Mutations in isocitrate dehydrogenase 1 and 2 (IDH1/2) have been discovered in several cancers, and these mutant enzymes exhibit neomorphic activity resulting in production of D2-hydroxyglutaric acid (D-2HG). Akbay et al. find that adult transgenic mice with conditionally activated IDH2R140Q and IDH2R172K alleles exhibit dilated cardiomyopathy and muscular dystrophy. These phenotypes were even more pronounced in embryos. Cardiac hypertrophy was also observed in nude mice implanted with IDH2R140Q-expressing xenografts. Silencing of IDH2R140Q in mice with an inducible transgene restored heart function by lowering 2HG levels.
Keywords: D-2HG, D2HGA, IDH2, cardiomyopathy, glycogen, myopathy, vacuolar leukoencephalopathy
Abstract
Mutations in isocitrate dehydrogenase 1 and 2 (IDH1/2) have been discovered in several cancer types and cause the neurometabolic syndrome D2-hydroxyglutaric aciduria (D2HGA). The mutant enzymes exhibit neomorphic activity resulting in production of D2-hydroxyglutaric acid (D-2HG). To study the pathophysiological consequences of the accumulation of D-2HG, we generated transgenic mice with conditionally activated IDH2R140Q and IDH2R172K alleles. Global induction of mutant IDH2 expression in adults resulted in dilated cardiomyopathy, white matter abnormalities throughout the central nervous system (CNS), and muscular dystrophy. Embryonic activation of mutant IDH2 resulted in more pronounced phenotypes, including runting, hydrocephalus, and shortened life span, recapitulating the abnormalities observed in D2HGA patients. The diseased hearts exhibited mitochondrial damage and glycogen accumulation with a concordant up-regulation of genes involved in glycogen biosynthesis. Notably, mild cardiac hypertrophy was also observed in nude mice implanted with IDH2R140Q-expressing xenografts, suggesting that 2HG may potentially act in a paracrine fashion. Finally, we show that silencing of IDH2R140Q in mice with an inducible transgene restores heart function by lowering 2HG levels. Together, these findings indicate that inhibitors of mutant IDH2 may be beneficial in the treatment of D2HGA and suggest that 2HG produced by IDH mutant tumors has the potential to provoke a paraneoplastic condition.
The metabolic diseases D2-hydroxyglutaric aciduria (D2HGA), L2-hydroxyglutaric aciduria (L2HGA), and combined D,L2-hydroxyglutaric aciduria (D,L-2HGA) are rare neurodegenerative disorders resulting from accumulation of 2HG isomers (Chalmers et al. 1980; Duran et al. 1980) that are produced as metabolic intermediates in reactions catalyzed by L-malate dehydrogenase and hydroxyacid–oxoacid transhydrogenase (Struys et al. 2005; Rzem et al. 2007). Underlying genetic causes have been discovered for nearly all patients for L2HGA and D2HGA; a majority of L2HGA patients have inactivating mutations in L2 hydroxyglutaric acid dehydrogenase (L2HGDH) and about half of D2HGA patients have mutations in D2-hydroxyglutaric acid dehydrogenase (D2HGDH), both of which are responsible for hydrolyzing 2HG to α-ketoglutarate (αKG) (Kranendijk et al. 2012). Inactivating mutations in SLC25A1 encoding the mitochondrial citrate carrier have recently been identified as the cause for the combined D,L-2HGA (Nota et al. 2013). L2HGA is a homogeneous disease, whereas D2HGA is clinically more heterogeneous with two subtypes, type I and type II, distinguished by the severity of the symptoms, time of onset, and inheritance pattern. Homozygous inactivating mutations in D2HGDH were detected in type I patients exhibiting mild phenotypes, whereas heterozygous mutations of isocitrate dehydrogenase 2 (IDH2) codon 140 are associated with the neonatal onset type II group (Kranendijk et al. 2010b). The IDH2 enzyme catalyzes the oxidative decarboxylation of isocitrate to αKG. A missense mutation at codon 140 of IDH2 (R140Q) is responsible for causing the neomorphic activity of reducing αKG to D2-hydroxyglutaric acid (D-2HG), a metabolite that is normally maintained at low intracellular concentrations. Both groups of D2HGA patients have elevated D-2HG in the body fluids (cerebrospinal fluid, serum, and urine) and exhibit developmental delay, epilepsy, hypotonia, dysmorphic features, and brain white matter abnormalities. Cardiomyopathy requiring treatment is frequently observed in type II D2HGA patients (Kranendijk et al. 2010b, 2012). Although 2HG levels fluctuate in the same individuals (van der Knaap et al. 1999), patients who have IDH2 mutations generally have twofold to eightfold higher 2HG levels than those who have mutations in D2HGDH (Kranendijk et al. 2010a).
Elevated 2HG resulting from recurrent mutations in the mitochondrial metabolic enzyme IDH2 (R140Q and R172K) or its cytosolic counterpart, IDH1, has also been associated with the development of several cancers: glioma, acute myeloid leukemia (AML), and subsets of chondrosarcoma, cholangiocarcinoma, acute lymphoblastic leukemia (ALL), prostate cancer, melanoma, and non-small-cell lung cancer (Balss et al. 2008; Kang et al. 2009; Yan et al. 2009; Abbas et al. 2010; Green and Beer 2010; Amary et al. 2011a,b; Sequist et al. 2011; Shibata et al. 2011; Borger et al. 2012; Ghiam et al. 2012). Oncology-focused studies suggest that 2HG accumulation inhibits αKG-dependent dioxygenases, including the TET family methylcytosine dioxygenases, and the JmjC domain-containing histone demethylases, thereby causing a global DNA and histone hypermethylation phenotype (Figueroa et al. 2010; Xu et al. 2011; Lu et al. 2012; Turcan et al. 2012). Furthermore, some metabolic processes are affected by 2HG accumulation, leading to alterations in the TCA cycle (Reitman et al. 2011) and branched chain amino acid metabolism (Tonjes et al. 2013).
Although it has been >30 years since the first report of DH2GA (Chalmers et al. 1980), the progression of the disease is still poorly understood, and there is no effective treatment (Kranendijk et al. 2012). Here, we report the development of a series of mouse models expressing mutant IDH2 that recapitulate the central nervous system (CNS) abnormalities and cardiomyopathy found in the human condition. Our studies using these models reveal that 2HG can cause myocyte death and neuropathy associated with mitochondrial damage and glycogen accumulation. Moreover, we show that silencing mutant IDH2 expression and reduction in 2HG levels can reverse at least some of these pathologic features.
