Abstract
In embryogenesis, p63 is essential to develop mammary glands. In the adult mammary gland, p63 is highly expressed in the basal cell layer that comprises myoepithelial and interspersed stem/progenitor cells, and has limited expression in luminal epithelial cells. In adult skin, p63 has a crucial role in the maintenance of epithelial stem cells. However, it is unclear whether p63 also has an equivalent role as a stem/progenitor cell factor in adult mammary epithelium. We show that p63 is essential in vivo for the survival and maintenance of parity-identified mammary epithelial cells (PI-MECs), a pregnancy-induced heterogeneous population that survives post-lactational involution and contain multipotent progenitors that give rise to alveoli and ducts in subsequent pregnancies. p63+/− glands are normal in virgin, pregnant and lactating states. Importantly, however, during the apoptotic phase of post-lactational involution p63+/− glands show a threefold increase in epithelial cell death, concomitant with increased activation of the oncostatin M/Stat3 and p53 pro-apoptotic pathways, which are responsible for this phase. Thus, p63 is a physiologic antagonist of these pathways specifically in this regressive stage. After the restructuring phase when involution is complete, mammary glands of p63+/− mice again exhibit normal epithelial architecture by conventional histology. However, using RosaLSL-LacZ;WAP-Cre transgenics (LSL-LacZ, lox-stop-lox β-galactosidase), a genetic in vivo labeling system for PI-MECs, we find that p63+/− glands have a 30% reduction in the number of PI-MEC progenitors and their derivatives. Importantly, PI-MECs are also cellular targets of pregnancy-promoted ErbB2 tumorigenesis. Consistent with their PI-MEC pool reduction, one-time pregnant p63+/− ErbB2 mice are partially protected from breast tumorigenesis, exhibiting extended tumor-free and overall survival, and reduced tumor multiplicity compared with their p63+/+ ErbB2 littermates. Conversely, in virgin ErbB2 mice p63 heterozygosity provides no survival advantage. In sum, our data establish that p63 is an important survival factor for pregnancy-identified PI-MEC progenitors in breast tissue in vivo.
Keywords: p63, mammary gland, involution, stem cells, PI-MECs, ErbB2 breast cancer
During embryonic development the transcription factor and p53 homolog p63 is absolutely essential for stratified epithelial morphogenesis, including limb buds, skin and mammary gland (a major skin appendage).1 Moreover, in adult skin and some other stratified epithelia p63 is highly expressed in basal stem cells where it is required as a master proliferative/differentiation and maintenance factor.2, 3, 4 Owing to early post-natal lethality of global p63−/− mice, it has been difficult to analyze the role of p63 during adult tissue homeostasis. Specifically, the functional role of p63 in adult breast has only begun to be explored and virtually no in situ studies exist, except for basic p63 immunohistochemistry data.
The mammary gland is a unique structure of ducts and acini composed of basal (myoepithelial) and luminal epithelial cells, which undergoes major changes throughout life.5, 6 Initially, the mammary epithelium forms in embryogenesis and undergoes allometric growth until puberty. Next, in response to sex hormones the epithelial tree undergoes major expansion and branching to fill the entire mammary fat pad and, in virgins, cycles with estrus. During pregnancy, massive cell proliferation and differentiation leads to the formation of alveoli, the milk-producing structural units.6 After lactation is complete and pups are weaned, post-lactational involution takes place, where the mammary gland returns to its nonlactating state, resembling again a virgin gland. Involution is regulated by environmental (milking or suckling) and hormonal factors, and has two distinct phases. During early involution, the now obsolete secretory epithelial cells get shed into the alveolar lumen where they undergo apoptosis.7, 8 This is followed by late involution (structural remodeling) when excessive cellular components (epithelial, myoepithelial, endothelial and basement membrane) are removed and replaced by adipose tissue.7 During lactation, the mammary epithelium is maintained by prosurvival signals (e.g., Akt and Jak2/Stat5), which are subsequently inactivated in involution (e.g., by leukemia inhibitory factor (LIF), Stat3 and TGFβ3 signaling). In addition, pro-apoptotic proteins (e.g., Bax and p53) are activated during the apoptotic phase of involution.7, 8
It has long been known that p63 is required for fetal mammary gland development, as p63 knockout (KO) mice lack mammary epithelium altogether.1 However, whether p63 also has a role in adult mammary has only begun to be elucidated. p63 protein exists in two major isoforms. TAp63 contains the N-terminal transactivation (TA) domain, whereas ΔNp63 lacks this TA domain, but retains a second internal TA domain.9 In addition, differential splicing at the C-terminus produces α-, β- (long) and γ- (short) isoforms.10 In the epidermis, abundantly expressed ΔNp63α protein is the predominant isoform and exerts its critical pro-proliferative/maintenance function in skin stem cells.10, 11, 12, 13, 14, 15 Selective genetic ablation of all ΔNp63 isoforms14 or of the long p63α/β isoforms (Wolff et al.16 and references therein) closely reproduces the phenotypes of global p63−/− mice. In contrast, TAp63 mRNA and protein, expressed at nearly undetectable levels in the skin, has a fine-tuning role in keratinocyte differentiation15 and helps maintain adult skin stem cells.17
In human and murine breast immunohistochemistry, ΔNp63 protein is highly expressed in the nuclei of basal myoepithelial cells, similar to other stratified epithelia.2, 18, 19, 20 In addition, TAp63 protein was reported in luminal cells in a few human samples.13 This is consistent with ΔNp63 expression in sorted CD29high/CD49fhigh (integrins β1/α6, marking basal and stem cells) and TAp63 expression in CD29low/CD49flow (marking luminal and progenitor cells) mouse mammary epithelial cells, respectively.21 Loss of epithelial p63 expression correlates with invasion and can be used as a prognostic marker in breast and prostate cancer.22 However, the biological role of p63 in healthy adult breast only begins to be elucidated. To this end, in primary adult human breast epithelial cells ΔNp63 was shown to be required for maintenance of the myoepithelial (CD10positive) compartment.23 Indeed, ΔNp63 silencing in CD10positive cells induces luminal markers (CK18 (cytokeratin 18) and CD24), but reduces basal markers (CK14 (cytokeratin 14) and CD29/CD49f) as well as proliferation. Conversely, forced ΔNp63 expression in CD10negative cells confers a basal phenotype and increases proliferation. This essential role of ΔNp63 in the balance between basal and luminal breast compartments is regulated by p63's antagonistic interaction with Notch signaling, which is active in luminal cells.23 In addition, humans born with frameshift mutations in exons 13 and 14 of p63 (corresponding to the p63 α- and β-isoforms) can develop Limb-mammary syndrome, which includes nipple and mammary gland hypoplasia,24 strongly suggesting an essential role of p63 in adult human breast.
To fully understand the role of p63 in adult breast in health and disease, pregnancy and lactation needs to be studied, as complete mammary differentiation (i.e., milk production) only occurs right after pregnancy. Moreover, both in humans and mice pregnancy strongly influences mammary tumorigenesis.25, 26 During pregnancy, p63 is also expressed in the basal cell layer,20 again with ΔNp63 predominating.27 Moreover, Ihh signaling is enhanced and likely contributes to the expansion of ΔNp63negative progenitors to allow mammary gland ‘elaboration'.21 In addition, cytopoplasmic p63 protein is detected in a small fraction (∼0.1%) of cells isolated from human breast milk, which increases to almost 100% in both adherent and non-adherent cells after 5 days in culture.28 On cell adherence, p63 translocates to the nucleus (which coincides with the expression of cell cycle arrest protein 14-3-3σ), followed by loss of p63 expression and differentiation into CK14positive (basal) and CK18positive (luminal) cells. On 14-3-3σ knockdown, p63 expression is prolonged and proliferation is enhanced.28 These circumstantial data suggest that p63 is expressed in human mammary stem/progenitor cells and serves to maintain their undifferentiated, highly proliferative state. Similarly, block of Notch signaling in mouse mammary gland during pregnancy stimulates p63 expression in luminal cells and increases the number of p63positive basal cells, which coincided with their hyperproliferation.29
Altogether, these data point to p63 as an important maintenance/proliferation factor of the basal epithelial compartment, where mammary stem/progenitor cells are known to reside.30, 31 However, whether p63 indeed regulates any of the multiple types of adult mammary stem/progenitor cells in vivo was unknown.32, 33, 34 Here we answer this question with a yes for parity-identified mammary epithelial cells (PI-MECs), a pregnancy-identified heterogeneous population that survives post-lactational involution and contain multipotent progenitors that give rise to alveoli and ducts in subsequent pregnancies.34 Moreover, p63 was shown to be a major regulator of cell adhesion in nontransformed human mammary MCF10A cells in vitro, consistent with the juxtaposition of basal cells to the basement membrane and their high expression of cell adhesion molecules.35 Indeed, ablation of all p63 isoforms caused MCF10A detachment, downregulation of adhesion molecules and death by apoptosis and anoikis, which could be rescued by ΔNp63α (but not by TAp63γ) or by certain ΔNp63 target genes.35 This suggests that p63 might regulate cell detachment and apoptosis during mammary gland involution in vivo. Here we also show this to be the case. Lastly, we show that indirectly, by maintaining the size of the PI-MEC pool, p63 promotes pregnancy-promoted ErbB2 oncogenesis.
