Abstract
Regeneration is the ability of multicellular organisms to replace damaged tissues and regrow lost body parts. This process relies on cell fate transformation that involves changes in gene expression as well as in the composition of the cytoplasmic compartment, and exhibits a characteristic age-related decline. Here, we present evidence that genetic and pharmacological inhibition of autophagy – a lysosome-mediated self-degradation process of eukaryotic cells, which has been implicated in extensive cellular remodelling and aging – impairs the regeneration of amputated caudal fins in the zebrafish (Danio rerio). Thus, autophagy is required for injury-induced tissue renewal. We further show that upregulation of autophagy in the regeneration zone occurs downstream of mitogen-activated protein kinase/extracellular signal-regulated kinase signalling to protect cells from undergoing apoptosis and enable cytosolic restructuring underlying terminal cell fate determination. This novel cellular function of the autophagic process in regeneration implies that the role of cellular self-digestion in differentiation and tissue patterning is more fundamental than previously thought.
Keywords: autophagy, regeneration, zebrafish, differentiation, cell death
Understanding the mechanism and regulation of tissue regeneration is a fundamental and fascinating problem in biology, with significant medical implications.1 Intensive research in the past decade provided evidence that animals have evolved three main mechanisms to restore damaged tissues: the reactivation of resident, multipotent adult stem cells, the dedifferentiation of mature somatic cells near the plane of injury into dividing unipotent progenitors, and, in specific instances, transdifferentiation, during which terminally specialized cells switch lineages to adopt fates different from the original ones.2, 3, 4 Zebrafish has emerged as a powerful vertebrate genetic model to study regeneration. It can replace up to one-fifth of its heart ventricle after injury,5, 6 and has a seemingly unlimited potential to regrow its caudal fin after amputation.7, 8
The zebrafish caudal fin has become a useful proxy to examine the paradigm of appendage regeneration. Similar to renewal of amphibian limbs, caudal fin regeneration starts from a highly proliferative tissue, the blastema, which is formed distal from the site of amputation via dedifferentiation and generates almost exclusively cells of the parent cell's type.9, 10, 11, 12 For example, dedifferentiating cells derived from mature osteoblasts in the zebrafish fin give rise to osteoblasts only. Of note, recent evidence suggests the existence of a putative second source of osteoblast precursors.13 In the case of the zebrafish fin, most major signalling pathways have been implicated in the regenerative process,14, 15, 16 including Fgf (fibroblast growth factor) signalling that occupies a prominent, upstream position in the regulatory network driving blastema formation.17, 18
Cell fate transformation (‘reprogramming') underlying dedifferentiation occurs in both the nucleus (changes in global gene expression) and the cytoplasm (changes in cytoplasmic constituents). Previous studies described profound alterations in the cellular composition of dedifferentiating osteoblasts.10 Extensive remodelling of the cytoplasmic compartment involves the synthesis of new proteins and, in parallel, the degradation of existing ones. The latter process occurs predominantly by autophagy (‘cellular self-eating'), during which parts of the cytoplasm are delivered to the lysosome for degradation by hydrolytic enzymes, in particular proteases.19 Consistent with this primordial role of autophagy in cellular remodelling, autophagic degradation has been implicated in diverse developmental and differentiation processes in divergent animal taxa ranging from nematodes to mammals.20, 21
Here we tested whether cytoplasmic reorganisation enabling dedifferentiation that precedes caudal fin regeneration in zebrafish requires autophagy. We provide evidence that autophagy is activated in the blastema during fin regeneration, and that an elevated level of autophagic activity is indispensable for fin regrowth. According to our data, inactivation of autophagy stimulates mesenchymal blastema cells to undergo apoptosis, and causes defects in the expression of differentiation markers characterising terminal cell fate determination. We conclude that autophagy is required for survival and differentiation of cells in this model of tissue regeneration, and may have an essential role in various regeneration processes such as wound healing or the generation of new neurons to combat neurodegenerative diseases in humans.
Results
Autophagy is upregulated in the blastema during zebrafish caudal fin regeneration
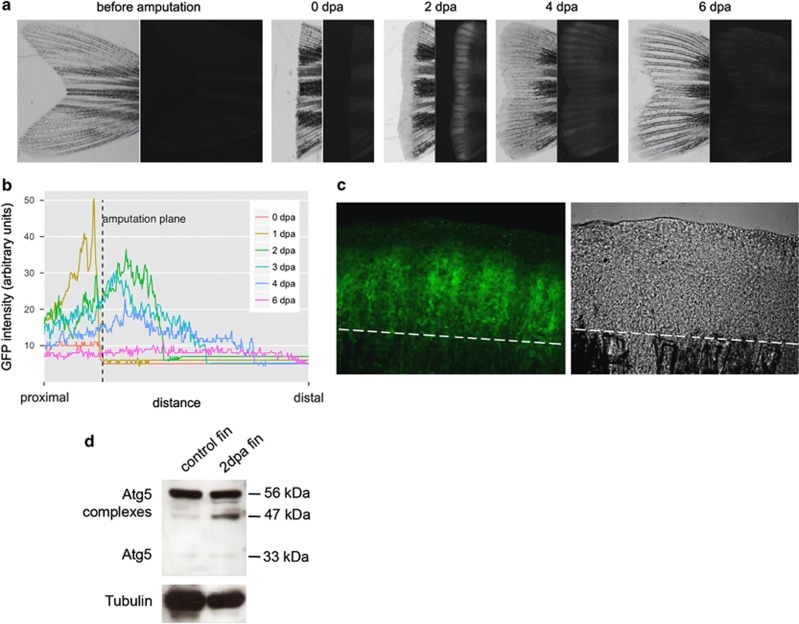
Dramatic changes in osteoblast morphology during dedifferentiation10 suggested the possibility that some regulated process of cytoplasm reorganisation is involved in zebrafish fin regeneration. Our aim was to assess whether this process involves autophagy, a highly conserved pathway of cellular self-degradation.19, 22 To test this hypothesis, we amputated the caudal fins of adult transgenic zebrafish carrying a green fluorescent protein-microtubule-associated protein 1 light chain 3 isoform (GFP-Lc3) reporter.23 Lc3 is a zebrafish orthologue of the yeast Atg8 (autophagy-related 8) protein and mammalian MAP1LC3 (microtubule-associated protein 1 light chain 3) proteins, and the Atg8/LC3 family of proteins is widely used to detect macroautophagic structures (e.g., autophagosomes and autolysosomes) in various species.22, 24 In zebrafish that underwent amputation, expression intensity of the GFP-Lc3 reporter was markedly upregulated in the fin, proximal to the injury plane and in the blastema 2 days post amputation (dpa), then gradually decreased to nearly normal levels during the rest of the regeneration process (Figures 1a–c). During this period, transgene expression could be observed in newly differentiated cells, with the highest levels in the proliferating zone, close to the tail tip. As these cells are descendants of previously dedifferentiated cells, reporter activity could reflect an increased autophagic activity during the dedifferentiation–redifferentiation process.