Results
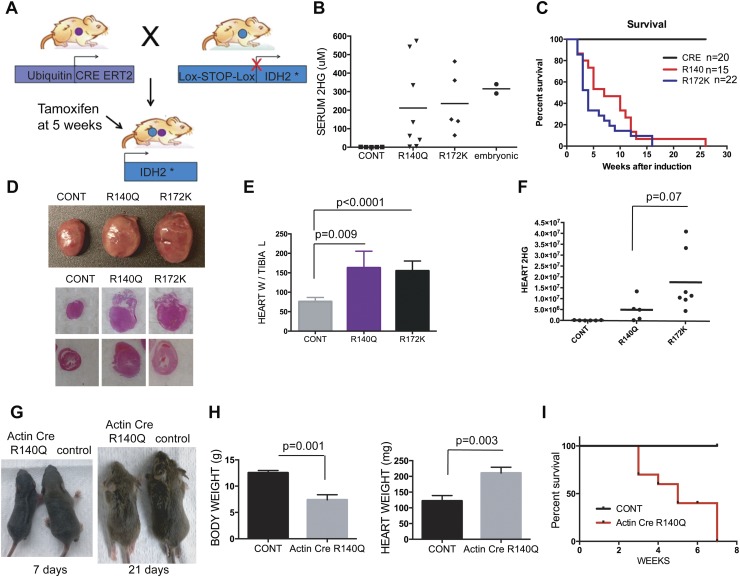
We developed knock-in transgenic mouse strains in which either IDH2R140Q or IDH2R172K is expressed under the control of the chicken β-actin promoter and a floxed stop cassette (Supplemental Fig. 1A). To study the impact of conditional activation of mutant IDH2 in the adults, we crossed the IDH2R140Q or IDH2R172K transgenic mice (subsequently referred to as R140Q and R172K mice) to the broadly expressed, tamoxifen-inducible Ubiquitin CreERT2 strain and administered tamoxifen at 5 wk of age (Fig. 1A). These mice exhibited induction of the mutant IDH2 in the lung, spleen, kidney, brain, and heart (Supplemental Fig. 1D,E) associated with a marked increase in serum 2HG as compared with controls, with levels ranging from 30 μM to 550 μM (Fig. 1B). Both mutant IDH2 mouse strains showed signs of lethargy and shortness of breath within 3–4 wk of tamoxifen administration and rapidly became moribund, requiring sacrifice (mean survival 7 wk for R140Q and 4 wk for R172K; P = 0.16) (Fig. 1C). Necropsy revealed significant enlargement of the hearts in IDH2 mutants (Fig. 1D,E). While serum levels of 2HG were comparable in the R140Q and R172K strains, there was a trend toward increased intracellular 2HG in hearts from the latter group (P = 0.07) (Fig. 1F). Examination of other organs, including the lung, kidney, liver, spleen, and testis/ovary, failed to reveal additional gross abnormalities, while they did show microscopic signs of circulatory congestion associated with cardiac failure (data not shown). Moreover, levels of glucose, blood urea nitrogen (BUN), potassium, and sodium were normal in the serum, indicating intact liver and kidney function in these animals (data not shown). Embryonic activation of IDH2R140Q resulting from crosses to the Actin-Cre strain resulted in more severe phenotypes with partially penetrant embryonic lethality (total offspring: 63, approximate expected Actin-Cre; R140Q: 15, observed: 8; P = 0.002). Surviving animals displayed runting (Fig. 1G,H), tremors, seizures, hydrocephalus, and cardiac hypertrophy, resulting in death between 3 and 7 wk (Fig. 1I). Levels of serum 2HG were comparable with those observed in adults with somatic activation of mutant IDH2 (Fig. 1B). Thus, the IDH2 mutant mice appear to recapitulate the full spectrum of D2HGA syndrome, with embryonic IDH2 expression provoking a more pronounced disease phenotype.
Figure 1.
Generation and characterization of conditional mutant IDH2 mice. (A) Schematic for the adult IDH2 mutant mouse model generated from crosses of Ubiquitin CreERT2 and Lox-stop-lox IDH2. (B–F) Mice of the indicated genotypes were examined for serum 2HG levels measured using the 2HG standards at 4 wk after tamoxifen administration (B), Kaplan-Meier survival analysis (C), gross appearance and H&E staining of the heart (D), heart weight (n = 5, 6, and 6 mice, respectively) (E), and heart 2HG levels (F), presented in relative arbitrary units. (G–I) Mice with embryonic activation of IDH2 R140Q (progeny from crossing Actin Cre and LSL-R140Q mice) and controls examined for gross appearance at 7 and 21 d (G), body weight and heart weight at ages 3 wk and 6 wk, respectively (n = 3 for both groups for both graphs) (H), and survival (n = 8 and 6 for Actin Cre LSL-R140Q and controls, respectively) (I).
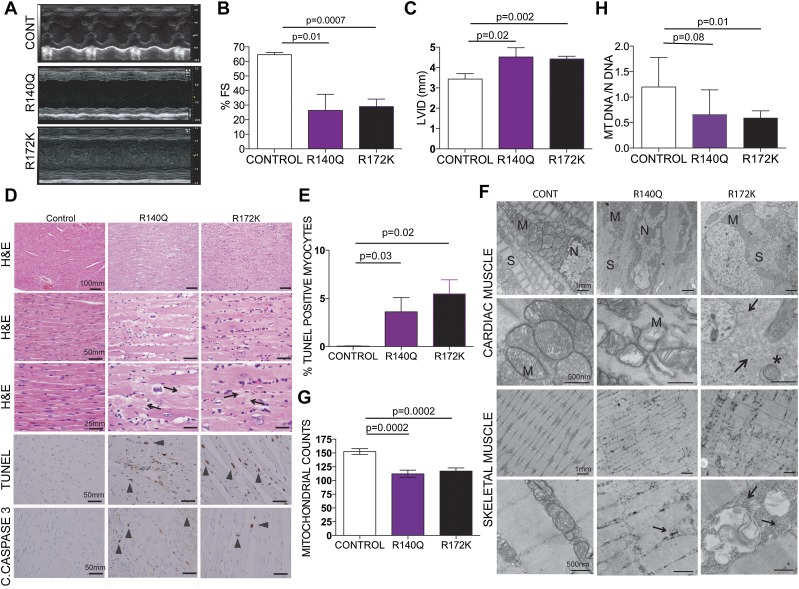
To begin to understand the functional consequences of the cardiac malformations in the R140Q and R172K mice, we performed echocardiography 4 wk after tamoxifen administration. These animals exhibited end-stage cardiomyopathy characterized by severely decreased wall motion and increased dilatation (Fig. 2A–C) without a change in heart rate (data not shown). Histological examination revealed sarcomere degeneration and myocyte dropout associated with myocyte hypertrophy and nuclear enlargement (Fig. 2D). Scarring of the hearts associated with accumulating collagen deposits was evident and likely contributed to the cardiac remodeling and cardiac failure (Fig. 2D). Cardiac defects were associated with high levels of apoptosis in cardiomyocytes as determined by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL), showing a rate of 4%–8% TUNEL-positive myocytes in IDH2 mutants as compared with <0.1% in controls (P = 0.03 and 0.02) (Fig. 2D,E), and cleaved caspase-3 staining (Fig. 2D).
Figure 2.