Results
p63 in the mammary gland inhibits cell detachment during early post-lactational involution
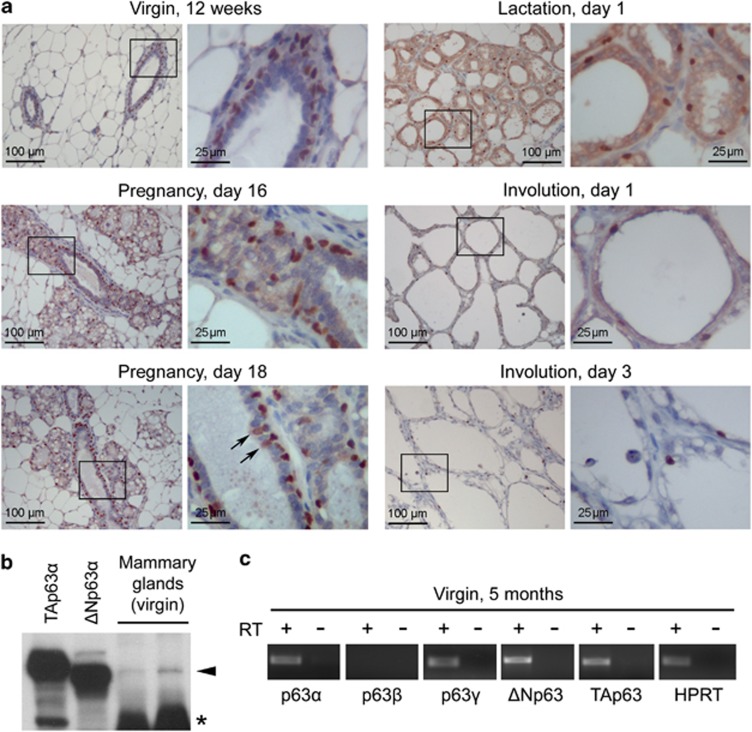
p63 is essential for the initial formation of mammary buds during embryogenesis.1 p63 protein is expressed in the adult mammary gland of wild-type (WT) mice at all stages of pregnancy, lactation and involution (Figure 1a). p63 protein is strongly expressed in the basal cell layer during all analyzed stages and weakly in the luminal layer at day 18 of pregnancy (Figure 1a). The ΔNp63α isoform is predominant (Figure 1b and Supplementary Figure 1A), and at the mRNA level both ΔNp63 and TAp63, as well as C-terminal isoforms p63α and p63γ, are also detected, as previously reported21 (Figure 1c). To determine whether p63 also has a functional role in the adult mammary gland, we made use of p63+/− mice (p63−/− mice die at birth) and compared them with their p63+/+ littermates. Analysis of gross mammary gland morphology and glandular epithelial structures showed that p63+/− mammary glands are normal during puberty, pregnancy and active lactation (Figures 2a and c and Supplementary Figures 1B-D). Moreover, p63+/− females have litter sizes comparable to their WT sisters and are competent to nurse their pups normally (data not shown). This indicates that p63 does not have a major role in the mammary gland before and during lactation.
Figure 1.
p63 expression in the mouse mammary gland during pregnancy, lactation and early involution. p63 protein is expressed in the adult mammary gland of WT mice in all stages. (a) Immunohistochemistry with pan-p63 H137 antibody detects p63 mainly in the basal cell layer at all stages analyzed. At day 18 of pregnancy, few luminal cells are stained as well (arrows), which was previously described for the TAp63 isoform using isoform-specific antibodies and cell sorting.13, 21 Similar results were obtained with pan-p63 4A4 antibody (not shown). Note that in lactation day 1, the cytoplasmic staining is nonspecific, as it was also seen when only secondary antibody was used. Images on the right are enlargements of the boxed areas in the corresponding images on the left. (b) Immunoblot analysis with 4A4 antibody shows that ΔNp63α is the predominant isoform in the mammary gland. Protein lysates of Phoenix E cells transfected with TAp63α (lane 1) or ΔNp63α (lane 2) plasmids are isoform-specific controls (see also Supplementary Figure 1A). (c) p63α, p63γ, ΔNp63 and TAp63 mRNAs are detected by reverse transcriptase (RT)-PCR in the virgin mammary gland. cDNA prepared in the absence of RT was used as a negative control. HPRT was a positive control
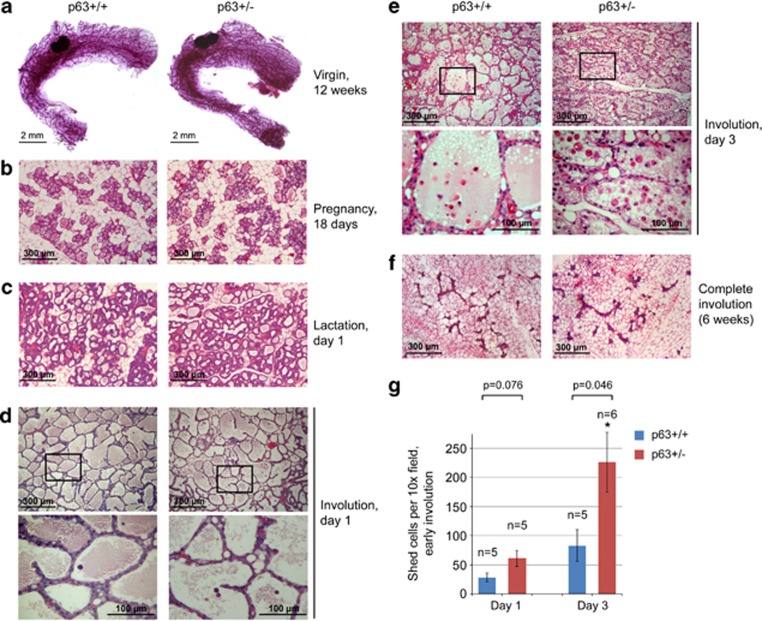
Figure 2.
p63 is required in the mammary gland to inhibit epithelial cell death during the apoptotic phase of involution. (a) Whole-mount carmine staining of mammary glands from 12-week-old virgin mice. Representative images from a sister pair are shown. (b–f) Hematoxylin and eosin staining of mammary glands at indicated stages. (d and e) Bottom images are enlargements of the boxed areas in the top images, for number of analyzed animals see g. (g) Quantification of cells shed into the alveolar lumen at days 1 and 3 of involution. Mean±S.E.M. N, number of animals analyzed (one mammary gland per animal), *P<0.05. (see also Supplementary Figure 1A)
Importantly, however, during early post-lactational involution p63+/− glands exhibit increased alveolar epithelial cell detachment, with more cell shedding into the distended lumen than in the littermate controls (Figures 2d, e and g). This trend was present already at day 1 of involution when p63+/− mammary glands exhibited 1.8 times more shed cells on average, although it did not reach statistical significance. At day 3 of involution, many more shed cells are apparent in p63+/− mammary glands, on average about three times more than in their p63+/+ littermate controls (P<0.05, Figures 2e and g). Yet, after involution is complete after 6 weeks, the overall morphology and epithelial structures in p63+/− glands are again indistinguishable from WT littermates (Figure 2f and Supplementary Figure 1E, also see Figure 5a). These data indicate that p63 has a specific role during the time window of early involution where it counteracts excessive cell death. Moreover, it suggests that p63 might suppress apoptosis, the principle mechanism of cell death at this stage.