Figure 1.
Autophagy is upregulated during zebrafish caudal fin regeneration. (a) Intense accumulation of the autophagy marker Lc3/Atg8 in the blastema tissue. Expression of GFP-Lc3 in a zebrafish caudal fin before, at the time of amputation and 2, 4 and 6 days thereafter (dpa). Left: light microscopy images; right: the corresponding fluorescent images. (b) Quantification of fluorescence intensity for a representative fin ray during regeneration. The plane of amputation is kept fixed, and fluorescence is shown relative to this position. Intensity was plotted for the same fin ray over the regeneration period using ImageJ. (c) Confocal images of a 2 dpa GFP-Lc3 blastema. Some GFP-positive foci labelling autophagic elements are interconnected by filamentous structures. The white broken line indicates the amputation plane. (d) Western blots of intact and regenerating tails indicate a strong increase in the amount of complexed Atg5. On average, the amount of Atg5 complexes is roughly 60% higher in 2 dpa regenerates than in control tissues, and in regenerating blastemas, we consistently detect a new ∼47-kDa complex, besides the more common ∼56-kDa one
Analysis of regenerating tails also showed elevated levels of complexes containing Atg5, a mediator of the lipidation and binding of soluble Lc3 (Lc3-I) to the phagophore membrane (Lc3-II),25 in the blastema (Figure 1d). Interestingly, independent experiments in regenerating fins repeatedly showed a stark increase in the amount of a 47-kDa complex, which is present only slightly above background levels in intact controls (Figure 1d). The level of different Atg5-containing complexes changed relative to γ-tubulin (Figure 1d), and thus provided evidence for a selective upregulation of the autophagic process, rather than for a nonselective elevation of protein synthesis. Indeed, a recent proteomic analysis of the regenerating caudal fin provided unequivocal evidence for the differential regulation of dozens of proteins.26
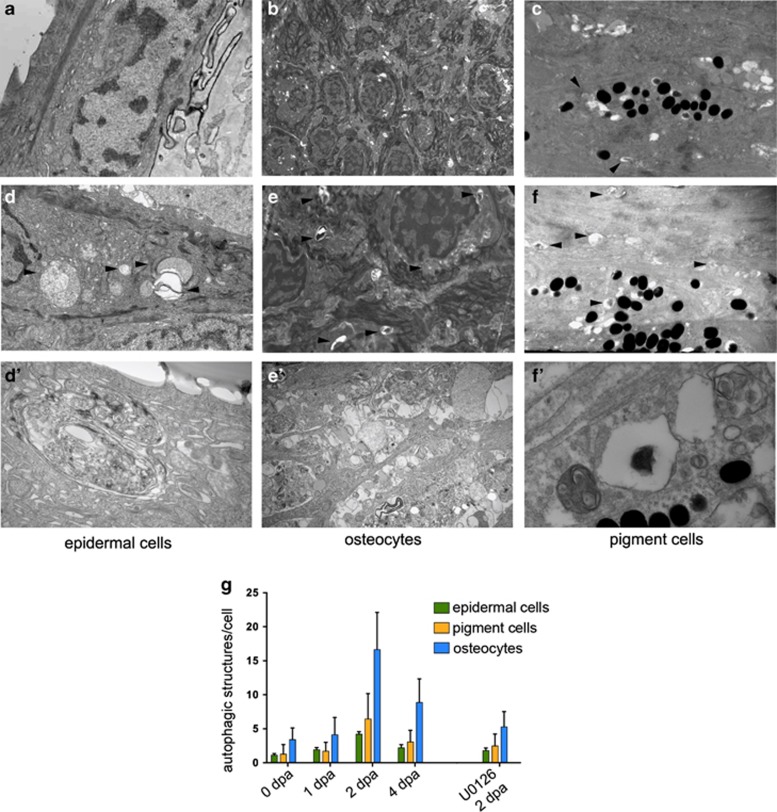
Consistent with the idea that the GFP-Lc3 expression data above reflect an increased rate of autophagy during early phases of regeneration, electron microscopy revealed an elevated number of autophagic structures in several cell types of the 2 dpa fins, compared with untreated control tissues (Figures 2a–g). As revealed by quantification of the autophagic structures (Figure 2g), the increase in autophagic activity in epidermal cells, in osteocytes and in pigment cells detected at the ultrastructural level mirrors the reporter-transgene activity observed in the GFP-Lc3 line. The number of autophagic structures was most abundant after 2 days of amputation and decreased by 4 dpa. Together, these results imply a massive upregulation of autophagy in the regenerating blastema tissue.
Figure 2.
Ultrastructural evidence for an increase in autophagy in the regenerating blastema tissue. Electron microscopy pictures of epidermal cells (a), osteocytes (b) and pigment cells (c) from the tail tip region of control animals show no sign or only low levels of autophagic activity. In the corresponding tissues of the regenerating zone of a 2-dpa fin, elevated numbers of autophagic structures (arrowheads) can be observed (d–f′). (g) The quantification of autophagic vesicles in our EM data set shows a temporary increase in the number of these structures, with the maximum observed at 2 dpa and a consequent decrease by 4 dpa (n=25 for 0-dpa, 1-dpa and 2-dpa samples, and n=15 for the 4-dpa data set). The difference is highly significant for all three examined cell types (P<0.005). In U0126-treated animals (n=25) at 2 dpa, the amount of autophagic structures is significantly lower (P<0.0001) (Student's t-test, error bars refer to S.E.M.)