Physiologic and structural defects in IDH mutant muscle. (A) Representative M mode images of the echocardiograms of the mice with the indicated genotypes. (B) Fractional shortening (%FS) measured from the M mode images of the echocardiograms plotted for each genotype (n = 3, 3, and 6 mice, respectively). (C) Left ventricular internal diameter in diastole (LVID) measured from the B mode images of the echocardiograms showing dilatation in the mutant IDH hearts (n = 3, 3, and 6, respectively). (D) Representative histology of the hearts from the indicated genotypes. H&E-stained longitudinal sections of IDH2 mutant cardiac muscle show myocyte hypertrophy characterized by fiber thickening and nuclear enlargement with increased pleomorphism. TUNEL staining and immunohistochemistry for cleaved caspase-3 on formalin-fixed tissues show increased apoptosis in the R140Q and R172K hearts compared with controls. Arrowheads point to fibrotic areas. Bars show the indicated lengths and are the same across the images on the same row. (E) Quantification of the TUNEL staining on the heart tissues (n = 3, 3, and 6, respectively). (F) Representative electron microscopy images of the hearts from the indicated genotypes of mice 4 wk after tamoxifen administration show defects in sarcomere organization, perturbed cardiac Z disks, a decrease in mitochondrial number, an increase in abnormal small mitochondria, and a substantial increase in glycogen deposits in mutant IDH hearts compared with controls. Arrows point to areas with massive glycogen deposits. Asterisks indicate autophagosomes. Bars show the indicated lengths and are the same across images on the same row. (M) Mitochondria; (N) nucleus; (S) sarcomere. (G,H) Mitochondrial quantification for the indicated genotypes. (G) Mitochondrial counts from the electron microscopy images; the number of images used for counts is 9, 6, and 11, respectively. (H) Real-time PCR for mitochondrial DNA in the hearts. All samples were normalized to genomic DNA of the same sample. n = 7 for all genotypes.
To further understand the cellular abnormalities in IDH2 mice, we performed ultrastructural examination of the cardiac muscles. Conforming to the histologic findings, electron microscopy showed sarcomere degeneration and severe disorganization in the R140Q and R72K mice (Fig. 2F). We also observed mitochondrial abnormalities characterized by disrupted cristae, smaller or swollen mitochondria, and decreases in the total mitochondrial counts and mitochondrial DNA content compared with controls (Fig. 2G,H). There was evidence for mitophagy, as reflected by the presence of double-membrane autophagosomes (Fig. 2F). 2HG accumulation has been previously shown to cause alterations in the mitochondrial TCA cycle (Reitman et al. 2011). Metabolite profiling of the R140Q and R172K hearts and controls has revealed significant decreases in the steady-state TCA cycle intermediates (Supplemental Fig. 2), likely disrupting transfer of electrons from TCA metabolites into the electron transport chain. Skeletal muscle of IDH2 mutants exhibited a comparable set of defects, including abnormalities in sarcomere organization, mitochondria, and glycogen accumulation, all of which are more severe in the R172K mutants (Fig. 2F).
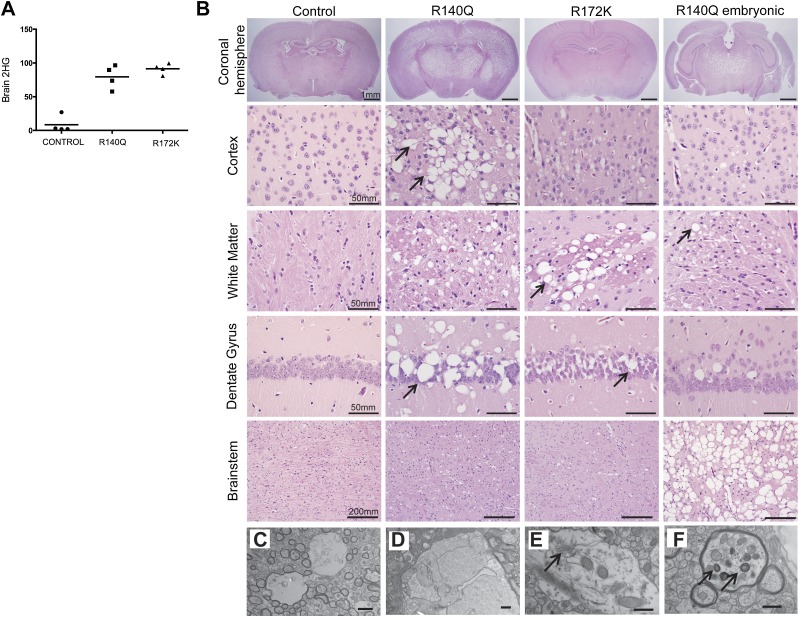
Neuropathological examination of the brains of both IDH mutants revealed diffuse vacuolar leukoencephalopathy in the white and gray matter throughout the CNS upon increased levels of brain 2HG (Fig. 3A,B; Supplemental Fig. 3A–D). Unlike in the muscle, we did not observe any evidence of apoptosis (using TUNEL assays) or glycogen accumulation (periodic acid-Schiff [PAS] staining) in R140Q or R172K brain tissue (Supplemental Fig. 3E–I). On the other hand, electron microscopy revealed the presence of vacuoles filled with membranous debris at both myelinated axons and nonmyelinated naked axons but not in the neuron bodies (Fig. 3C,F; Supplemental Fig. 3A,B). The presence of double-membraned vesicles (Fig. 3F) suggests that, as in the muscle, there is an elevation in autophagy and mitochondrial abnormalities in the axons of the IDH2 mutant brain. Thus, 2HG causes intracellular toxicity in the neurons that leads to vacuolarization in the brain parenchyma.
Figure 3.
Histological and ultrastructural analysis of mutant IDH2 mouse brains. (A) 2HG levels from the whole brains of control, R140Q, and R172K mice are presented in relative arbitrary units. (B) Representative H&E sections of control, R140Q, and R172K mice 4 wk after tamoxifen administration and of embryonic R140Q mice (6 wk old). IDH2 mutants show a diffuse vacuolar leukoencephalopathy of variable penentrance involving multiple CNS compartments, including the cortex, white matter, hippocampus, and brain stem. Bars show the indicated lengths and are the same across images on the same row. (C–F) Electron micrographs of the R140Q mice brain. (C) Vacuoles appear mostly in the axons; the myelinated axon is on the left, and the naked axon is on right. (D) Membranous debris in the vacuoles. (E) Arrow points to endoplasmic reticulum. (F) Degenerated axon; arrows point to organelles in autophagosomes. Bars: C,D, 2 µm; E,F, 500 nm.