p63 suppresses pro-apoptotic pathways during early involution
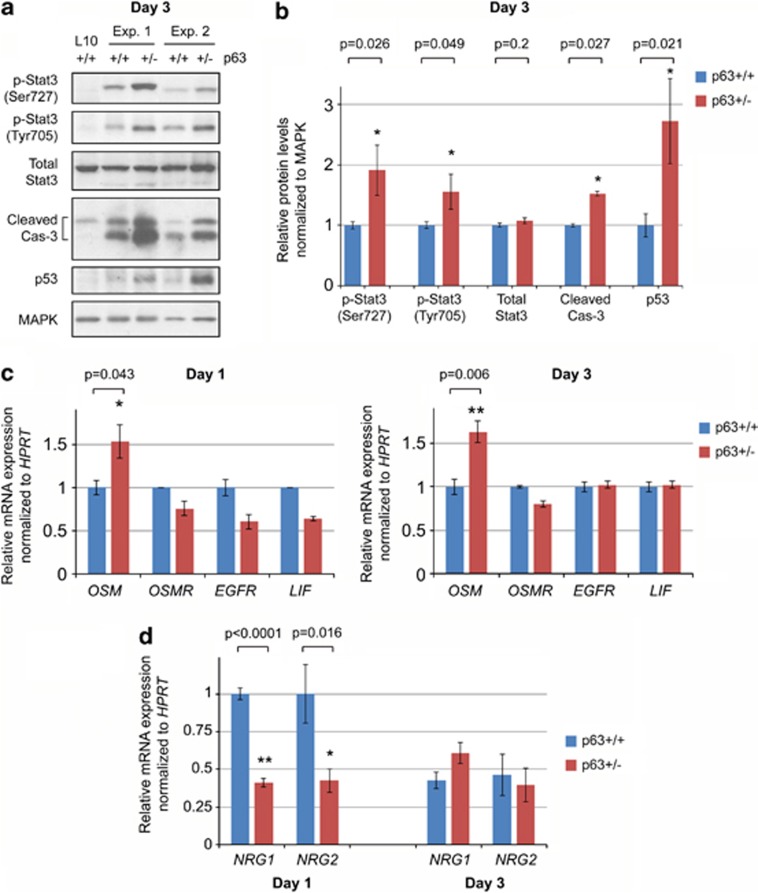
The hallmark apoptosis of early involution is known to be mediated by two principal pathways.7, 8, 36 First, activated Stat3 is a central mediator of apoptosis at this stage.7, 37, 38 Second, p53, which induces apoptosis in a transcription-dependent and -independent manner,39 has also been strongly implicated in the regulation of mammary involution.37, 40, 41, 42 Both of these pathways are enhanced in p63+/− mammary glands. Indeed, two phosphoactivated forms of Stat3 (p-Ser727 and p-Tyr705) are upregulated, whereas total Stat3 is unchanged (Figures 3a and b). Stat3 phosphorylation during involution is initiated by the interleukin-6 homologs LIF (signaling through the LIF receptor) and oncostatin M (OSM, signaling through its receptor OSM receptor (OSMR)/gp130 and EGFR).38 Thus, we tested whether p63 – being a transcription factor – regulates mRNA expression of components of these pathways. Indeed, OSM (but not OSMR, EGFR or LIF) mRNA is upregulated in p63+/− glands at both day 1 and day 3 of involution (Figure 3c). This provides a mechanistic link for the observed p63-mediated suppression of pStat3 in WT glands. On the other hand, there is a known negative interplay between Stat3 and its prosurvival homolog Stat5.16 pStat5 is essential in the mammary gland during the prior stages of pregnancy and progressively declines during lactation and early involution.38, 43 pStat5 is activated by upstream Neuregulin/ErbB signaling.44, 45 In agreement, at involution day 1 neuregulin 1 and 2 (NRG1 and NRG2) genes are repressed in p63+/− glands, but no longer at day 3 when their expression has declined even in WT glands (Figure 3d). The observed NRG1/2 downregulation likely contributes to pStat3 activation in p63+/− mammaries via suppression of pStat5.
Figure 3.
p63 suppresses pro-apoptotic pathways during early involution. (a and b) Immunoblot analysis of apoptosis-related proteins in whole mammary glands at day 3 of involution. Two representative experiments out of four (a) and quantification from all four independent experiments analyzing a total of seven p63+/+ and five p63+/− mice (b) are shown. Mean±S.E.M., *P<0.05. L10, lactation day 10, serves as a negative control for apoptosis. (c and d) Upregulation of oncostatin M (OSM), but not other known activators of pStat3, as well as downregulation of NRG1 and NRG2, known activators of the ErbB/pStat5 axis, in p63+/−compared with p63+/+ mammary epithelial cells. qRT-PCR analysis was done on mammary epithelial cells freshly isolated from littermate females at day 1 and day 3 of involution. Mean±S.E.M., *P<0.05, **P<0.01
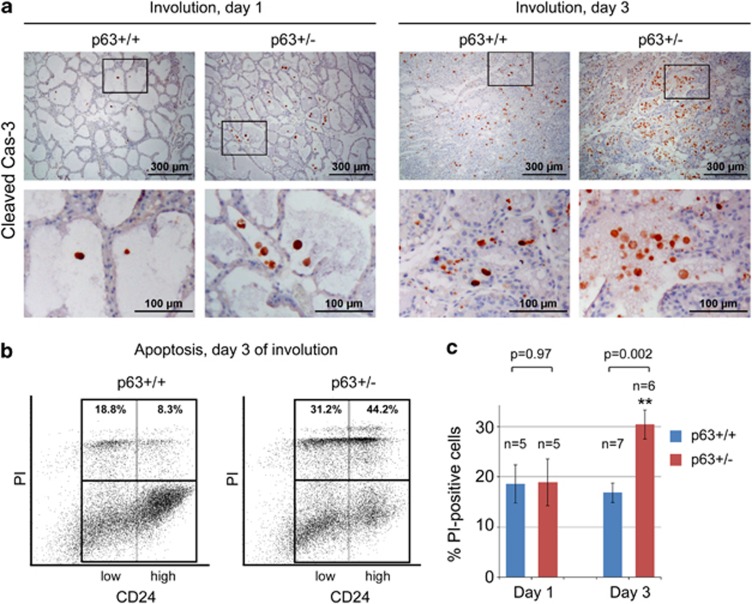
As a second pro-apoptotic mechanism, p53 protein is stabilized in the p63+/− glands compared with that in WT littermates (Figures 3a and b). In addition, there is a tendency in p63+/− glands for upregulated p21 protein, which is a p53 target gene (Supplementary Figure 2A). However, this difference was not statistically significant (data not shown), which can be due to high p21 levels already present during lactation (Supplementary Figure 2A, lane 1). With regard to p53 in the p63+/− background, we considered the possibility that it might be stabilized nonspecifically as a compensatory mechanism between family members.46 However, p53 upregulation is specific, as it is not upregulated in any prior or later stage other than early involution, and is not even upregulated in homozygous p63−/− mouse embryonic fibroblasts (Supplementary Figure 2B). Furthermore, as a prominent readout of apoptosis during involution,7 enhanced cleavage of caspase 3 is observed in p63+/− mammary glands (Figures 3a, b and 4a). In addition, fluorescence-activated cell sorting (FACS) shows an increased incorporation of propidium iodide (PI) marking dead cells in both CD24low (myoepithelial) and CD24high (luminal) mammary epithelial cells.47 Specifically, there is a 45% increase in total CD24positive/PIpositive cells in p63+/− glands (P=0.002) at involution day 3 (Figures 4b and c). In contrast, there is no change at day 1 (Figure 4c). Altogether, these data indicate that in adult mammary glands p63 acts as a counteracting prosurvival factor during the apoptotic phase of post-lactational involution, opposing the physiological cell death mediated by the pro-apoptotic Stat3 and p53 pathways.
Figure 4.