Autophagy is required for caudal fin regeneration
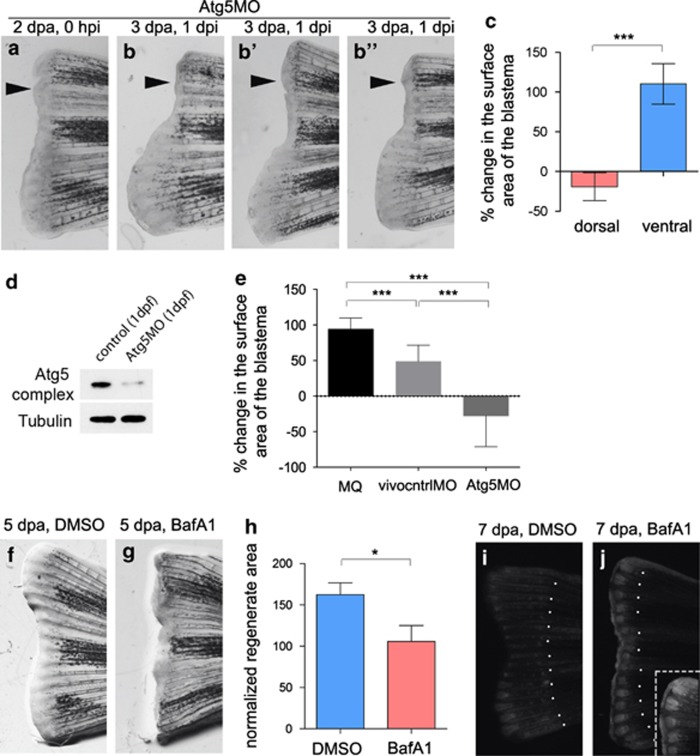
To test whether autophagy is necessary for caudal fin regeneration, we used several strategies to block the process. First, we injected synthetic antisense morpholino oligonucleotide (MO) targeting the translation site of atg5 (Atg5MO) into the dorsal part of the fin stump at 2 dpa (Figures 3a and b′). Depletion of Atg5 was able to effectively block regeneration in the injected fin area, and even induce the degeneration of the existing blastema (Figures 3a–c). As these effects are significantly different from those observed when MilliQ water or a standard control MO were used for injections (Figure 2e), and the same morpholino caused specific defects during embryonic development (Supplementary Figure S1), similar to phenotypes described before for an overlapping Atg5 morpholino,27 its antiregenerative effect is not merely due to a general toxicity of the reagent.
Figure 3.
Macroautophagy is required for caudal fin regeneration. (a–c) When Atg5MO was injected into dorsal areas of 2-dpa blastemas, regeneration was severely compromised 1 day after the injection, compared with uninjected ventral fins (black arrowheads indicate site of injection). On panels b′ and b′′, additional examples of 3 dpa, 1 day post injection regenerates are shown. Statistical analysis (c) was performed for n=9 fins, P<0.001 (indicated by ***), paired t-test. (d) The efficacy of the Atg5MO was tested in western blots. In 1-dpf embryos the intensity of the band demarking the Atg12-Atg5 complex was reduced to less than 20%, indicating a potent inhibition of the Atg5 translation. (e) The effectiveness of the Atg5MO in inhibiting regeneration was significant compared with MilliQ water (MQ) and a standard control vivo MO (vivocntrlMO). (Statistical analysis was performed for n=5 fins from isogenic wild-type tue background fish for each treatment, P<0.001 for each pairwise comparison (indicated by ***), unpaired t-test.) (f–h) Regenerating fins were treated with either DMSO (f) or bafilomycin A1 (g), a potent inhibitor of autophagosome-autolysosome fusion. The latter treatment significantly impaired regeneration (h) at 5 dpa (n=6 fins, P<0.05, unpaired t-test). (i and j) When the fins of GFP-Lc3 transgenic fish were amputated, treatment with bafilomycin A1 led to a massive accumulation of the reporter in the blastema (i), as compared with DMSO controls (i). Inset in (j) shows the dorsal area of the representative fin at higher magnification. Dotted lines indicate amputation sites. Dpa, days post amputation; hpi, hours post injection; dpi, days post injection. Error bars refer to S.E.M.
To further confirm these results, animals that underwent amputation were also treated with the late-stage autophagy inhibitor bafilomycin A1, which blocks both starvation-induced and non-starvation-dependent autophagy.28 Bafilomycin A1-treated animals also lost their ability to regenerate their caudal fin (Figures 3f–j). In addition, the treatment provoked a massive activity of GFP-Lc3 reporter in the blastema (Figure 3j), supporting previous findings that showed an inhibitory effect of bafilomycin A1 on autophagosome-autolyosome fusion,28 and further suggesting an important role for autophagy in the regeneration process. Interestingly, a similar phenotype was observed when fish were incubated with chloroquine, another small-molecule inhibitor of the autophagic process (Supplementary Figure S2), suggesting that the effects of bafilomycin A1 were not merely due to non-specific effects of the drug.
Compromised autophagy triggers apoptotic cell death and blocks differentiation in the blastema
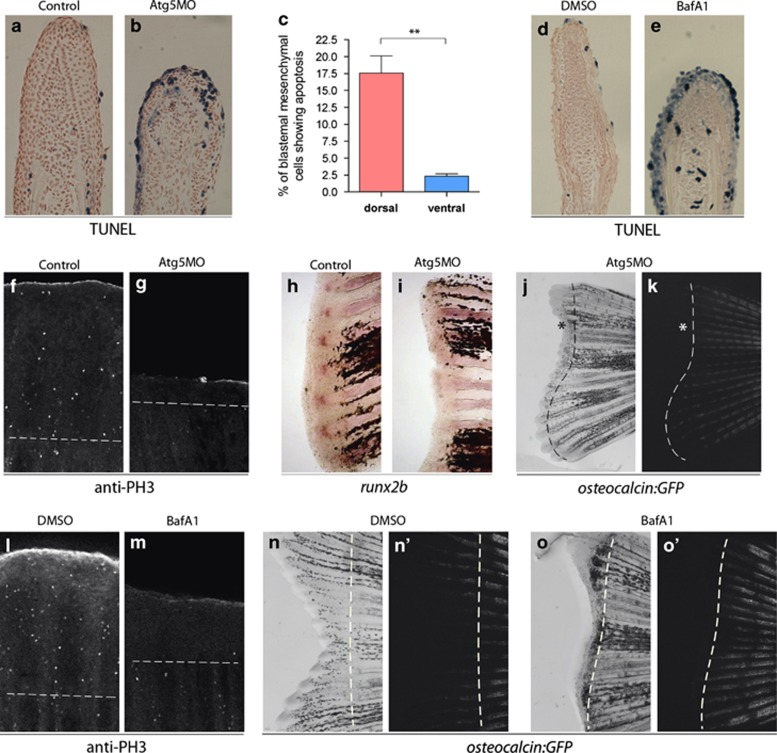
As inhibition of autophagy triggers apoptosis in some in vivo models,29, 30 we monitored whether the reduced regenerative capacity of Atg5MO-injected or bafilomycin A1-treated fin tissues is due, at least in part, to an increase in apoptotic activity. Using TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick end labelling) staining that specifically marks fragmented DNA in apoptotic cells, we examined apoptosis in the blastema of Atg5MO-injected dorsal fins and found an increased rate of cell death at 6 h post injection (hpi), compared with control ventral fins (Figures 4a–c). Quantification of TUNEL-positive cells in autophagy-defective dorsal fin tissues showed elevated levels of cell death compared with control fins (Figure 4c). Increased numbers of apoptotic cells were also observable in bafilomycin A1-treated samples (Figures 4d and e). Atg5-specific morpholino and bafilomycin A1 treatments also reduced the proliferative capacity of the blastema, as revealed by anti-phospho-histone 3 staining on 3 dpa regenerates31 (Figures 4f, g, l, m). These results suggest that impaired autophagy prevents both survival and proliferation of dedifferentiating cells at early stages of the regeneration process.