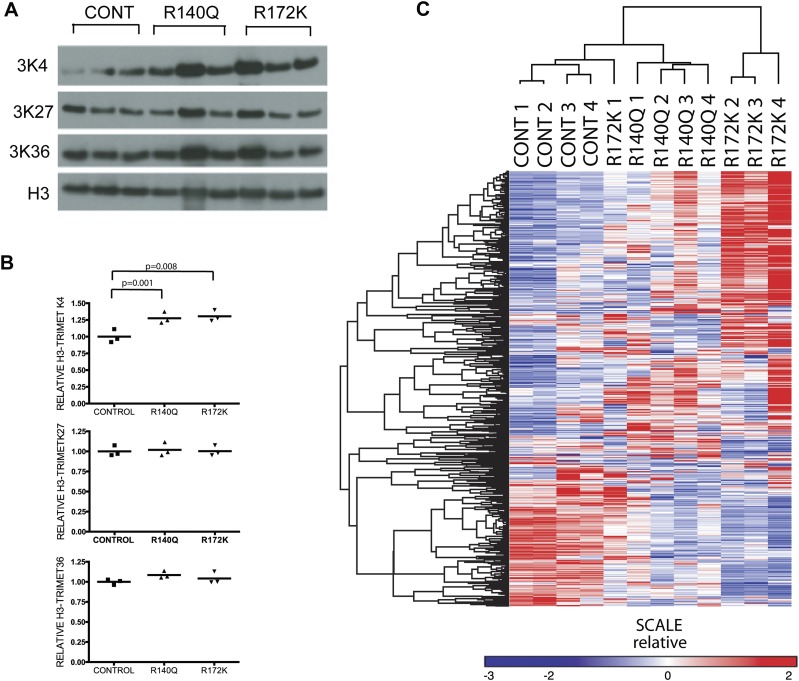
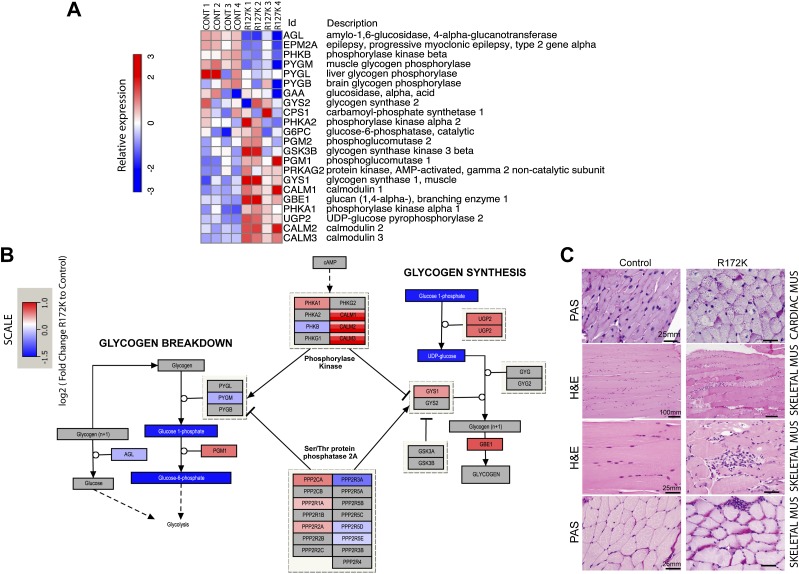
2HG accumulation is associated with global hypermethylation and changes in gene expression in glioma and leukemia (Lu et al. 2012; Turcan et al. 2012) and has been shown to directly modulate chromatin states by inhibiting the function of both TET family enzymes involved in DNA demethylation (Figueroa et al. 2010; Xu et al. 2011; Losman et al. 2013) and the Jumonji family of histone demethylases (Lu et al. 2012). Analysis of global histone marks in heart tissue revealed that in comparison with controls, the IDH2 mutants had increased levels of trimethylated H3K4 but no changes in trimethylated H3K27 or trimethylated H3K36 (Fig. 4A,B). Furthermore, microarray expression profiling showed that there were broad transcriptional differences involving several metabolic pathways between the IDH2 mutant and control mice (Fig. 4C; Supplemental Tables 1–4). Notably, in keeping with the histological findings, we detected changes in the expression of genes involved in glycogen synthesis, with marked up-regulation of key biosynthetic enzymes (glycogen synthase 1 [GYS1], UDP [uridine diphosphate] glucose phosphorylase [UGP2], and phosphoglucomutase 1 and 2 [PGM1, PGM2]) and down-regulation of enzymes involved in the regulation of glycogen breakdown (epilepsy progressive myoclonus type 2-laforin [EPM2A], phosphorylase kinase β [PHKB], muscle glycogen phosphorylase [PYGM], phosphorylase glycogen [PYGL], and amylo-α-1,6-glucosidase [AGL]) in R172K hearts compared with controls (Fig. 5A,B; Supplemental Fig. 4B). Although glycogen synthesis is controlled by a complex network of signals (Hardie 2012; Roach et al. 2012), increased expression of biosynthesis genes could contribute to the increase in cardiac glycogen deposition. Normal adult hearts use fatty acids as their main source of energy (Strauss and Johnson 1996), whereas there is a shift toward glycolysis in the context of heart failure (Allard et al. 1994). Metabolite profiling of the hearts expressing mutant IDH2 R172K indicated a significant decrease in intermediate metabolites involved in glycogen biosynthesis, including UDP glucose and glucose 1 phosphate (G1P) (Fig. 5B). These changes were more prominent in the R172K mutants compared with R140Q. (Supplemental Fig. 4A–C). We also performed chemical staining for lipid accumulation (Oil red O) and glycogen (PAS) to extend our investigation of potential metabolic alterations in the end-stage IDH2 mutant hearts. Consistent with our ultrastructural observations (Fig. 2F), the R172K cardiac and skeletal muscle showed elevation in PAS staining, indicating substantial glycogen deposition as compared with age-matched controls (Fig. 5C). Oil Red O staining was comparable in all animals (data not shown). Thus, cardiomyopathy in IDH2 mutant mice is associated with mitochondrial defects and elevations in glycogen levels, suggesting roles for impairment in nutrient utilization and metabolism in the disease phenotype.
Figure 4.
Histone modifications and gene expression profiling of the IDH2 mutant hearts. (A) Western blots for trimethyl-H3K4, trimethyl-H3K27, and trimethyl-H3K36 on the hearts from the mice with the indicated genotypes. Each lane is from an individual mouse, and results represent two independent experiments. (B) Relative quantification of H3-3MetK4, H3-3MetK27, and H3-3MetK36 normalized to H3. (C) Gene expression profiles were filtered to exclude probe sets with signals present at background noise levels and for probe sets that do not vary significantly across samples (more than threefold over the entire sample set). After filtering, probes representing the same genes were collapsed into a single value and clustered using average linkage method with the Pearson centered distance correlation metric using Gene-e software. Each lane represents a different mouse.
Figure 5.
Transcriptional and metabolic changes in IDH2 mutant hearts compared with controls. (A) Expression of the genes involved in glycogen biosynthesis and degradation in R172K mutant hearts compared with controls. (B) Pathway map of enzymes, metabolites, and polysaccharides in the glycogen biosynthesis pathway. Color scales indicating RNA levels of the enzymes are the same as in A. The color scale for metabolites indicates log2 fold change of levels in R172K mutant hearts (n = 9) relative to controls (n = 10). Metabolites that were not measured or had no detectable change are shown in gray. P-values for UDP glucose and G1P are P < 0.05 and P < 0.007, respectively. (C) Histology of the cardiac and skeletal muscles. PAS-stained sections of cardiac muscle showing increased collections of diastase-sensitive glycogen in R172K animals compared with control. H&E-stained sections of skeletal muscle show myopathic changes, including increased variation in fiber size, increased numbers of internalized nuclei, and increased endomysial fibrosis. Myofiber regeneration, degeneration, and necrosis are present in association with multifocal myophagocytosis. PAS stains highlight increased glycogen deposition in IDH2 mutant skeletal muscles compared with control. Bars show the indicated lengths and are the same across images on the same row.