Enhanced apoptosis in p63+/− mammary glands during early involution. (a) Immunostaining of cleaved caspase-3 in p63+/+ and p63+/− mammary glands at day 1 and day 3 of involution. Bottom images are enlargements of the boxed areas in the corresponding top images. One out of two independent experiments with similar results is shown. (b and c) FACS analysis of CD24positive cells stained for PI (propidium iodide) at day 1 and day 3 post involution. A representative analysis at day 3 (b) and quantification from four (day 1) and six (day 3) independent experiments (c) is shown. See also Supplementary Figure 3A for gating before the analysis shown here. Mean±S.E.M. N, number of animals analyzed, **P<0.01
p63 promotes survival of PI-MEC progenitor cells after mammary gland involution
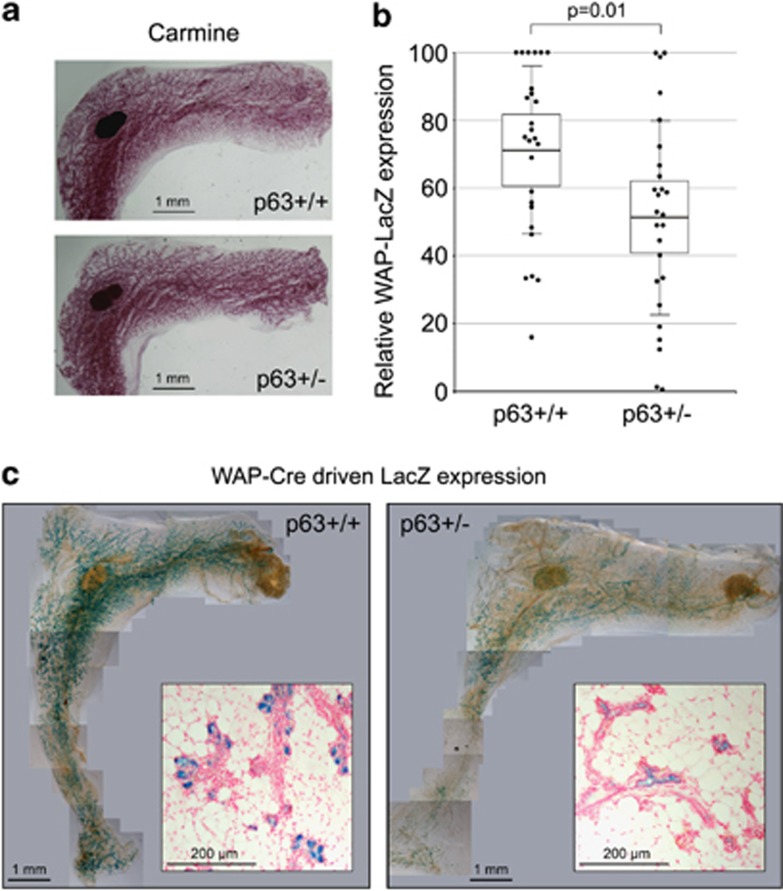
After involution is complete at 6 weeks post lactation, p63+/− and p63+/+ mammary glands had regained comparable epithelial structures (Figures 2f and 5a and Supplementary Figure 1E), suggesting that p63 has no role in late-stage involution that is marked by tissue remodeling. However, as p63 is a major regulator of stem/progenitor cells in the epidermis and other stratified epithelia, we wanted to test whether p63 heterozygosity also affects specific pools of mammary stem/progenitor cells after involution. An important type of mammary progenitors that are greatly affected by pregnancy and are traceable in vivo are the PI-MECs.34, 48, 49 Although in virgin mice PI-MECs comprise only 0.8–4% of mammary epithelial cells, they increase to 20–30% after pregnancy and differentiate into milk-producing alveolar cells that express late milk protein genes.48, 50 However, unlike other differentiated epithelial cells that get eliminated by shedding, a significant pool of PI-MECs survives post-lactational involution in vivo to give rise to new alveoli in subsequent pregnancies.49, 51 Moreover, PI-MECs can self-renew and give rise to fully functional mammary glands in transplantation studies, which establishes them as true pregnancy-related mammary stem cells.49, 50, 51, 52 PI-MECs can be readily identified in situ using the transgenic RosaLSL-LacZ;WAP-Cre reporter mouse (LSL-LacZ, lox-stop-lox β-galactosidase), as the whey acidic protein (WAP) promoter is specifically and only temporarily activated in late pregnancy to drive Cre expression. This removes the upstream Lox-Stop-Lox codon and permanently activates LacZ only in the surviving PI-MEC cells and their progeny.49, 50, 52 We used this system to test whether p63 heterozygosity affects PI-MECs and their survival after involution. Again, there was no difference during pregnancy and lactation (data not shown). However, after complete involution p63+/−RosaLSL-LacZ;WAP-Cre mammary glands exhibited on average 30% lower LacZ expression – marking PI-MECs and their progeny – than matching p63+/+RosaLSL-LacZ;WAP-Cre glands (Figures 5b and c; combining 8 independent experiments with a total of 13 p63+/− and 13 p63+/+ animals, 2 mammary glands per mouse). We confirmed that LacZ exclusively stained epithelial cells by sectioning representative glands, which again showed a significant reduction in LacZpositive cells in the end buds and terminal ducts of p63+/− glands (Figure 5c, insets). Together with the enhanced apoptosis of p63+/− epithelial cells during the apoptotic phase of involution, these data indicate that the physiological role of p63 in adult mammary gland is to promote survival of PI-MECs in post-parous mammary glands.
Figure 5.
Reduction of PI-MECs in p63+/− mammary glands. (a) Overall morphology of p63+/− and p63+/+ mammary glands at 6 weeks post involution analyzed by whole-mount carmine staining. Representative images from 7 independent experiments, analyzing 10 p63+/+ and 12 p63+/− mammary glands in total are shown. (b and c) p63+/+RosaLSL-LacZ; WAP-Cre (p63+/+) and p63+/−RosaLSL-LacZ;WAP-Cre (p63+/−) animals analyzed for β-galactosidase expression in whole mammary glands at 6 weeks post involution. Quantification from 8 independent experiments, analyzing 13 p63+/+ and p63+/− mice each (two mammary glands per animal) is shown (b). Error bars represent S.D. and the boxes represent the 95% confidence interval. (c) Representative images of LacZ-stained mammary glands from a sister pair analyzed in b. Insets show cross-sections of LacZ-stained mammary glands counterstained with nuclear fast red
p63 heterozygosity reduces tumorigenesis and mortality in the parous HER2 breast cancer model
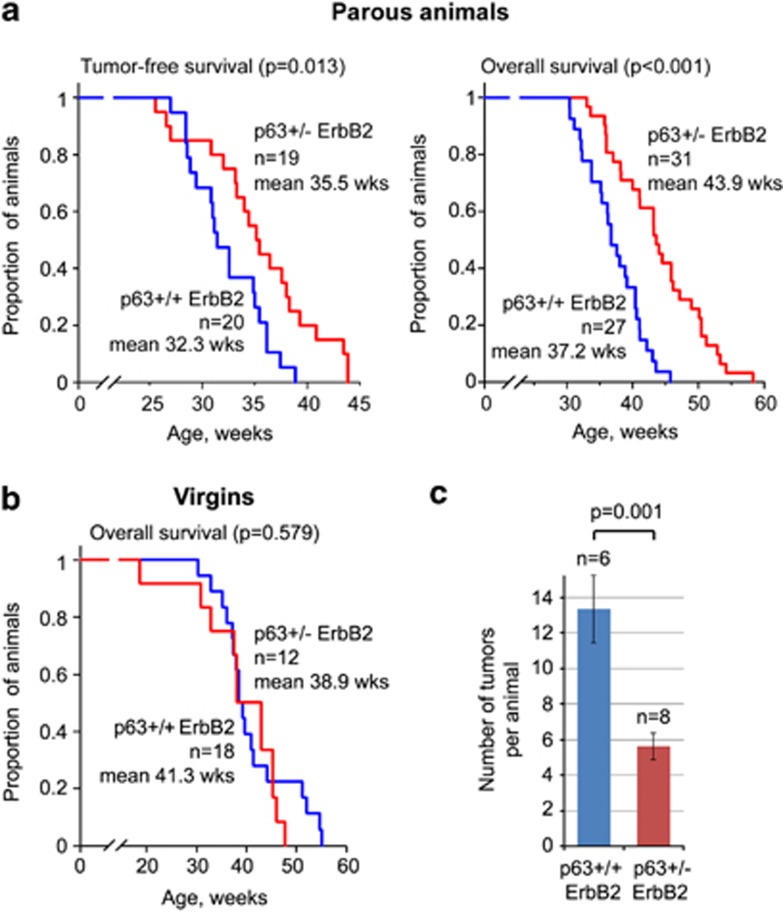
HER2-positive tumors comprise 20–25% of all human breast cancers and are associated with poor clinical outcome. Using the mouse model of HER2 breast cancer, mouse mammary tumor virus (MMTV)-driven ErbB2 (homolog of human HER2/neu),53 PI-MECs were shown to be cellular targets of HER2-mediated oncogenesis.54, 55, 56 Indeed, ErbB2 tumorigenesis in mice increases with parity, and targeted removal of PI-MECs in vivo reduced tumor incidence in ErbB2 mice.55, 56, 57 Likewise, clinical studies demonstrate that women during or right after pregnancy (when the number of PI-MECs is highest) have increased susceptibility for HER2 breast cancer.58, 59 Thus, we hypothesized that p63 hemizygosity would suppress ErbB2-mediated tumorigenesis as a consequence of a reduced PI-MEC pool. Indeed, we found that tumor onset was delayed by over 3 weeks (P<0.05) and the survival was extended by over 6 weeks (P<0.001) in one-time pregnant p63+/− ErbB2 mice compared with their p63+/+ ErbB2 littermates (Figure 6a). However, no difference in tumorigenesis or survival was found between virgin p63+/− ErbB2 and p63+/+ ErbB2 animals (Figure 6b). Of note, as previously reported, pregnancy itself increased the breast cancer mortality of p63+/+ (WT) ErbB2 mice (P=0.041).55 In addition, the number of visible tumor nodules was 58% lower in one-time pregnant p63+/− ErbB2 mice compared with their p63+/+ ErbB2 sisters (Figure 6c). Taken together, our data establish p63 as an essential prosurvival factor of the PI-MEC progenitor pool in adult mammary gland after pregnancy. Moreover, because of its role as a regulator of the PI-MEC pool, p63 is indirectly implicated as a factor promoting HER2 breast carcinogenesis.