Figure 4.
Reduced autophagic activity impairs cell survival, proliferation and differentiation during regeneration. (a–c) When dorsal 2-dpa blastema cells were injected with Atg5MO and fixed at 6 hpi, TUNEL staining showed increased cell death in the blastemal mesenchyme of treated fin halves, suggesting that autophagy is required for the survival of dedifferentiating cells. (For the statistical analysis apoptotic and non-apoptotic cells were counted on 62 ventral and 49 dorsal sections, **P<0.01, paired t-test.) (d and e) Chronic treatment with bafilomycin A1 also results in reduced regeneration and increased cell death at 2 dpa in the blastemal mesenchyme. (f and g) At 3 dpa significant amount of mitotic cells, marked by anti-phospho-histone 3 staining can be detected in a control blastema, whereas the number of proliferating cells is significantly reduced in Atg5MO-injected blastemas. (h and i) Six hours post injection 2-dpa caudal fins were fixed and stained for runx2b expression. Whereas the uninjected ventral half shows runx2b staining in the blastema at the tip of the individual fin rays (h), in injected dorsal blastema no staining could be observed (i), suggesting that the survival of pre-osteoblasts was impaired upon morpholino treatment. (j and k) An amputated adult fish shows no osteocalcin:GFP expression at 6 dpa in the Atg5MO-treated dorsal areas. White lines indicate the extent of GFP expression in the bright field (left) and the corresponding fluorescent image (right). The asterisk indicates parts of the blastema tissue that fail to express osteocalcin. (l and m) Chronic treatment with bafilomycin A1 decreases the number of mitotic cells at 3 dpa. (n–o′) Long-term impairment of autophagic activity with bafilomycin A1 suppresses (re)differentiation as shown by the osteocalcin:GFP reporter line in 6 dpa regenerates. Unlike in controls (n′), in treated fish no new osteocalcin-positive cells could be observed at 6 dpa in the fin (o′) (Dashed lines denote the plane of amputation.)
Furthermore, we found that surviving blastema cells in Atg5MO-treated fin tissues are not able to differentiate properly, as they showed impaired expression of the preosteoblast marker runx2b, normally expressed at the top of the growing fin rays10 (Figure 4h). According to our data, runx2b was completely absent from the regenerative zone of caudal fins in the Atg5MO genetic background (Figure 4i). The expression of the differentiated osteoblast reporter osteocalcin:GFP10 was also compromised in Atg5MO-treated animals (Figures 4j and k). Impaired expression of osteocalcin:GFP was also evident upon chronic bafilomycin A1 treatment (Figures 4n–o′). Thus, proliferating cells behind the amputation plane in an autophagy-defective background failed to differentiate into mature osteoblasts. We conclude that autophagy promotes both cell survival and proliferation, and mediates differentiation in the regenerative zone during the regrowth of zebrafish caudal fin.
Mapk/Erk signalling is essential for and regulates autophagy during caudal fin regeneration
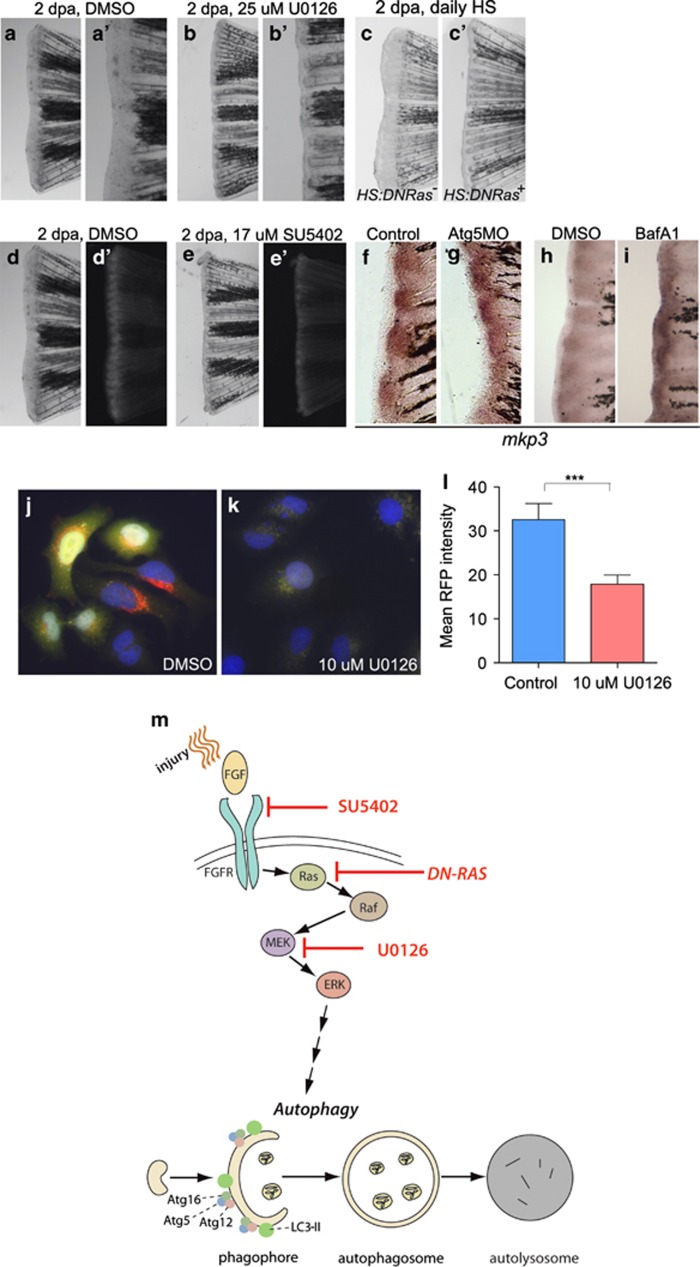
Fgf signalling has a well-established role in zebrafish fin regeneration, and, according to previous reports, inhibition of Fgf signalling blocks the dedifferentiation and concomitant cytoplasmic reorganisation of osteoblasts.10, 18, 32 As one of the effector (downstream) mechanisms of Fgf signalling is the mitogen-activated protein kinase/extracellular signal-regulated kinase (Mapk/Erk)-mediated signal transduction system,33 which has an important role in the regulation of autophagy,34, 35 we tested whether the impairment of Mapk/Erk influences the regeneration capacity of amputated caudal fins. Using the Mapk/Erk inhibitor U0126,36 we observed an almost complete halt of the regeneration process in animals defective for Mapk/Erk activity (Figures 5a–b′). This effect was comparable to treatments with SU5402, a small-molecule inhibitor of the Fgf receptor, that also impairs regeneration of surgically removed fins (Figures 5d–e′).10, 18
Figure 5.