Several isoforms of collagen are expressed in the heart (the most important ones being type I and type III), and collagen content has an impact on the motility of the myofibers (Bishop and Laurent 1995). Global expression analysis revealed a broad up-regulation of genes involved in collagen biosynthesis and organization (Supplemental Tables 2, 4) and significant increases in specific isoforms of collagens I, III, IV, V, and VI (Supplemental Fig. 5A–C), findings that are in accord with previous reports from dilated cardiomyopathy patients (Bishop et al. 1990; Gunja-Smith et al. 1996). Unlike recent findings in IDH1 mutant mice (Sasaki et al. 2012), we did not observe significant changes in the maturation of collagen type IV in the IDH2 mutant hearts (data not shown).
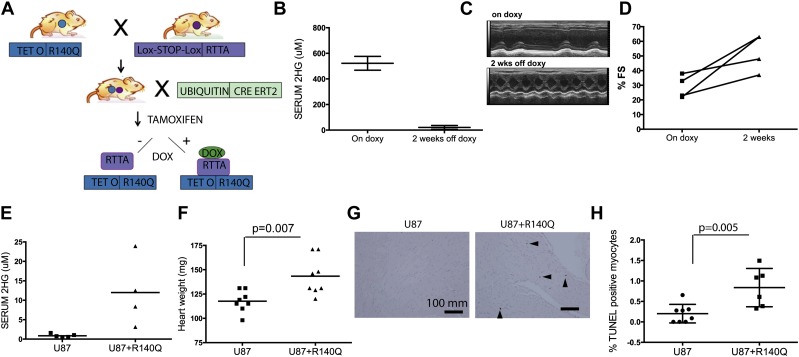
We next sought to examine whether the cardiomyopathy induced by mutant IDH2 is an irreversible degenerative condition or can be ameliorated by extinction of mutant IDH2 expression. To this end, we generated mice with a doxycyline-inducible IDH2R140Q (TetOp R140Q) allele as previously described (Beard et al. 2006) and intercrossed them with the Rosa26-LSL-rtTA strain (which expresses the reverse tetracycyline transactivator line after a Lox-stop-Lox cassette) and the Ubiquitin CreERT2 strain (Fig. 6A). We induced mutant IDH expression at 5 wk of age by tamoxifen administration and doxycycline diet. These mice showed induction of IDH2R140Q expression, with the highest levels in the liver and kidney (Supplemental Fig. 6A,B), and had serum 2HG levels that were comparable with those of the constitutive IDH2 mice (Figs. 1B, 6B). These mice developed cardiomyopathy phenotypes resembling those in the constitutive model but with longer latency (14–16 wk), likely due to lower levels of transgene expression in the heart (Supplemental Fig. 6A,B). Importantly, upon doxycycline withdrawal and extinction of transgene expression, we observed a reduction in serum 2HG levels in 2 wk (Fig. 6B) accompanied by significantly improved heart function, as reflected by increased fractional shortening and overall survival (P-value = 0.0009) (Fig. 6C,D; Supplemental Fig. 6C).
Figure 6.
Paracrine cardiomyopathy effect of 2HG and reversibility of the cardiomyopathy phenotype in inducible R140Q transgenic mice. (A) Schematic for inducible IDH2 R140Q mouse model. (B–D) Inducible IDH2 R140Q mice were examined for heart function while under doxycycline administration for 16 wk (On doxy) or 2 wk after doxycycline withdrawal for serum 2HG levels (B), M mode short axis view of echocardiography (C), and percent fractional shortening (%FS) (D). (E–H) Nude mice carrying tumors of U87 cells or U87 cells expressing IDH2 R140Q examined for serum 2HG levels (E), heart weights (n = 8 and 9, respectively) (F), TUNEL staining of the fixed heart sections (G), and quantification of the percentage of TUNEL-positive myocytes (n = 8 and 6, respectively) (H).
2HG is present at high levels in the circulation of our IDH2 mutant mice and in patients with D2HGA or with IDH2 mutant cancers, suggesting the possibility that 2HG can elicit disease phenotypes in a paracrine manner. To test this question, we analyzed the hearts of nude mice carrying subcutaneous tumors of IDH2R140-expressing U87 cells, which have fivefold to 20-fold increased serum 2HG levels compared with mice carrying control U87 tumors (Fig. 6E). Remarkably, mice with R140Q-expressing tumors had significantly enlarged hearts relative to those with control tumors (Fig. 6F) and exhibited a corresponding increase in apoptotic myocytes (Fig. 6G,H). These results indicate a potential role for circulating 2HG in causing cardiac hypertrophy and suggest that patients with IDH mutant tumors may develop cardiac abnormalities as a paraneoplastic condition.
Discussion
Results from both genetically engineered mice and nude mice carrying tumors expressing mutant IDH2 suggest that 2HG has a previously unrecognized role of development of cardiomyopathy. The cardiomyopathy and vacuolarization phenotypes specifically observed in mice embryonically expressing IDH2R140Q model the disease progression in the D2HGA patients. While differences in response were observed in different tissues, it is notable that the tissues with the highest mitochondrial activity (cardiac/skeletal muscle and brain) were most vulnerable to the 2HG-related metabolic insult. Importantly, although our mouse models express mutant IDH2 from a transgenic promoter, the resulting serum 2HG levels are comparable with those in type II D2HGA patients (99–757 μM) (Kranendijk et al. 2012), supporting the physiologic relevance of our findings.
Differences between the 2HG production potential of mutant IDH1 and IDH2 alleles have been investigated in in vitro systems and attributed to distinctions in intracellular localization and access to substrate (αKG) (Ward et al. 2013). These findings likely account for the dramatic differences in serum 2HG and life span between the previously reported IDH1R132H knock-in mice (Sasaki et al. 2012) and mice globally expressing mutant IDH2.
Mice globally expressing mutant IDH2 succumbed to cardiomyopathy in <3 mo. Upon detailed full body pathologic analysis, there was no evidence for a premalignant phenotype. Similar to D2HGA patients (Kranendijk et al. 2010b), the lack of malignant phenotypes in the mice globally expressing the IDH2 mutant alleles may be due to shortened life span. Our findings are in line with a prior report showing that expression of mutant IDH1 specifically in the bone marrow compartment fails to induce tumors even in aged mice (Sasaki et al. 2012). Notably, nearly all IDH mutations in human tumors are invariably concurrent with additional genetic alterations (such as inactivation of TP53 or P14ARF) or deletion of 1p/19q in glioma patients (Ichimura et al. 2009) and with NRAS, ASXL1, or FLT3 mutations in leukemia patients (Rocquain et al. 2010). We found that mice engineered to express mutant IDH2 in specific tissues eventually develop carcinomas although only with very long latencies (data not shown). Thus, we believe that 2HG has transforming capability in vivo after very long exposures and may cooperate with other mutations to cause malignant tumors.