Figure 6.
p63 heterozygosity extends tumor-free and overall survival, and decreases tumor multiplicity in parous ErbB2 animals. (a) Kaplan–Meier curves of tumor-free survival and overall survival of p63+/− ErbB2 and p63+/+ ErbB2 one-time parous females; n, number of animals. (b) No difference in tumor-free and overall survival of virgin p63+/+ ErbB2 and p63+/− ErbB2 females. (c) Tumor multiplicity at endpoint (time when animal had to be killed) of p63+/− ErbB2 and p63+/+ ErbB2 females impregnated at 20 weeks. Mean±S.E.M.; n, number of animals
Discussion
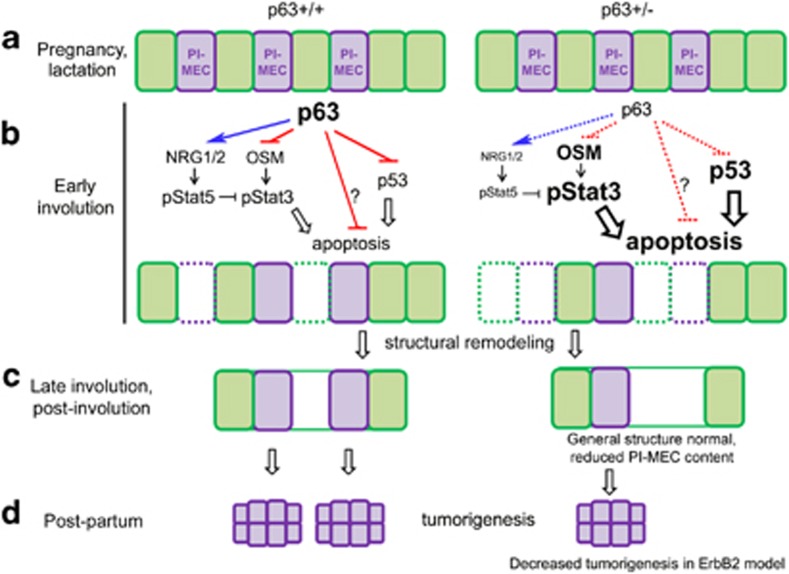
This study set to define a functional role of p63 in the adult mammary gland. Our main findings are as follows (see model Figure 7): (1) p63 (predominantly ΔNp63α) is expressed in the adult mammary gland during all physiologic stages, that is, in the virgin and pregnant state, lactation, post-lactational early involution and completely involuted gland. Yet, p63 heterozygosity has no effect on mammary gland development or milk production (Figure 7a). (2) However, a prominent effect of p63 deficiency appears in the early apoptotic phase of post-lactational involution, where p63 acts as a prosurvival factor for mammary epithelium by antagonizing the OSM/pStat3 and the p53 apoptotic pathways (Figure 7b). (3) Functionally, this prosurvival role of p63 specifically and strongly affects PI-MECs (Figure 7b, purple cells), whose pool is reduced in parous p63+/− glands after involution is completed, whereas the overall morphology of the gland is unaffected (Figure 7c). (4) The reduced PI-MEC pool in parous p63+/− mice is likely the cause of their partial protection from HER2-driven mammary tumorigenesis, revealed by delayed tumor onset, extended survival and reduced tumor multiplicity, as PI-MECs are a known tumor-initiating cell type for HER2 breast oncogenesis (Figure 7d). The fact that p63 heterozygosity does not exert tumor protection in virgin HER2 mice also supports the last conclusion, as p63+/− and WT virgins are predicted to have similar and very small (i.e., pre-pregnancy) PI-MEC pools.
Figure 7.
Proposed model, the role of p63 in adult mammary gland. (a) The mammary glands develop normally in p63+/− females and have normal structure and function during pregnancy and lactation. (b) During early involution, p63 antagonizes apoptosis in WT mammary glands by suppressing p53 and pStat3 pathways. Specifically, p63 inhibits expression of OSM, an upstream positive regulator of pStat3, and activates neuregulins 1 and 2 (NRG1/2) known to activate the prosurvival pStat5 pathway that can oppose pStat3. In addition, p63 possibly suppresses other apoptosis-related genes (question mark), besides p53. In p63+/− mammary glands, these mechanisms are weakened, leading to increased Stat3 phosphorylation/activation (at Tyr705 and Ser727) and increased p53 activity and, as a net result, to enhanced apoptosis in PI-MECs and other cell types, including terminally differentiated milk-producing cells. (c) In late involution, after structural tissue remodeling WT and p63+/− mammary glands appear to have an overall similar morphology by conventional histology. However, the number of PI-MEC progenitor and derivative cells is significantly reduced in p63+/− mammary glands. (d) PI-MECs are known cellular targets of ErbB2 tumorigenesis. As a result, p63+/− females bearing activated ErbB2 are more resistant to pregnancy-enhanced ErbB2-driven mammary carcinoma. Purple, PI-MECs. Green, other epithelial cell types, including terminally differentiated milk-producing luminal cells
ΔNp63 likely exerts its prosurvival function in the mammary gland via multiple mechanisms. First, p63 represses the OSM/pStat3 axis, known to be the major mediator of apoptosis during early involution.38 Second, similar to that in other epithelial systems ΔNp63 most likely interferes with the pro-apoptotic activity of p53 or TAp73 that are also expressed in mammary cells.11, 12, 22, 60, 61 Furthermore, ΔNp63 has been shown to directly regulate transcription of apoptosis-related genes. Specifically, together with HDAC1/2 it can suppress the pro-apoptotic gene PUMA.62 p63 can also activate anti-apoptotic genes Hsp7063 and the caspase 8 inhibitor FLIP.64 In addition, ΔNp63α was shown by Carroll et al.35 to be a direct transcriptional activator of the cell adhesion program in human mammary nontransformed MCF10A cells and in primary mouse mammary epithelial cells. This suggests that ΔNp63α could also promote cell adhesion, and thus prevent shedding, during post-lactational involution of the mammary gland. Indeed, one of the identified ΔNp63 target genes, integrin α6 (CD49f), is highly expressed in PI-MECs.51, 56 The study by Carroll et al.35 also highlights the crosstalk between p63-dependent cell adhesion and apoptosis. Specifically, on ΔNp63 knockdown MCF10A cells exhibit detachment and cell death via anoikis and apoptosis, whereas re-expression of integrin β4 partially rescues both cell adhesion and apoptosis. In addition, we re-examined the published microarray data35 and found that the pro-apoptotic program triggered in MCF10A on ΔNp63 knockdown may also be activated at the transcriptional level (Supplementary Figure 4). Specifically, seven out of nine differentially regulated pro-apoptotic genes are upregulated (e.g., Bax and Caspase 8), whereas all seven differentially regulated prosurvival genes (e.g., Jak2, Stat5b and Bcl2) are downregulated. Whether p63 also regulates adhesion and/or apoptosis at the transcriptional level in mammary gland in vivo remains to be elucidated (Figure 7b, question mark).
It is unclear how ΔNp63 affects shedding of luminal layer while being expressed predominantly in the basal layer. Most likely, ΔNp63 affects luminal cells indirectly by affecting attachment of underlying basal cells. This is supported by increased cell death not only in CD24low cells (luminal) but also in CD24high cells (basal myoepithelial) in p63+/− glands (Figure 4b). Consistently, PI-MECs can be found in both the basal and the luminal sorted compartments.33, 51, 56 In addition, we cannot exclude the possibility that TAp63, alone or as a supporting factor, contributes to epithelial survival during involution. Indeed, TAp63 is localized in the luminal cells,13, 21 which are known to contain PI-MECs and some other mammary progenitors.33 Moreover, TAp63 has been implicated as a prosurvival factor in adult dermal and epidermal progenitors.17 Thus, a decrease of TAp63 in p63+/− post-lactation glands would be consistent with the observed decreased survival of PI-MECs. Future studies using isoform-specific p63 KO mice will be needed to elucidate the precise roles of ΔNp63 and TAp63 isoforms in the adult mammary gland.
Our study used a haploinsufficient global p63 model, with one remaining functional p63 allele, as p63 KO animals die at birth. Thus, this model uncovered the most robust functions of ΔNp63 in the adult mammary gland. Further studies using conditional global p63 KO mice (which are not yet publicly available65) might uncover additional functions of p63 in the adult mammary gland.
