10.1038/nm.2074. [DOI] [PubMed] [Google Scholar]
- Kim DS, Kim SY, Chung JH, Kim KH, Eun HC, Park KC. Delayed ERK activation by ceramide reduces melanin synthesis in human melanocytes. Cell Signal. 2002;14:779–785. doi: 10.1016/s0898-6568(02)00024-4. [DOI] [PubMed] [Google Scholar]
- Cruz J, Reis-Filho JS, Silva P, Lopes JM. Expression of c-met tyrosine kinase receptor is biologically and prognostically relevant for primary cutaneous malignant melanomas. Oncology. 2003;65:72–82. doi: 10.1159/000071207. [DOI] [PubMed] [Google Scholar]
- McGill GG, Haq R, Nishimura EK, Fisher DE. c-Met expression is regulated by Mitf in the melanocyte lineage. J Biol Chem. 2006;281:10365–10373. doi: 10.1074/jbc.M513094200. [DOI] [PubMed] [Google Scholar]
- Nakai N, Kishida T, Shin-Ya M, Imanishi J, Ueda Y, Kishimoto S, et al. Therapeutic RNA interference of malignant melanoma by electrotransfer of small interfering RNA targeting Mitf. Gene Ther. 2007;14:357–365. doi: 10.1038/sj.gt.3302868. [DOI] [PubMed] [Google Scholar]
- Cheli Y, Giuliano S, Fenouille N, Allegra M, Hofman V, Hofman P, et al. Hypoxia and MITF control metastatic behaviour in mouse and human melanoma cells. Oncogene. 2012;31:2461–2470. doi: 10.1038/onc.2011.425. [DOI] [PubMed] [Google Scholar]
- Halaban R. Pigmentation in melanomas: changes manifesting underlying oncogenic and metabolic activities. Oncol Res. 2002;13:3–8. doi: 10.3727/096504002108747908. [DOI] [PubMed] [Google Scholar]
- Adler MJ, White CR., Jr Amelanotic malignant melanoma. Semin Cutan Med Surg. 1997;16:122–130. doi: 10.1016/s1085-5629(97)80006-5. [DOI] [PubMed] [Google Scholar]
- Jimbow K, Lee SK, King MG, Hara H, Chen H, Dakour J, et al. Melanin pigments and melanosomal proteins as differentiation markers unique to normal and neoplastic melanocytes. J Invest Dermatol. 1993;100:259S–268S. doi: 10.1111/1523-1747.ep12470103. [DOI] [PubMed] [Google Scholar]
- Bennett DC, Dexter TJ, Ormerod EJ, Hart IR. Increased experimental metastatic capacity of a murine melanoma following induction of differentiation. Cancer Res. 1986;46:3239–3244. [PubMed] [Google Scholar]
- Kameyama K, Vieira WD, Tsukamoto K, Law LW, Hearing VJ. Differentiation and the tumorigenic and metastatic phenotype of murine melanoma cells. Int J Cancer. 1990;45:1151–1158. doi: 10.1002/ijc.2910450627. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Choe C, Shin YS, Jeon MJ, Choi SJ, Lee J, et al. Human lung cancer-associated fibroblasts enhance motility of non-small cell lung cancer cells in co-culture. Anticancer Res. 2013;33:2001–2009. [PubMed] [Google Scholar]
- Aggarwal R, Lu J, Kanji S, Das M, Joseph M, Lustberg MB, et al. Human Vgamma2Vdelta2 T cells limit breast cancer growth by modulating cell survival-, apoptosis-related molecules and microenvironment in tumors. Int J Cancer. 2013;133:2133–2144. doi: 10.1002/ijc.28217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YJ, Lee CH, Lin YP, Huang JY, Su CC, Chang WT, et al. Caspase-3 enhances lung metastasis and cell migration in a protease-independent mechanism through the ERK pathway. Int J Cancer. 2008;123:1278–1285. doi: 10.1002/ijc.23592. [DOI] [PubMed] [Google Scholar]
- Jenkins RW, Clarke CJ, Canals D, Snider AJ, Gault CR, Heffernan-Stroud L, et al. Regulation of CC ligand 5/RANTES by acid sphingomyelinase and acid ceramidase. J Biol Chem. 2011;286:13292–13303. doi: 10.1074/jbc.M110.163378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenova KA, Deevska GM, Karakashian AA, Nikolova-Karakashian MN. Studies on the role of acid sphingomyelinase and ceramide in the regulation of tumor necrosis factor alpha (TNFalpha)-converting enzyme activity and TNFalpha secretion in macrophages. J Biol Chem. 2010;285:21103–21113. doi: 10.1074/jbc.M109.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Damsky WE, Curley DP, Santhanakrishnan M, Rosenbaum LE, Platt JT, Rothberg BEG, et al. beta-catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell. 2011;20:741–754. doi: 10.1016/j.ccr.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima I, Kumasaka MY, Thang ND, Goto Y, Takeda K, Iida M, et al. Molecular network associated with MITF in skin melanoma development and progression. J Skin Cancer. 2011;2011:730170. doi: 10.1155/2011/730170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, Denat L, et al. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433:764–769. doi: 10.1038/nature03269. [DOI] [PubMed] [Google Scholar]