Mutations in IDH1/2 may provide a selective advantage at the initial stages of cellular transformation by altering epigenetic reprogramming (Figueroa et al. 2010; Turcan et al. 2012); somewhat paradoxically, GBM patients with IDH mutations have a better prognosis compared with patients with wild-type IDH (31 mo vs. 15 mo) (Parsons et al. 2008; Yan et al. 2009). Along those lines, cultured mouse embryonic fibroblasts (MEFs) expressing mutant IDH2 proliferate slower compared with normal MEFs (data not shown). This supports the previous glioma cell line studies that reported extinction of IDH mutation when xenografted or slower growth compared with IDH wild-type glioma cell lines (Kelly et al. 2010; Luchman et al. 2012), suggesting a 2HG-related cellular toxicity. Correspondingly, in brain tumors, it appears that IDH may have context-specific oncogenic activity restricted to specific cell lineages and stages of differentiation (Sturm et al. 2012).
Widespread changes in the epigenetic landscape resulting from IDH mutations have been previously reported by several groups (Parsons et al. 2008; Figueroa et al. 2010; Lu et al. 2012; Turcan et al. 2012). We observed altered global histone modifications—particularly an increase in the levels of trimethylated H3K4. IDH2 mutant hearts exhibited a specific increase in genes involved in the glycogen biosynthesis pathway. Excessive glycogen may be responsible for causing impairment in myofiber motility similar to glycogen storage diseases (Arad et al. 2005). In-depth future work would be required to explore the possibility that DNA methylation changes together with histone modifications provoked by IDH mutations may have a role in these global transcriptional changes.
Existing data point to the potential impact of IDH mutations on hypoxia response. Initial reports found increases in HIF1α levels in gliomas with IDH1 mutations compared with IDH wild-type tumors (Zhao et al. 2009; Xu et al. 2011), while a later study showed a decrease in HIF1α activation through the 2HG-mediated activation of αKG-dependent prolyl hydroxylases (PHDs or EglNs) (Koivunen et al. 2012). In our mouse models, hearts expressing mutant IDH2 showed slight decreases in HIF, which correlated with serum 2HG levels (data not shown). Moreover, as in mice with expression of an IDH1R132H knock-in allele in the hematopoietic cells (Sasaki et al. 2012), the cardiomyocytes from IDH2 mutant mice did not show altered expression of hypoxia-responsive genes (Liu et al. 2012) upon microarray analysis (data not shown). These findings suggest that changes in HIF activity are unlikely to account for the observed cardiomyocyte defects in the IDH2 mutant mice.
In a previously reported study, Maffucci patients who carry tumors with no detectable IDH mutations along with the tumors with detectable IDH mutations raised the possibility that 2HG can influence cells nonautonomously (Amary et al. 2011b). Results from the xenograft study demonstrated that 2HG can act in a paracrine fashion to cause cardiotoxicity. Although the capacity of paracrine 2HG to promote malignant transformation needs validation, our results suggest patients who have high circulating 2HG levels due to IDH mutations could develop cardiotoxicity. Thus, some cases of idiopathic cardiomyopathy or neuromuscular disorders may result from IDH mutations or 2HG accumulation. Given the reversibility of the cardiomyopathy in the inducible mutant mice, novel treatments that specifically target the mutant IDH2 proteins to lower the 2HG levels could be effective in the treatment of patients with type II D2HGA.
Materials and methods
Mouse husbandry and breeding
Mice were housed in pathogen-free animal facilities, and all experiments were performed with the approval of the Animal Care and Use Committee at Harvard Medical School and Dana-Farber Cancer Institute.
Generation and maintenance of transgenic mice
Point mutations R140Q and R172K were introduced to human IDH2 cDNA (Origene, #SC319226) using the QuickChange kit (Agilent, #200521) per the manufacturer’s instructions. cDNAs were cloned into transgenic targeting vectors and coelectroporated into the v6.5 C57BL/6(F) × 129/sv(M) embryonic stem cells (Open Biosystems, #MES 1402) with plasmid expressing FLP recombinase as described (Beard et al. 2006). Embryonic stem cells were screened for integration of the transgene by PCR. Correctly targeted embryonic stem cells were injected into Black 6 blastocysts, and the resulting chimeras were bred with BALB/c strain wild-type mice for germline transmission of the transgenes. IDH2 transgenic mice were crossed with Ubiquitin CreERT2 (Ruzankina et al. 2007) or ActinCre mice (Jackson Laboratories). Mice were maintained in a mixed background strain, and double-mutant mice were given 2 mg of 4-hydroxytamoxifen (4-OHT) (Sigma, #1054029-01) at 5 wk of age for 3 d, skipping a day between treatments. In all of the experiments, hemizygous IDH2 mice were used, and mice were included in the survival curves when they either died or reached disease burden euthanasia criteria.
Echocardiography
Echocardiography was performed as previously described (Moslehi et al. 2010). Left ventricular internal diameter in diastole was measured from the B mode images on the long axis, and fractional shortening was calculated from the M mode images from the short axis using VEVO 2100 software (Visual Sonics).
Histology and electron microscopy
Tissues were fixed overnight in 10% buffered formalin and paraffin-embedded (FFPE). PAS stainings were performed on 5-μm sections as described (Moslehi et al. 2010). Oil red O staining was performed on the OCT-embedded and frozen tissue sections using the Oil red O dye (Polysciences #06317-25). Monoclonal cleaved caspase-3 antibody (Cell Signaling Technology #9664) was used for immunohistochemistry on the FFPE sections. TUNEL assay was performed per the manufacturer's instructions (Millipore, #17-141). Three different representative areas on each heart section were used for TUNEL quantifications. For the electron microscopy analysis, tissues were fixed in Bouin's fixative (Polysciences, #866) and formaldhyde/glutaraldehyde mixture (EMS, #15949) in a 4:1 ratio overnight at 4°C and processed and imaged as described (Moslehi et al. 2010).
U87 xenograft experiments
U87 cells (1.8 × 106) or 1.6× U87 cells expressing IDH2 R140Q were injected into both flanks of 10 6-wk-old Nude (Nu/Nu) mice. Mice that grew tumors were sacrificed at 6–7 wk after injections and analyzed.
Real-time PCRs
Total RNA was extracted from different tissues using Trizol (Invitrogen, #15596018) followed by RNA cleanup (Qiagen, #74204). cDNA were prepared from total RNA preparations using the high-capacity RNA-cDNA kit (Invitrogen, #4377474). Real-time assays were done using TaqMan real-time probes (Invitrogen) for human IDH2 (HS_00271858) and mouse GAPDH (Mm01197443_m1) using 80 ng of cDNA. Triplicates were run for each sample. GAPDH was used as internal control, and ΔΔCT method was used for relative mRNA calculations.
Gene expression microarray
Total RNA was extracted using Trizol (Invitrogen) followed by RNA cleanup (Qiagen, #74204). Samples were hybridized to Affymetrix MoGene-1_0-st-v1 chips. Data analysis was conducted using R (http://www.r-project.org). Probe-level data were summarized using RMA (Irizarry et al. 2003).