Materials and Methods
Mouse strains
The p63 global KO mouse, a generous gift from F McKeon, on the enriched 129SVJ background, was previously described.1, 66 RosaLSL-LacZ/LSL-LacZ animals were purchased from Jackson Labs (Bar Harbor, ME, USA) (strain B6.129S4-Gt(ROSA)26Sortm1Sor/J). WAP-Cre transgenic mice67 were obtained from the NCI mouse repository. p63;RosaLSL-LacZ/+;WAP-Cre females were generated by crossing the RosaLSL-LacZ allele to homozygosity into the p63+/− background and then crossing p63+/−RosaLSL-LacZ/LSL-LacZ animals with WAP-Cre to generate p63+/−RosaLSL-LacZ/+;WAP-Cre and p63+/+RosaLSL-LacZ/+;WAP-Cre female littermates on a mixed C57Bl6-129SVJ background. A diagram outlining this generation is described in Supplementary Figure 3B. The MMTV-ErbB2 mouse model with activated ErbB2 (Muller et al.68) was obtained from Jackson Labs (strain FVB-Tg(MMTV-Erbb2)NK1Mul/J). Littermate p63-ErbB2 females were on a 50 : 50 129SVJ:FVBN background.
For timed pregnancies and involution studies, all females were bred at 12 weeks of age. For ErbB2 pregnancy studies, p63-ErbB2 females were bred at 12–20 weeks of age. Plug day was counted as day 0.5 of pregnancy. For timed involution, females were normalized to eight pups per litter and allowed 7 to 10 days of lactation before pup removal. Time of pup removal was considered hour zero of involution. p63-ErbB2 mice were monitored twice weekly and euthanized when the largest palpable tumor reached 2.0 cm. All animals were treated humanely and according to guidelines approved by the Institutional Animal Care and Use Committee at Stony Brook University.
Whole-mount morphological analysis
All morphological studies were done using mammary glands 4 or 9 that were flattened on filter paper before fixation in glass vials. For whole-mount carmine-alum staining, glands were fixed in 4% paraformaldehyde overnight before defatting in acetone (three times 1 h each). After defatting, glands were rehydrated and stained in carmine-alum (1 g carmine (Sigma, St. Louis, MO, USA; C1022) with 4.6 g aluminum potassium sulfate dodecahydrate (Sigma, A7162)) overnight at room temperature and then dehydrated in ethanol before clearing in xylene and storing in methyl salicylate for imaging. Images were taken with a Nikon Digital Sight DS Fi-1 camera (Nikon, Melville, NY, USA) and NIS-Elements F3.0 software (Nikon) through an Olympus SZ61 microscope (Olympus, Center Valley, PA, USA) for whole-mount images or through an Nikon Labophot-2 microscope (Nikon) for sections. Shed epithelial cells were counted in 10–12 non-overlapping fields for each mammary gland at × 10 magnification and the average number of cells per field per gland was calculated.
Histology and immunohistochemistry
Mammary glands were fixed in formalin, embedded in paraffin and sectioned (5 μm). For immunohistochemical staining of p63 (H-137, Santa Cruz, Santa Cruz, CA, USA; sc-8343) and cleaved caspase-3 (D175, Cell Signaling, Danvers, MA, USA; 9661), slides were deparaffinized and boiled in citrate buffer (10 mM, pH 6.0, 35 min) for antigen retrieval, blocked in 10% goat serum and incubated with primary antibody (1 : 500) for 2 h at room temperature. After washing, slides were incubated with biotinylated secondary antibody and HRP-Streptavidin using the Histostain SP Broad Spectrum kit (Invitrogen, Carlsbad, CA, USA; 959943B) and stained with DAB substrate with hematoxylin counterstain.
β-Galactosidase (X-gal) staining
X-gal staining was done as described.49 Briefly, glands were flattened, fixed for 1 h (2% paraformaldehyde, 0.25% glutaraldehyde, 0.01%NP-40 in PBS) and washed two times for 1 h each (2 mM MgCl2, 0.01% Na-deoxycholate, 0.02% NP-40 in PBS). Glands were stained for 48 h at room temperature in the dark with 0.5 mg/ml X-gal (Roche, Indianapolis, IN, USA; 10745740001) in staining buffer (30 mM K3Fe(CN)6, 30 mM K4Fe(CN)6.3H2O, 2 mM MgCl2, 0.01% Na-deoxycholate, 0.02% NP-40, pH 7.2–7.4 in PBS). After PBS rinse, glands were defatted in acetone (two times 1 h each), rehydrated and cleared in glycerol before imaging.
Images were taken for whole mounts or for sections as described above. Whole-mount images were reconstructed in Adobe Photoshop. The intensity of LacZ staining was quantified using the ImageJ (U. S. National Institutes of Health, Bethesda, MD, USA) software (at 65% of autothreshold in the red channel of RGB stack). To normalize for possible differences in processing on different days, the X-gal intensity of the most stained gland (whether from a p63+/+ or a p63+/− animal) within each simultaneously processed set was set to 100 and the X-gal intensities in the other glands from the same set were calculated relative to it.
For sections, X-gal-stained glands were dehydrated and processed for paraffin embedding. Sections (6 μm) were counterstained with Nuclear Fast Red (Sigma, N8002). Images were taken using the Nikon Digital Sight DS Fi-1 camera and NIS-Elements F3.0 software with a Nikon Labophot-2 microscope.
Immunoblot analysis
Mammary glands were minced and resuspended in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% Deoxycholate, 0.1% SDS and 50 mM Tris HCl, pH 8) with protease inhibitors (Roche) as described. After sonication, the suspension was spun down and fat was removed before protein quantification. Immunoblots were performed using 20–60 μg of protein. The following antibodies were used: p63 (4A4, Santa Cruz, sc-8431), p-Stat3 (Ser727, Cell Signaling, 9134), p-Stat3 (Tyr705) (3E2, Cell Signaling, 9138), total Stat3 (124H6, Cell Signaling, 9139), cleaved caspase-3 (D175, Cell Signaling, 9661), p53 (FL-393, Santa Cruz, sc-6243) and MAPK2 (Millipore, Billerica, MA, USA; 1B3B9). Band densities were quantified by ImageJ software.
FACS analysis
For single cell suspension of mammary epithelial cells, mammary tissue was digested in Collagenase/Hyaluronidase enzymatic cocktail (Stem Cell Tech, Vancouver, BC, Canada; 07912) at 37 °C for 2 h with shaking and then digested in 0.05% Trypsin, followed by DNaseI treatment before further digestion with Dispase (Stem Cell Tech, 07923). The cell suspension was then treated with Red Blood Cell lysis buffer, rinsed with PBS and passed through a 40-μm mesh after resuspending in Opti-Mem (Invitrogen). Cell clumps were mechanically disrupted by triturating with a glass pipette and cell numbers were determined using the Beckman Coulter Z1 Counter (Beckman Coulter Inc., Brea, CA, USA). For flow cytometry, cells were stained with APC-labeled lineage markers CD31, CD45 and Ter119 (BD Pharmingen, San Diego, CA, USA) and FITC-labeled mammary epithelial marker CD24 (BD Pharmingen). After incubation with the antibodies, cells were washed and stained with PI (Sigma, P4864) and analyzed with a FACScalibur flow cytometer (BD Biosciences, San Jose, CA, USA) and Cyflogic (CyFlo Ltd, Turku, Finland) software (v1.2.1).
Reverse transcription PCR
Total RNA was extracted from mammary epithelial cells (freshly isolated as described above for FACS analysis) using Trizol reagent (Invitrogen). cDNA was prepared with SuperScript II reverse transcriptase (Invitrogen). The PCR primers for total p63, ΔNp63 and TAp63 were previously described.16 The other primers were: p63α-F 5′-CACAGGCAGCTGCACGACT-3′, p63α-R 5′-TCATTCTCCTTCCTCTTTGATACGC-3′ p63β-F 5′-TCCCTCAGCACACGATCGA-3′, p63β-R 5′-ACTTGCCAAATCCTGACAATGC-3′ p63γ-F 5′-TCCCTCAGCACACGATCGA-3′, p63γ-R 5′-GACGTCAGACTGTGTCGGAGC-3′ OSM-F 5′-ATGCAGACACGGCTTCTAAGA-3′, OSM-R 5′-TTGGAGCAGCCACGATTGG-3′ OSMR-F 5′-GCATCCCGAAGCGAAGTCTT-3′, OSMR-R 5′-GGGCTGGGACAGTCCATTCTA-3′ EGFR-F 5′-CCCATGCGGAACTTACAGGAA-3′, EGFR-R 5′-TTGGATCACATTTGGGGCAAC-3′ LIF-F 5′-AACCAGATCAAGAATCAACTGGC-3′, LIF-R 5′-TGTTAGGCGCACATAGCTTTT-3′ NRG1-F 5′-GCAAAGAAGGCAGAGGCAAG-3′, NRG1-R 5′-GGTTCAGCTCATTCCCGTTC-3′ and NRG2-F 5′-TCGAAAGGAACCAGCGCTAC-3′, NRG2-R 5′-CCTCCACCCTCACTTTGTTG-3′. Real-time PCR was performed using the QuantiTect SYBR Green PCR kit (Qiagen, Valencia, CA, USA) on an Opticon 2 instrument (MJ Research Inc., Foster City, CA, USA) and analyzed with Opticon Monitor software (Bio-Rad, Hercules, CA, USA). The PCR conditions were: 94 °C for 1 min, 60 °C for 1 min and 72 °C for 1.5 min (40 cycles). Samples were analyzed in duplicates and normalized to HPRT. Expression of each gene was set to 1 in WT mammary cells.