Inhibition of the Mapk/Erk signalling pathway blocks regeneration and impairs autophagy induction in the blastema. (a and b) Inhibition of Mapk/Erk signalling using U0126 impaired caudal fin regeneration (panels a′ and b′ are magnifications from the central fin areas of the samples on panels a and b, respectively). (c and c′) Inhibition of Ras-MAPK signalling using a transgenic hs:DNRas reporter line also impaired regeneration and blastema formation. (d and e) Regeneration of caudal fin can be inhibited with SU5402, which also results in a decrease in GFP-Lc3 activity. (f–i) mkp3 expression does not depend on autophagic activity. Atg5MO-injected dorsal fin blastemas express mkp3 at levels comparable to that in uninjected ventral fins (f and g). Similarly, mkp3 staining displays a similar extent of labelling in DMSO-treated control (h) and autophagy-defective, bafilomycin A1 (BafA1)-treated backgrounds (i). (j–l) Serum-starved, DMSO-treated GFP-mRFP-LC3 HeLa cells show significant levels of autophagic activity, as denoted by the yellow and red puncta (autophagosomes and autolysosomes, respectively) (j and l). When 10 μM or 100 μM U0126 was added to the medium, we observed a drop in the autophagic activity (k and l). At concentrations comparable to our treatments, the effect of U0126 was highly significant (l). n=43 cells measured in both control and treated groups, ***P<0.001, unpaired t-test, error bars refer to S.E.M. (m) Schematic diagram showing the proposed interaction between the Fgf-Mapk/Erk signalling axis and autophagy. Injury activates Fgf signalling in the fin, which leads to the phosphorylation of Mapk/Erk. Activated Mapk/Erk regulates the sequestration step of autophagy, an essential step in the degradation of the cytosolic component. The small molecule reagents and the MO used in this study inhibit the process at the indicated steps. The three arrows from ‘ERK' to ‘Autophagy' indicate an indirect link between Mapk/Erk signalling and autophagy
The Ras-Mapk/Erk signalling pathway was identified previously as an important stimulator of the autophagic process.34, 35 Therefore, we decided to test whether the impairment of Mapk/Erk signalling influences autophagic activity in amputated caudal fins. Our electron microscopy analysis revealed reduced numbers of autophagic structures in the distal zone of caudal fins of 2 dpa animals treated with U0126 (Figure 1g). This implies that Mapk/Erk activity is required for upregulation of autophagy during caudal fin regeneration.
To test this hypothesis further, we took advantage of a transgenic fish line expressing a dominant-negative form of Ras (DNRas) under a heat shock promoter (hs:DNRAS).37 Ectopic expression of DNRas impaired regeneration and blastema formation (Figures 5c and c′). We also found that treatment with SU5402 led to basal levels of fluorescence in GFP-Lc3 blastemas (Figures 5e and e′). Similar results were obtained from transgenic animals treated with U0126 (data not shown).
We also examined the expression of the Mapk/Erk signalling marker mkp3 (mitogen-activated protein kinase phosphatase 3) in the blastema, and found that it was not affected in autophagy-defective fish, as compared with normal, non-treated animals (Figures 5f–i). To further confirm that autophagy acts downstream of Mapk/Erk signalling, serum-starved human (HeLa) cells transgenic for a GFP::RFP::Atg8/LC3 reporter were treated with U0126 and analysed with fluorescence microscopy. Untreated cells contained significantly more yellow and red foci (autophagosomes and mature autolysosomes, respectively) than the treated cells (Figures 5j–l). Together, our data are in accordance with a possible model where autophagy acts downstream of Fgf-Mapk/Erk signalling to mediate cellular reorganisation and promote cell survival during the regeneration of amputated caudal fins in zebrafish (Figure 5m, Supplementary Figure S3).
Discussion
Most regeneration studies hitherto concentrated on transcriptional events occurring during tissue restoration, yet ultrastructural observations already provided clear evidence that the cytoplasmic component of dedifferentiating cells is dramatically reorganised during heart and caudal fin regeneration in the zebrafish.5, 10 In this study, we observed increased autophagic activity in the blastema during regeneration of amputated caudal fins (Figure 1), suggesting a previously unaccounted biological role for a lysosome-mediated intracellular degradation process in this model of tissue restoration. Supporting the novelty of these results, none of the recent major in-depth reviews of vertebrate regeneration2, 3, 4 cover autophagy to any extent, clearly showing that the role of this process has been hitherto undervalued, if appreciated at all.
An increase in autophagic activity was also reported during planarian regeneration,38 a process reliant on a specialized pool of stem-cell-like cNeoblasts.4 However, a general requirement for autophagy in that tissue replacement model was not tested by genetic tools. Therefore, we could not answer the fundamental question of whether upregulation of autophagy was a simple consequence of cellular stress caused by the injury or an underlying causative mechanism of tissue renewal, which may provide energy and building blocks for proliferating cells or remove specific cytoplasmic components to allow cellular differentiation.
In caudal fin blastemas of autophagy-defective zebrafish, significant levels of apoptotic cell death were observed (Figures 3a–c). It is unlikely that such a massive amount of cells with apoptotic features could be generated as a simple consequence of failure in the heterophagic elimination of apoptotic cell corpses, as was suggested earlier.19 Thus, our present data further support an active role for autophagy in preventing intensively proliferating and differentiating cells from undergoing apoptosis, similarly to what was shown in mammalian cell lines and nematodes.29, 30 These results suggest that (re)patterning of amputated caudal fin in zebrafish depends on the death-suppressing function of the autophagic process.
Furthermore, surviving blastema cells from the autophagy-defective zebrafish failed to express differentiation markers, such as runx2b and osteocalcin (Figure 4),10 and were eventually unable to regrow surgically removed caudal fins (Figure 3). Thus, autophagic degradation also appears to be required for cellular remodelling that underlies cell fate reprogramming during injury-induced regeneration. Our present study provides the first evidence that autophagy acts as a prerequisite for the (re)formation of a complete organ, the zebrafish caudal fin.