Western blotting
Tissues were lysed in 1× lysis buffer (Cell Signaling Technology, #9803) with protease inhibitor cocktail (EMD Biosciences, 539131). Mouse monoclonal IDH2 antibody (Abcam, #ab55271) was used at 1 μg/mL dilution, rabbit monoclonal β-actin antibody (Cell Signaling Technology, #4970) was used at 1:2000 dilution, GYS1 antibody (Cell Signaling Technology, #3893) was used at 1:1000 dilution, and UGP2 antibody (Abcam, ab157473) was used at 1:1000 dilution.
Histone extraction and H3 Western blots
Heart tissues were homogenized in TEB buffer (0.5% Triton X-100, 2 mM PMSF, 0.02% NaN3 in PBS) at 200 mg/mL. Homogenates were centrifuged at 3000 rpm for 5 min at 4°C, and supernatant was removed. Pellets were resuspended in 3 vol of extraction buffer (0.5N HCL, 10% glycerol) and incubated for 30 min on ice. Mixture was centrifuged at 12,000 rpm for 5 min at 4°C, and supernatants were removed and mixed with 8 vol of acetone and incubated overnight at −20°C. Samples were centrifuged at 12,000 rpm for 5 min, and resulting pellets/histones were air-dried. Isolated histones were dissolved in distilled water, quantified, and used for Western blots. The antibodies used were H3 (Abcam, #AB1791), H3-3K4 (Millipore #07-473), H3-3K27 (Millipore, #07-449), and H3-3K36 (Abcam, #AB9050) per the manufacturers' instructions. Relative quantifications of histone marks were performed using ImageJ software.
Mitochondrial counts
Mitochondrial counts were performed by manually counting the number of total mitochondria on the images taken at 2900× magnification from the different areas of the tissues from two different mice.
Mitochondrial DNA real-time PCR
Genomic DNA was extracted using the DNA minikit (Qiagen) per the manufacturer's instructions. Ten nanograms of genomic DNA was used in the real-time reaction with SYBR Green master mix (Invitrogen) using primers previously described (Moslehi et al. 2010).
Statistical tests
T-tests were performed for the statistical analysis of the graphs. A χ2 test was performed on the breeding results. All bar graphs represent mean + standard error of the mean (SEM).
Serum 2HG measurements
Metabolites from 100 μL of sera were extracted in prechilled 80% methanol, and 2HG was measured using liquid chromatography-tandem mass spectrometry (LC-MS/MS) as described (Yuan et al. 2012). Pure 2HG (Sigma, #H8378) was used as standard to determine serum 2HG concentrations.
Tissue metabolite profiling
Reagents of LC-MS-grade were used for both extraction and LC-MS/MS applications. Tissues were extracted by homogenization in prechilled 80% aqueous methanol HPLC-grade (−80°C, 500 uL). After centrifugation at 14,000g for 15 min, the supernatant was removed and evaporated under reduced pressure. In order to detect metabolites in the central carbon pathways, extracts from 1 mg of tissue were analyzed for relative abundance of metabolites by using scheduled selective reaction monitoring (SRM) for each metabolite of interest, with the detector set to negative mode as detailed (Munger et al. 2008). LC separation was achieved by reverse-phase ion-pairing chromatography as described (Buescher et al. 2010).
Accession numbers
Array data have been deposited in Gene Expression Omnibus under accession number GSE54838.
Competing interest statement
K.M.S., S.M., W.L., C.G., S.C., M.K., and K.E.Y. are employees of Agios pharmaceuticals. W.G.K. is a scientific advisor for Agios, and A.T.F. served on an advisory board for Agios Pharmaceuticals and Seattle Genetics. K.K.W. received sponsored research grants from Takeda, AstraZeneca, Roche, Agios, and Infinity Pharmaceuticals and is a consultant for G1 Therapeutics.
Acknowledgments
We thank Ronglih Liao for cardiology expertise; Brigham and Women's Hospital Cardiac Physiology core for help with the physiology experiments; Dana-Farber Microarray Core for microarray gene expression profiling; Beth Israel Deaconess Medical Center Mass Spectrometry Core, Min Yuan, and Susanne Breitkopf for 2HG measurements; Brigham and Women's Hospital Transgenic Facility for blastocyst injections; Jinhee K. Oh for help with the maintenance of mouse colonies; Harvard Medical School Electron Microscopy Core for help with electron microscopy; Christine Lam for tissue processing; and Jacob B. Reibel for reading the manuscript. This work was supported by National Institute of Health (NIH) grants CA122794, CA140594, CA163896, CA166480, and CA154303 to K.K.W.; NIH grant 5T32HL007627-28 to S.H.R.; NIH 5P30CA006516 and 5P01CA120964 to J.M.A.; and NIH grant P50CA1270003, TargetCancer Foundation, and the Gallagher Chair in GI Cancer Research to N.B.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.231233.113.
Freely available online through the Genes & Development Open Access option.
References
- Abbas S, Lugthart S, Kavelaars FG, Schelen A, Koenders JE, Zeilemaker A, van Putten WJ, Rijneveld AW, Lowenberg B, Valk PJ 2010. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: Prevalence and prognostic value. Blood 116: 2122–2126 [DOI] [PubMed] [Google Scholar]
- Allard MF, Schonekess BO, Henning SL, English DR, Lopaschuk GD 1994. Contribution of oxidative metabolism and glycolysis to ATP production in hypertrophied hearts. Am J Physiol 267: H742–H750 [DOI] [PubMed] [Google Scholar]
- Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O'Donnell P, Grigoriadis A, Diss T, et al. 2011a. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol 224: 334–343 [DOI] [PubMed] [Google Scholar]
- Amary MF, Damato S, Halai D, Eskandarpour M, Berisha F, Bonar F, McCarthy S, Fantin VR, Straley KS, Lobo S, et al. 2011b. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet 43: 1262–1265 [DOI] [PubMed] [Google Scholar]
- Arad M, Maron BJ, Gorham JM, Johnson WH Jr, Saul JP, Perez-Atayde AR, Spirito P, Wright GB, Kanter RJ, Seidman CE, et al. 2005. Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N Engl J Med 352: 362–372 [DOI] [PubMed] [Google Scholar]
- Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A 2008. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol 116: 597–602 [DOI] [PubMed] [Google Scholar]
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R 2006. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44: 23–28 [DOI] [PubMed] [Google Scholar]
- Bishop JE, Laurent GJ 1995. Collagen turnover and its regulation in the normal and hypertrophying heart. Eur Heart J 16: 38–44 [DOI] [PubMed] [Google Scholar]
- Bishop JE, Greenbaum R, Gibson DG, Yacoub M, Laurent GJ 1990. Enhanced deposition of predominantly type I collagen in myocardial disease. J Mol Cell Cardiol 22: 1157–1165 [DOI] [PubMed] [Google Scholar]
- Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, Schenkein DP, Hezel AF, Ancukiewicz M, Liebman HM, et al. 2012. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 17: 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher JM, Moco S, Sauer U, Zamboni N 2010. Ultrahigh performance liquid chromatography-tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Anal Chem 82: 4403–4412 [DOI] [PubMed] [Google Scholar]
- Chalmers RA, Lawson AM, Watts RW, Tavill AS, Kamerling JP, Hey E, Ogilvie D 1980. D-2-hydroxyglutaric aciduria: Case report and biochemical studies. J Inherit Metab Dis 3: 11–15 [DOI] [PubMed] [Google Scholar]