- Luo C, Merz PR, Chen Y, Dickes E, Pscherer A, Schadendorf D, et al. MiR-101 inhibits melanoma cell invasion and proliferation by targeting MITF and EZH2. Cancer Lett. 2013;341:240–247. doi: 10.1016/j.canlet.2013.08.021. [DOI] [PubMed] [Google Scholar]
- Borgdorff V, Rix U, Winter GE, Gridling M, Muller AC, Breitwieser FP, et al. A chemical biology approach identifies AMPK as a modulator of melanoma oncogene MITF Oncogene 2013. e-pub ahead of print 3 June 2013doi: 10.1038/onc.2013.185 [DOI] [PubMed]
- Vazquez F, Lim JH, Chim H, Bhalla K, Girnun G, Pierce K, et al. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Dong XD, Chen X, Yao S, Wang L, Wang J, et al. Role of microRNA-182 in posterior uveal melanoma: regulation of tumor development through MITF, BCL2 and cyclin D2. PLoS One. 2012;7:e40967. doi: 10.1371/journal.pone.0040967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige E, Yokoyama S, Levy C, Khaled M, Igras V, Lin RJ, et al. Hypoxia-induced transcriptional repression of the melanoma-associated oncogene MITF. Proc Natl Acad Sci USA. 2011;108:E924–E933. doi: 10.1073/pnas.1106351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, et al. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006;20:3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, et al. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 2004;6:565–576. doi: 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Hertzman Johansson C, Azimi A, Frostvik Stolt M, Shojaee S, Wiberg H, Grafstrom E, et al. Association of MITF and other melanosome-related proteins with chemoresistance in melanoma tumors and cell lines Melanoma Res 2013. e-pub ahead of print 5 August 2013doi: 10.1097/CMR.0b013e328362f9cd [DOI] [PubMed]
- Cheli Y, Giuliano S, Botton T, Rocchi S, Hofman V, Hofman P, et al. Mitf is the key molecular switch between mouse or human melanoma initiating cells and their differentiated progeny. Oncogene. 2011;30:2307–2318. doi: 10.1038/onc.2010.598. [DOI] [PubMed] [Google Scholar]
- Kenessey I, Keszthelyi M, Kramer Z, Berta J, Adam A, Dobos J, et al. Inhibition of c-Met with the specific small molecule tyrosine kinase inhibitor SU11274 decreases growth and metastasis formation of experimental human melanoma. Curr Cancer Drug Targets. 2010;10:332–342. doi: 10.2174/156800910791190184. [DOI] [PubMed] [Google Scholar]
- Bedikian AY, Millward M, Pehamberger H, Conry R, Gore M, Trefzer U, et al. Bcl-2 antisense (oblimersen sodium) plus dacarbazine in patients with advanced melanoma: the Oblimersen Melanoma Study Group. J Clin Oncol. 2006;24:4738–4745. doi: 10.1200/JCO.2006.06.0483. [DOI] [PubMed] [Google Scholar]
- Jiang Y, DiVittore NA, Kaiser JM, Shanmugavelandy SS, Fritz JL, Heakal Y, et al. Combinatorial therapies improve the therapeutic efficacy of nanoliposomal ceramide for pancreatic cancer. Cancer Biol Ther. 2011;12:574–585. doi: 10.4161/cbt.12.7.15971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morad SA, Cabot MC. Ceramide-orchestrated signalling in cancer cells. Nat Rev Cancer. 2013;13:51–65. doi: 10.1038/nrc3398. [DOI] [PubMed] [Google Scholar]
- Passini MA, Bu J, Fidler JA, Ziegler RJ, Foley JW, Dodge JC, et al. Combination brain and systemic injections of AAV provide maximal functional and survival benefits in the Niemann-Pick mouse. Proc Natl Acad Sci USA. 2007;104:9505–9510. doi: 10.1073/pnas.0703509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orfanelli U, Wenke AK, Doglioni C, Russo V, Bosserhoff AK, Lavorgna G. Identification of novel sense and antisense transcription at the TRPM2 locus in cancer. Cell Res. 2008;18:1128–1140. doi: 10.1038/cr.2008.296. [DOI] [PubMed] [Google Scholar]
- Medic S, Ziman M. PAX3 expression in normal skin melanocytes and melanocytic lesions (naevi and melanomas) PLoS One. 2010;5:e9977. doi: 10.1371/journal.pone.0009977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Noble WS. Matrix2png: a utility for visualizing matrix data. Bioinformatics. 2003;19:295–296. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.