Fgf signalling has been demonstrated to be essential for initiating both zebrafish fin regeneration and homoeostatic regeneration.18, 39 However, the effector (downstream) mechanisms that mediate Fgf receptor activity in these regeneration processes remain elusive. Previously, the Ras-Mapk/Erk pathway, one of the signalling modules downstream of Fgf receptor activation, has been linked to the regulation of autophagy. There is some ambiguity as to how exactly Mapk/Erk regulates autophagy, but links to BECN1 and G-protein-type regulators of the autophagic machinery have been proposed,40, 41 as well as a role in the control of autophagosome maturation.34 We showed here that a specific inhibitor of Mapk/Erk, which is able to downregulate the autophagic process in other organisms35 and in human cells (Figures 5j–l), effectively blocks fin regeneration in the zebrafish model (Figure 1g, Figures 5a–b′). Furthermore, impairment of Ras activity also inhibits effective regeneration (Figures 5c and c′). Overall, our results support a model, where the activation of the Fgf-Ras-Mapk/Erk signalling axis, following amputation, induces concomitant activation of a conserved genetic dedifferentiation programme and reorganisation of the cytoplasmic milieu in differentiated cells close to the amputation plane. The profound changes in the cytoplasmic component are dependent on autophagy and required for the generation of proliferating unipotent progenitor cells of the blastema (Supplementary Figure S3). Such a potential role for autophagy during regeneration is also highlighted by a recent study, where reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) was significantly enhanced by autophagy-promoting drugs.42
The ultimate goal of regenerative medicine is to enable the regrowth of damaged or lost tissues in humans. For example, restoring neurons damaged in an accident, or cardiac muscle lost due to heart attack, has the potential to dramatically increase survival and life quality in the affected individuals. Our present finding that autophagy is required for zebrafish caudal fin regeneration may help to develop innovative strategies to stimulate regeneration by modulating autophagic activity in the damaged tissues. Finally, the importance of autophagic degradation in trauma-induced tissue renewal also raises the possibility that autophagy functions in homoeostatic regeneration, the natural replacement of cells that become lost due to minor damage that occurs every day and during aging. This may reflect a novel aspect of aging control by autophagy.43
Materials and Methods
Fish care
Wild-type fish of the ekwill (ekw) strain and transgenic zebrafish outbred into the same genetic background of 4–12 months of age were used. Fin amputations were performed according to the standard protocols of the field.44 After surgery, the fish were kept at 28.5 °C. During heat shock experiments, water temperature was gradually increased to 38 °C. After 40-min incubation the fish were placed back at 28.5 °C. Heat shocks were delivered after amputation and at 1 and 2 dpa, respectively. All fish stocks were maintained in the Animal Facility of Eötvös Loránd University. All protocols used in this study were approved by the Hungarian National Food Chain Safety Office (Permit Number: XIV-I-001/515–4/2012).
Electron microscopy
Tail tissues were fixed overnight in 2% paraformaldehyde (PFA) (Sigma-Aldrich, St. Louis, MO, USA, 158127), 2% glutaraldehyde (TAAB, G011), 1% sucrose, 0.028% CaCl2 (Sigma-Aldrich, C5670) in 0.1 M sodium cacodylate (Sigma-Aldrich, C0250) (pH 7.4), then rinsed with cacodylate for 3 days at 4 °C. Samples were post fixed with 0.5% OsO4 (Sigma-Aldrich, 419494) and embedded in Durcupan (Electron Microscopy Sciences, Hatfield, PA, USA, 14020) plastic resin. Ultrathin sections were collected on formvar-coated grids and stained with lead citrate and uranyl acetate. The samples were observed with a JEOL CM-II 100 electron microscope.
Morpholino experiments
To block the activity of atg5 transcripts, synthetic morpholino antisense oligonucleotides (MOs) were synthesised (GeneTools, Philomath, OR, USA) to mask the translational start site. The Atg5MO sequence was as follows (5′ to 3′): CCTTGTCATCTGCCATTATCATCGT. The morpholino was vivo-porter coupled, as this system was shown previously to be highly effective in zebrafish balstemas.45 MOs were injected into regenerates as described previously45, 46 or at the 1–2 cell stage, into the yolk, under the cytoplasm. In the case of the osteocalcin:GFP reporter line, dorsal fin halves were injected every day from 2 dpa on. As a control, Gene Tools' standard vivo control MO was used.
In situ hybridisation
Digoxigenin-labelled antisense probes for runx2b47 and mkp348 were synthesised as described previously.49 Detection of mRNA transcripts was performed as follows: Fin fragments were fixed in 4% PFA (PFA; Taab Laboratories Equipment Ltd, Reading, UK) in phosphate-buffered saline containing 0.1% Tween-20 (PBST) overnight at 4 °C, washed briefly in PBST and then transferred into methanol and stored at −20°C. Samples were rehydrated at room temperature through a 10 min wash in 50% methanol-PBST mix, followed by four 5-min washes in PBST Then, fins were incubated with 10 μg/ml proteinase K in PBST for 40 min, followed by a 20-min refixation in 4% PFA in PBST After five more washes of 5 min in PBST, samples were prehybridized for 3 h at 65 °C in 50% formamide, 5x SSC, 0.1% Tween-20, 50 μg/ml heparin and 500 μg/ml torula yeast RNA, adjusted to pH 6.0 with citric acid, and then hybridised overnight in the same buffer including a 1 : 400 dilution of the digoxigenin-labelled probes. The second day, the samples were rinsed in 75% formamide/25% 2x SSC solution, followed by a 1 h wash in 50% formamide/50% 2x SSC solution at 65 °C. Two 10-min washes with 2x SSC in PBST followed at room temperature, and a 10 min wash in 0.1 M NaCl, 0.01 M Tris-HCl (pH 8.0), 0.1% Tween-20 (RNase buffer). Next, samples were incubated for 10 min with 20 μg/ml RNase A and 10 U/ml RNase T1 in RNase buffer, and washed for another 10 min with 2x SSC in PBST Samples were returned to 65 °C into the 50% formamide/50% SSC buffer for 1 h, followed by 15 min washes with 2x SSC in PBST, 0.2x SSC in PBST and PBST, all at room temperature. Samples were incubated for 1 h in Blocking Reagent (Roche, 11096176001) and with a 1 : 4000 dilution of an alkaline phosphatase conjugated anti-digoxigenin antibody (Roche Applied Science, Penzberg, Germany, 11093274910) overnight at 4 °C. In the last day, after three rinses with PBST, samples were washed six times for 15 min in PBST Before detection, three 5-min washes in an alkaline phosphatase buffer (100 mM Tris-HCl (pH 9.5), 50 mM MgCl2, 100 mM NaCl, 0.1% Tween-20) followed. 1 × NBT/BCIP (combination of nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate) substrate was used as chromogen. Samples were kept in the dark during the staining reaction. The reaction was stopped with three brief rinses in PBST, followed by a post-fixation with 4% PFA in PBST Samples were stored in 80% glycerol at 4 °C. For imaging, fin fragments were mounted on a standard glass slide.