Acknowledgments
This work was funded by grants to UMM from the National Cancer Institute (CA-060664 and CA-093853), the New York Stem Cell Science Program (NYSTEM N08T-040) and the Carol Baldwin Breast Cancer Research Fund (to UMM and NDM). EMA was a recipient of NIDDK T32 NIH-sponsored Postdoctoral Fellowship (DK007521) and ARY was a recipient of NCI T32 NIH-sponsored Postdoctoral Fellowship (CA917633).
Glossary
- CK14/18
cytokeratin 14/18
- FACS
fluorescence-activated cell sorting
- KO
knockout
- LIF
leukemia inhibitory factor
- LSL-LacZ
lox-stop-lox β-galactosidase
- MMTV
mouse mammary tumor virus
- NRG1/2
neuregulin 1/2
- OSM
oncostatin M
- OSMR
OSM receptor
- PI-MEC
parity-identified mammary epithelial cell
- WAP
whey acidic protein
- WT
wild type
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
Supplementary Material
References
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Crum CP, McKeon FD. p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu Rev Pathol. 2010;5:349–371. doi: 10.1146/annurev-pathol-121808-102117. [DOI] [PubMed] [Google Scholar]
- Pellegrini G, Dellambra E, Golisano O, Martinelli E, Fantozzi I, Bondanza S, et al. p63 identifies keratinocyte stem cells. Proc Natl Acad Sci USA. 2001;98:3156–3161. doi: 10.1073/pnas.061032098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat Rev Mol Cell Biol. 2005;6:715–725. doi: 10.1038/nrm1714. [DOI] [PubMed] [Google Scholar]
- Richert MM, Schwertfeger KL, Ryder JW, Anderson SM. An atlas of mouse mammary gland development. J Mammary Gland Biol Neoplasia. 2000;5:227–241. doi: 10.1023/a:1026499523505. [DOI] [PubMed] [Google Scholar]
- Watson CJ, Kreuzaler PA. Remodeling mechanisms of the mammary gland during involution. Int J Dev Biol. 2011;55:757–762. doi: 10.1387/ijdb.113414cw. [DOI] [PubMed] [Google Scholar]
- Baxter FO, Neoh K, Tevendale MC. The beginning of the end: death signaling in early involution. J Mammary Gland Biol Neoplasia. 2007;12:3–13. doi: 10.1007/s10911-007-9033-9. [DOI] [PubMed] [Google Scholar]
- Duijf PH, Vanmolkot KR, Propping P, Friedl W, Krieger E, McKeon F, et al. Gain-of-function mutation in ADULT syndrome reveals the presence of a second transactivation domain in p63. Hum Mol Genet. 2002;11:799–804. doi: 10.1093/hmg/11.7.799. [DOI] [PubMed] [Google Scholar]
- Vanbokhoven H, Melino G, Candi E, Declercq W. p63, a story of mice and men. J Invest Dermatol. 2011;131:1196–1207. doi: 10.1038/jid.2011.84. [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Candi E, Dinsdale D, Rufini A, Salomoni P, Knight RA, Mueller M, et al. TAp63 and DeltaNp63 in cancer and epidermal development. Cell Cycle. 2007;6:274–285. doi: 10.4161/cc.6.3.3797. [DOI] [PubMed] [Google Scholar]
- Nylander K, Vojtesek B, Nenutil R, Lindgren B, Roos G, Zhanxiang W, et al. Differential expression of p63 isoforms in normal tissues and neoplastic cells. J Pathol. 2002;198:417–427. doi: 10.1002/path.1231. [DOI] [PubMed] [Google Scholar]
- Romano RA, Smalley K, Magraw C, Serna VA, Kurita T, Raghavan S, et al. DeltaNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development. 2012;139:772–782. doi: 10.1242/dev.071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Talos F, Palacios G, Beyer U, Dobbelstein M, Moll UM. The alpha/beta carboxy-terminal domains of p63 are required for skin and limb development. New insights from the Brdm2 mouse which is not a complete p63 knockout but expresses p63 gamma-like proteins. Cell Death Differ. 2009;16:1108–1117. doi: 10.1038/cdd.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell. 2009;5:64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiRenzo J, Signoretti S, Nakamura N, Rivera-Gonzalez R, Sellers W, Loda M, et al. Growth factor requirements and basal phenotype of an immortalized mammary epithelial cell line. Cancer Res. 2002;62:89–98. [PubMed] [Google Scholar]
- Wang X, Mori I, Tang W, Nakamura M, Nakamura Y, Sato M, et al. p63 expression in normal, hyperplastic and malignant breast tissues. Breast Cancer. 2002;9:216–219. doi: 10.1007/BF02967592. [DOI] [PubMed] [Google Scholar]
- Li H, Cherukuri P, Li N, Cowling V, Spinella M, Cole M, et al. Nestin is expressed in the basal/myoepithelial layer of the mammary gland and is a selective marker of basal epithelial breast tumors. Cancer Res. 2007;67:501–510. doi: 10.1158/0008-5472.CAN-05-4571. [DOI] [PubMed] [Google Scholar]
- Li N, Singh S, Cherukuri P, Li H, Yuan Z, Ellisen LW, et al. Reciprocal intraepithelial interactions between TP63 and hedgehog signaling regulate quiescence and activation of progenitor elaboration by mammary stem cells. Stem Cells. 2008;26:1253–1264. doi: 10.1634/stemcells.2007-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergholz J, Xiao ZX. Role of p63 in development, tumorigenesis and cancer progression. Cancer Microenviron. 2012;5:311–322. doi: 10.1007/s12307-012-0116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin-Ozuysal O, Fiche M, Guitierrez M, Wagner KU, Raffoul W, Brisken C. Antagonistic roles of Notch and p63 in controlling mammary epithelial cell fates. Cell Death Differ. 2010;17:1600–1612. doi: 10.1038/cdd.2010.37. [DOI] [PubMed] [Google Scholar]
- van Bokhoven H, McKeon F. Mutations in the p53 homolog p63: allele-specific developmental syndromes in humans. Trends Mol Med. 2002;8:133–139. doi: 10.1016/s1471-4914(01)02260-2. [DOI] [PubMed] [Google Scholar]
- Britt K, Ashworth A, Smalley M. Pregnancy and the risk of breast cancer. Endocr Relat Cancer. 2007;14:907–933. doi: 10.1677/ERC-07-0137. [DOI] [PubMed] [Google Scholar]
- Tiede B, Kang Y. From milk to malignancy: the role of mammary stem cells in development, pregnancy and breast cancer. Cell Res. 2011;21:245–257. doi: 10.1038/cr.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuliere J, Faraldo MM, Deugnier MA, Shtutman M, Ben-Ze'ev A, Thiery JP, et al. Targeted activation of beta-catenin signaling in basal mammary epithelial cells affects mammary development and leads to hyperplasia. Development. 2005;132:267–277. doi: 10.1242/dev.01583. [DOI] [PubMed] [Google Scholar]
- Thomas E, Zeps N, Cregan M, Hartmann P, Martin T. 14-3-3sigma (sigma) regulates proliferation and differentiation of multipotent p63-positive cells isolated from human breastmilk. Cell Cycle. 2011;10:278–284. doi: 10.4161/cc.10.2.14470. [DOI] [PubMed] [Google Scholar]
- Buono KD, Robinson GW, Martin C, Shi S, Stanley P, Tanigaki K, et al. The canonical Notch/RBP-J signaling pathway controls the balance of cell lineages in mammary epithelium during pregnancy. Dev Biol. 2006;293:565–580. doi: 10.1016/j.ydbio.2006.02.043. [DOI] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Stingl J, Raouf A, Emerman JT, Eaves CJ. Epithelial progenitors in the normal human mammary gland. J Mammary Gland Biol Neoplasia. 2005;10:49–59. doi: 10.1007/s10911-005-2540-7. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Smith GH. Murine mammary epithelial stem cells: discovery, function, and current status. Cold Spring Harb Perspect Biol. 2011;3:a004879. doi: 10.1101/cshperspect.a004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll DK, Carroll JS, Leong CO, Cheng F, Brown M, Mills AA, et al. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol. 2006;8:551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- Stein T, Salomonis N, Gusterson BA. Mammary gland involution as a multi-step process. J Mammary Gland Biol Neoplasia. 2007;12:25–35. doi: 10.1007/s10911-007-9035-7. [DOI] [PubMed] [Google Scholar]