Western blots
Western blots were performed as described previously.50 Anti-γ-tubulin antibody, used at 1 : 10 000 dilution was obtained from Sigma-Aldrich (T5326), and anti-Atg5 antibody, diluted to 1 : 1000 was purchased from Novus Biologicals (Littleton, CO, USA, NB110–53818). Quantification of the bands was done with ImageJ software (http://rsbweb.nih.gov/ij/).
Immunohistochemistry
Anti-phospho-histone 3 staining on 3 dpa regenerates were performed as described previously.31
Cell death assays
TUNEL staining on fin samples was performed using an ApopTag kit (Chemicon/Millpore, Billerica, MA, USA, S7105, S7106, S7107), following the manufacturer's protocol. Samples were fixed, rehydrated, blocked and developed as described for the in situ hybridisation protocol. Samples were embedded in JB-4 resin and sectioned. Sections were counterstained with 0.002% Neutral Red for 15 min. Slides were subsequently dried and covered. After imaging, cells were counted manually in the blastemal mesenchyme.
Treatments with small molecule reagents
Bafilomycin A1 (Sigma-Aldrich, B1793) was dissolved in DMSO to create a 100 μM stock solution and then diluted to a concentration of 20 nM in fish water. U0126 (Promega, Fitchburg, WI, USA, V1121) was dissolved in DMSO and then diluted in E3 embryo medium to 25 μM final concentration. SU5402 (Merck Millipore, Damstadt, Germany, 572631) was used at 17 μM concentration. Control animals (exposed to 0.1% DMSO) and treated counterparts were always kept at 28.5 °C, with their medium changed daily. The GFP-RFP-LC3 HeLa cell line51 was cultured in DMEM (Dulbecco's Modified Eagle's medium; Sigma-Aldrich, D7777) containing 4500 mg/l glucose, 10% heat-inactivated fetal calf serum (Merck, Damstadt, Germany), 50 μg/ml gentamycin (Hungaropharma, Budapest, Hungary) and 600 μg/ml G418 (Sigma-Aldrich, G8168). Cells (2 × 104) were plated onto 10-mm diameter poly-D-lysine coated coverslips in 24-well plates (Greiner Bio-One, Frickenhausen, Germany) 10 h before the treatment. Autophagy was induced by serum starvation in fetal calf serum-free medium while cells were exposed to 10 μM U0126 or 0.1% DMSO as control for 14 h. Cells were fixed in 4% PFA and mounted in bis-benzimide (Sigma-Aldrich) containing mowiol 4.88 (Polysciences, Warrington, PA, USA). Epifluorescent images were taken of each condition using a BX51 microscope (Olympus, Tokyo, Japan) fitted with a FluoViewII camera and the AnalysisPro software (Olympus), using a × 60/1.4 oil Plan objective and the appropriate filter sets (DAPI: BP330–385/DM400/BA420; GFP: BP460–500/DM505/BP510–560; RFP: BP480–550/DM570/BA590). Images were analysed by ImageJ software. The quantity of RFP-positive autolysosomes (autophagosomes processed in the degradation pathway) was compared by measuring the RFP mean intensity in U0126-treated and control cells.
Imaging, fin area measurements and analysis
Bright field and fluorescence pictures of live fin regenerates were taken with a Zeiss AxioZoom stereomicroscope. Whole mount in situ hybridisation pictures were taken with a Nikon E1000 microscope. Changes in the size of regenerates were calculated using pictures taken right after injection and 1 day post injection. Relative areas of dorsal and ventral regenerates were calculated using the ImageJ software. The same programme was used to calculate regenerate sizes of DMSO- and bafilomycin A1-treated fins, where normalisation was done by dividing the area of the regenerate by the width of the fin. Statistical analysis (paired and unpaired t-test) was performed using the Prism (GraphPad) software.
Acknowledgments
This work is supported by the Hungarian Scientific Research Funds NK78012 and K109349. MV, KT-V and TV are grantees of the János Bolyai scholarship. DJK is supported by NIH grant GM053396. We thank Steve Wilson and members of the Wilson lab for technical help, Gilbert Weidinger and Alex Nechiporuk for the osteocalcin:GFP and hs:DNRas lines, David Rubinsztein for the GFP::RFP::Atg8/LC3 HeLa cell line and Kara Cerveny and Julianna Vig for helpful comments and suggestions regarding the manuscript.
Glossary
- Atg
autophagy-related
- DNRas
dominant-negative form of Ras
- Dpa
days post amputation
- Erk
extracellular signal-regulated kinase
- Fgf
fibroblast growth factor
- GFP
green fluorescent protein
- GFP-Lc3
GFP fused at the N terminus to Lc3
- Hpi
hours post injection
- Lc3
microtubule-associated protein 1 light chain 3
- Mapk
mitogen-activated protein kinase
- mkp3
mitogen-activated protein kinase phosphatase 3
- MO
synthetic antisense morpholino oligonucleotide
- PBST
phosphate-buffered saline containing 0.1% Tween-20
- PFA
paraformaldehyde
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick end labelling
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by E Baehrecke
Supplementary Material
References
- Berthiaume F, Maguire TJ, Yarmush ML. Tissue engineering and regenerative medicine: history, progress, and challenges. Annu Rev Chem Biomol Eng. 2011;2:403–430. doi: 10.1146/annurev-chembioeng-061010-114257. [DOI] [PubMed] [Google Scholar]
- Jopling C, Boue S, Izpisua Belmonte JC. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat Rev Mol Cell Biol. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- Poss KD. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat Rev Genet. 2010;11:710–722. doi: 10.1038/nrg2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka EM, Reddien PW. The cellular basis for animal regeneration. Dev Cell. 2011;21:172–185. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Martí M, Raya A, Izpisúa Belmonte JC, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, et al. Primary contribution to zebrafish heart regeneration by gata4+cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo AS, Sousa S, Jacinto A, Saude L. An amputation resets positional information to a proximal identity in the regenerating zebrafish caudal fin. BMC Dev Biol. 2012;12:24. doi: 10.1186/1471-213X-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itou J, Kawakami H, Burgoyne T, Kawakami Y. Life-long preservation of the regenerative capacity in the fin and heart in zebrafish. Biology Open. 2012;1:739–746. doi: 10.1242/bio.20121057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- Knopf F, Hammond C, Chekuru A, Kurth T, Hans S, Weber CW, et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell. 2011;20:713–724. doi: 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Sousa S, Afonso N, Bensimon-Brito A, Fonseca M, Simões M, Leon J, et al. Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development. 2011;138:3897–3905. doi: 10.1242/dev.064717. [DOI] [PubMed] [Google Scholar]
- Tu S, Johnson SL. Clonal analyses reveal roles of organ founding stem cells, melanocyte stem cells and melanoblasts in establishment, growth and regeneration of the adult zebrafish fin. Development. 2011;137:3931–3939. doi: 10.1242/dev.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SP, Holdway JE, Poss KD. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell. 2012;22:879–886. doi: 10.1016/j.devcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum N, Begemann G. Retinoic acid signaling controls the formation, proliferation and survival of the blastema during adult zebrafish fin regeneration. Development. 2012;139:107–116. doi: 10.1242/dev.065391. [DOI] [PubMed] [Google Scholar]
- Laforest L, Brown CW, Poleo G, Géraudie J, Tada M, Ekker M, et al. Involvement of the sonic hedgehog, patched 1 and bmp2 genes in patterning of the zebrafish dermal fin rays. Development. 1998;125:4175–4184. doi: 10.1242/dev.125.21.4175. [DOI] [PubMed] [Google Scholar]
- Smith A, Avaron F, Guay D, Padhi BK, Akimenko MA. Inhibition of BMP signaling during zebrafish fin regeneration disrupts fin growth and scleroblasts differentiation and function. Dev Biol. 2006;299:438–454. doi: 10.1016/j.ydbio.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310:1957–1960. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- Poss KD, Shen J, Nechiporuk A, McMahon G, Thisse B, Thisse C, et al. Roles for Fgf signaling during zebrafish fin regeneration. Dev Biol. 2000;222:347–358. doi: 10.1006/dbio.2000.9722. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Bartholomew CR, Zhou W, Klionsky DJ. Assaying autophagic activity in transgenic GFP-Lc3 and GFP-Gabarap zebrafish embryos. Autophagy. 2009;5:520–526. doi: 10.4161/auto.5.4.7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–170. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S, Singh SK, Lakshmi MG, Meghah V, Bhatti B, Swamy CV, et al. Proteomic analysis of zebrafish caudal fin regeneration. Mol Cell Proteomics. 2012;11:014118. doi: 10.1074/mcp.M111.014118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Zhang J, Zhang Q. Expression pattern and functions of autophagy-related gene atg5 in zebrafish organogenesis. Autophagy. 2011;7:1514–1527. doi: 10.4161/auto.7.12.18040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundeshagen P, Hamacher-Brady A, Eils R, Brady NR. Concurrent detection of autolysosome formation and lysosomal degradation by flow cytometry in a high-content screen for inducers of autophagy. BMC Biol. 2011;9:38. doi: 10.1186/1741-7007-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, González-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs-Vellai K, Vellai T, Puoti A, Passannante M, Wicky C, Streit A, et al. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr Biol. 2005;15:1513–1517. doi: 10.1016/j.cub.2005.07.035. [DOI] [PubMed] [Google Scholar]
- Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development. 2002;129:5141–5149. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–5183. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- Dailey L, Ambrosetti D, Mansukhani A, Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 2005;16:233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Corcelle E, Djerbi N, Mari M, Nebout M, Fiorini C, Fénichel P, et al. Control of the autophagy maturation step by the MAPK ERK and p38: lessons from environmental carcinogens. Autophagy. 2007;3:57–59. doi: 10.4161/auto.3424. [DOI] [PubMed] [Google Scholar]
- Kang C, You YJ, Avery L. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev. 2007;21:2161–2171. doi: 10.1101/gad.1573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- Lee Y, Lee Y, Hami D, De Val S, Kagermeier-Schenk B, Wills AA, et al. Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins. Dev Biol. 2009;331:270–280. doi: 10.1016/j.ydbio.2009.05.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Estevez C, Felix DA, Aboobaker AA, Salo E. Gtdap-1 promotes autophagy and is required for planarian remodeling during regeneration and starvation. Proc Natl Acad Sci USA. 2007;104:13373–13378. doi: 10.1073/pnas.0703588104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills AA, Kidd AR, III, Lepilina A, Poss KD. Fgfs control homeostatic regeneration in adult zebrafish fins. Development. 2008;135:3063–3070. doi: 10.1242/dev.024588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Whiteman MW, Lian H, Wang G, Singh A, Huang D, et al. A non-canonical MEK/ERK signaling pathway regulates autophagy via regulating Beclin 1. J Biol Chem. 2009;284:21412–21424. doi: 10.1074/jbc.M109.026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogier-Denis E, Pattingre S, El Benna J, Codongo P. Erk1/2-dependent phosphorylation of Gα-interacting protein stimulates its GTPase accelerating activity and autophagy in human colon cancer cells. J Biol Chem. 2000;275:39090–39095. doi: 10.1074/jbc.M006198200. [DOI] [PubMed] [Google Scholar]
- Chen T, Shen L, Yu J, Wan H, Guo A, Chen J, et al. Rapamycin and other longevity-promoting compounds enhance the generation of mouse induced pluripotent stem cells. Aging Cell. 2011;10:909–911. doi: 10.1111/j.1474-9726.2011.00722.x. [DOI] [PubMed] [Google Scholar]
- Vellai T, Takács-Vellai K, Sass M, Klionsky DJ. The regulation of aging: does autophagy underlie longevity. Trends Cell Biol. 2009;19:487–494. doi: 10.1016/j.tcb.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Shen J, Keating MT. Induction of lef1 during zebrafish fin regeneration. Dev Dyn. 2000;219:282–286. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1045>3.3.co;2-3. [DOI] [PubMed] [Google Scholar]
- Chablais F, Jazwinska A. IGF signaling between blastema and wound epidermis is required for fin regeneration. Development. 2010;137:871–879. doi: 10.1242/dev.043885. [DOI] [PubMed] [Google Scholar]
- Hyde DR, Godwin AR, Thummel R. In vivo electroporation of morpholinos into the regenerating adult zebrafish tail fin. J Vis Exp. 2012;29:e3632. doi: 10.3791/3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Felber K, Elks P, Croucher P, Roehl HH. Tracking gene expression during zebrafish osteoblast differentiation. Dev Dyn. 2009;238:459–466. doi: 10.1002/dvdy.21838. [DOI] [PubMed] [Google Scholar]
- Tsang M, Maegawa S, Kiang A, Habas R, Weinberg E, Dawid IB, et al. A role for MKP3 in axial patterning of the zebrafish embryo. Development. 2004;131:2769–2779. doi: 10.1242/dev.01157. [DOI] [PubMed] [Google Scholar]
- Maegawa S, Varga M, Weinberg ES. FGF signaling is required for β-catenin-mediated induction of the zebrafish organizer. Development. 2006;133:3265–3276. doi: 10.1242/dev.02483. [DOI] [PubMed] [Google Scholar]
- Link V, Shevchenko A, Heisenberg CP. Proteomics of early zebrafish embryos. BMC Dev Biol. 2006;6:1. doi: 10.1186/1471-213X-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.