- Chapman RS, Lourenco PC, Tonner E, Flint DJ, Selbert S, Takeda K, et al. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev. 1999;13:2604–2616. doi: 10.1101/gad.13.19.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haricharan S, Li Y. STAT signaling in mammary gland differentiation, cell survival and tumorigenesis. Mol Cell Endocrinol. 2013;382:560–569. doi: 10.1016/j.mce.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochim Biophys Acta. 2009;1787:414–420. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerry DJ, Dickinson ES, Roberts AL, Said TK. Regulation of apoptosis during mammary involution by the p53 tumor suppressor gene. J Dairy Sci. 2002;85:1103–1110. doi: 10.3168/jds.S0022-0302(02)74171-4. [DOI] [PubMed] [Google Scholar]
- Jerry DJ, Kuperwasser C, Downing SR, Pinkas J, He C, Dickinson E, et al. Delayed involution of the mammary epithelium in BALB/c-p53null mice. Oncogene. 1998;17:2305–2312. doi: 10.1038/sj.onc.1202157. [DOI] [PubMed] [Google Scholar]
- Matthews JR, Clarke AR. p53 mediates a default programme of mammary gland involution in the absence of STAT3. Oncogene. 2005;24:3083–3090. doi: 10.1038/sj.onc.1208512. [DOI] [PubMed] [Google Scholar]
- Bednorz NL, Brill B, Klein A, Gabel K, Groner B. Tracking the activation of Stat5 through the expression of an inducible reporter gene in a transgenic mouse line. Endocrinology. 2011;152:1935–1947. doi: 10.1210/en.2011-0053. [DOI] [PubMed] [Google Scholar]
- Britsch S. The neuregulin-I/ErbB signaling system in development and disease. Adv Anat Embryol Cell Biol. 2007;190:1–65. [PubMed] [Google Scholar]
- Long W, Wagner KU, Lloyd KC, Binart N, Shillingford JM, Hennighausen L, et al. Impaired differentiation and lactational failure of Erbb4-deficient mammary glands identify ERBB4 as an obligate mediator of STAT5. Development. 2003;130:5257–5268. doi: 10.1242/dev.00715. [DOI] [PubMed] [Google Scholar]
- Suliman Y, Opitz OG, Avadhani A, Burns TC, El-Deiry W, Wong DT, et al. p63 expression is associated with p53 loss in oral-esophageal epithelia of p53-deficient mice. Cancer Res. 2001;61:6467–6473. [PubMed] [Google Scholar]
- Sleeman KE, Kendrick H, Ashworth A, Isacke CM, Smalley MJ. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 2006;8:R7. doi: 10.1186/bcr1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KU, Smith GH. Pregnancy and stem cell behavior. J Mammary Gland Biol Neoplasia. 2005;10:25–36. doi: 10.1007/s10911-005-2538-1. [DOI] [PubMed] [Google Scholar]
- Wagner KU, Boulanger CA, Henry MD, Sgagias M, Hennighausen L, Smith GH. An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development. 2002;129:1377–1386. doi: 10.1242/dev.129.6.1377. [DOI] [PubMed] [Google Scholar]
- Booth BW, Boulanger CA, Smith GH. Alveolar progenitor cells develop in mouse mammary glands independent of pregnancy and lactation. J Cell Physiol. 2007;212:729–736. doi: 10.1002/jcp.21071. [DOI] [PubMed] [Google Scholar]
- Matulka LA, Triplett AA, Wagner KU. Parity-induced mammary epithelial cells are multipotent and express cell surface markers associated with stem cells. Dev Biol. 2007;303:29–44. doi: 10.1016/j.ydbio.2006.12.017. [DOI] [PubMed] [Google Scholar]
- Boulanger CA, Wagner KU, Smith GH. Parity-induced mouse mammary epithelial cells are pluripotent, self-renewing and sensitive to TGF-beta1 expression. Oncogene. 2005;24:552–560. doi: 10.1038/sj.onc.1208185. [DOI] [PubMed] [Google Scholar]
- Andrechek ER, Hardy WR, Siegel PM, Rudnicki MA, Cardiff RD, Muller WJ. Amplification of the neu/erbB-2 oncogene in a mouse model of mammary tumorigenesis. Proc Natl Acad Sci USA. 2000;97:3444–3449. doi: 10.1073/pnas.050408497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth BW, Boulanger CA, Anderson LH, Smith GH. The normal mammary microenvironment suppresses the tumorigenic phenotype of mouse mammary tumor virus-neu-transformed mammary tumor cells. Oncogene. 2011;30:679–689. doi: 10.1038/onc.2010.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry MD, Triplett AA, Oh KB, Smith GH, Wagner KU. Parity-induced mammary epithelial cells facilitate tumorigenesis in MMTV-neu transgenic mice. Oncogene. 2004;23:6980–6985. doi: 10.1038/sj.onc.1207827. [DOI] [PubMed] [Google Scholar]
- Jeselsohn R, Brown NE, Arendt L, Klebba I, Hu MG, Kuperwasser C, et al. Cyclin D1 kinase activity is required for the self-renewal of mammary stem and progenitor cells that are targets of MMTV-ErbB2 tumorigenesis. Cancer Cell. 2010;17:65–76. doi: 10.1016/j.ccr.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis MD, Seachrist DD, Abdul-Karim FW, Keri RA. Sustained trophism of the mammary gland is sufficient to accelerate and synchronize development of ErbB2/Neu-induced tumors. Oncogene. 2006;25:3325–3334. doi: 10.1038/sj.onc.1209365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps AI, Buist DS, Malone KE, Barlow WE, Porter PL, Kerlikowske K, et al. Reproductive history and risk of three breast cancer subtypes defined by three biomarkers. Cancer Causes Control. 2011;22:399–405. doi: 10.1007/s10552-010-9709-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed W, Sandstad B, Holm R, Nesland JM. The prognostic impact of hormone receptors and c-erbB-2 in pregnancy-associated breast cancer and their correlation with BRCA1 and cell cycle modulators. Int J Surg Pathol. 2003;11:65–74. doi: 10.1177/106689690301100201. [DOI] [PubMed] [Google Scholar]
- Leong CO, Vidnovic N, DeYoung MP, Sgroi D, Ellisen LW. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J Clin Invest. 2007;117:1370–1380. doi: 10.1172/JCI30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yan W, Jung YS, Chen X. Mammary epithelial cell polarity is regulated differentially by p73 isoforms via epithelial-to-mesenchymal transition. J Biol Chem. 2012;287:17746–17753. doi: 10.1074/jbc.M112.358143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey MR, He L, Forster N, Ory B, Ellisen LW. Physical association of HDAC1 and HDAC2 with p63 mediates transcriptional repression and tumor maintenance in squamous cell carcinoma. Cancer Res. 2011;71:4373–4379. doi: 10.1158/0008-5472.CAN-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Osada M, Guo Z, Fomenkov A, Begum S, Zhao M, et al. DeltaNp63alpha up-regulates the Hsp70 gene in human cancer. Cancer Res. 2005;65:758–766. [PubMed] [Google Scholar]
- Borrelli S, Candi E, Alotto D, Castagnoli C, Melino G, Vigano MA, et al. p63 regulates the caspase-8-FLIP apoptotic pathway in epidermis. Cell Death Differ. 2009;16:253–263. doi: 10.1038/cdd.2008.147. [DOI] [PubMed] [Google Scholar]
- Mills AA, Qi Y, Bradley A. Conditional inactivation of p63 by Cre-mediated excision. Genesis. 2002;32:138–141. doi: 10.1002/gene.10067. [DOI] [PubMed] [Google Scholar]
- Holembowski L, Schulz R, Talos F, Scheel A, Wolff S, Dobbelstein M, et al. While p73 is essential, p63 is completely dispensable for the development of the central nervous system. Cell Cycle. 2011;10:680–689. doi: 10.4161/cc.10.4.14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.