- Duran M, Kamerling JP, Bakker HD, van Gennip AH, Wadman SK 1980. L-2-Hydroxyglutaric aciduria: An inborn error of metabolism? J Inherit Metab Dis 3: 109–112 [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiam AF, Cairns RA, Thoms J, Dal Pra A, Ahmed O, Meng A, Mak TW, Bristow RG 2012. IDH mutation status in prostate cancer. Oncogene 31: 3826. [DOI] [PubMed] [Google Scholar]
- Green A, Beer P 2010. Somatic mutations of IDH1 and IDH2 in the leukemic transformation of myeloproliferative neoplasms. N Engl J Med 362: 369–370 [DOI] [PubMed] [Google Scholar]
- Gunja-Smith Z, Morales AR, Romanelli R, Woessner JF Jr 1996. Remodeling of human myocardial collagen in idiopathic dilated cardiomyopathy. Role of metalloproteinases and pyridinoline cross-links. Am J Pathol 148: 1639–1648 [PMC free article] [PubMed] [Google Scholar]
- Hardie DG 2012. Organismal carbohydrate and lipid homeostasis. Cold Spring Harb Perspect Biol 4: a006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Pearson DM, Kocialkowski S, Backlund LM, Chan R, Jones DT, Collins VP 2009. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro-oncol 11: 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, Lee JY, Yoo NJ, Lee SH 2009. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer 125: 353–355 [DOI] [PubMed] [Google Scholar]
- Kelly JJ, Blough MD, Stechishin OD, Chan JA, Beauchamp D, Perizzolo M, Demetrick DJ, Steele L, Auer RN, Hader WJ, et al. 2010. Oligodendroglioma cell lines containing t(1;19)(q10;p10). Neuro Oncol 12: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, et al. 2012. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 483: 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranendijk M, Struys EA, Gibson KM, Wickenhagen WV, Abdenur JE, Buechner J, Christensen E, de Kremer RD, Errami A, Gissen P, et al. 2010a. Evidence for genetic heterogeneity in D-2-hydroxyglutaric aciduria. Hum Mutat 31: 279–283 [DOI] [PubMed] [Google Scholar]
- Kranendijk M, Struys EA, van Schaftingen E, Gibson KM, Kanhai WA, van der Knaap MS, Amiel J, Buist NR, Das AM, de Klerk JB, et al. 2010b. IDH2 mutations in patients with D-2-hydroxyglutaric aciduria. Science 330: 336. [DOI] [PubMed] [Google Scholar]
- Kranendijk M, Struys EA, Salomons GS, Van der Knaap MS, Jakobs C 2012. Progress in understanding 2-hydroxyglutaric acidurias. J Inherit Metab Dis 35: 571–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Shen SM, Zhao XY, Chen GQ 2012. Targeted genes and interacting proteins of hypoxia inducible factor-1. Int J Biochem Mol Biol 3: 165–178 [PMC free article] [PubMed] [Google Scholar]
- Losman JA, Looper R, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley G, Root D, Ebert BL, Kaelin WG Jr 2013. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 339: 1621–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. 2012. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483: 474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchman HA, Stechishin OD, Dang NH, Blough MD, Chesnelong C, Kelly JJ, Nguyen SA, Chan JA, Weljie AM, Cairncross JG, et al. 2012. An in vivo patient-derived model of endogenous IDH1-mutant glioma. Neuro Oncol 14: 184–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moslehi J, Minamishima YA, Shi J, Neuberg D, Charytan DM, Padera RF, Signoretti S, Liao R, Kaelin WG Jr 2010. Loss of hypoxia-inducible factor prolyl hydroxylase activity in cardiomyocytes phenocopies ischemic cardiomyopathy. Circulation 122: 1004–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD 2008. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol 26: 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nota B, Struys EA, Pop A, Jansen EE, Fernandez Ojeda MR, Kanhai WA, Kranendijk M, van Dooren SJ, Bevova MR, Sistermans EA, et al. 2013. Deficiency in SLC25A1, encoding the mitochondrial citrate carrier, causes combined D-2- and L-2-hydroxyglutaric aciduria. Am J Hum Genet 92: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, He Y, Bigner DD, Vogelstein B, Yan H 2011. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci 108: 3270–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach PJ, Depaoli-Roach AA, Hurley TD, Tagliabracci VS 2012. Glycogen and its metabolism: Some new developments and old themes. Biochem J 441: 763–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocquain J, Carbuccia N, Trouplin V, Raynaud S, Murati A, Nezri M, Tadrist Z, Olschwang S, Vey N, Birnbaum D, et al. 2010. Combined mutations of ASXL1, CBL, FLT3, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2 and WT1 genes in myelodysplastic syndromes and acute myeloid leukemias. BMC Cancer 10: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ 2007. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1: 113–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzem R, Vincent MF, Van Schaftingen E, Veiga-da-Cunha M 2007. L-2-hydroxyglutaric aciduria, a defect of metabolite repair. J Inherit Metab Dis 30: 681–689 [DOI] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, Harris IS, Holmes R, Wakeham A, Haight J, et al. 2012. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature 488: 656–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequist LV, Heist RS, Shaw AT, Fidias P, Rosovsky R, Temel JS, Lennes IT, Digumarthy S, Waltman BA, Bast E, et al. 2011. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol 22: 2616–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Kokubu A, Miyamoto M, Sasajima Y, Yamazaki N 2011. Mutant IDH1 confers an in vivo growth in a melanoma cell line with BRAF mutation. Am J Pathol 178: 1395–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss AW, Johnson MC 1996. The genetic basis of pediatric cardiovascular disease. Semin Perinatol 20: 564–576 [DOI] [PubMed] [Google Scholar]
- Struys EA, Verhoeven NM, Ten Brink HJ, Wickenhagen WV, Gibson KM, Jakobs C 2005. Kinetic characterization of human hydroxyacid-oxoacid transhydrogenase: Relevance to D-2-hydroxyglutaric and γ-hydroxybutyric acidurias. J Inherit Metab Dis 28: 921–930 [DOI] [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, et al. 2012. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22: 425–437 [DOI] [PubMed] [Google Scholar]
- Tonjes M, Barbus S, Park YJ, Wang W, Schlotter M, Lindroth AM, Pleier SV, Bai AH, Karra D, Piro RM, et al. 2013. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med 19: 901–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. 2012. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483: 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap MS, Jakobs C, Hoffmann GF, Duran M, Muntau AC, Schweitzer S, Kelley RI, Parrot-Roulaud F, Amiel J, De Lonlay P, et al. 1999. D-2-hydroxyglutaric aciduria: Further clinical delineation. J Inherit Metab Dis 22: 404–413 [DOI] [PubMed] [Google Scholar]
- Ward PS, Lu C, Cross JR, Abdel-Wahab O, Levine RL, Schwartz GK, Thompson CB 2013. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J Biol Chem 288: 3804–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. 2009. IDH1 and IDH2 mutations in gliomas. N Engl J Med 360: 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Breitkopf SB, Yang X, Asara JM 2012. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc 7: 872–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, et al. 2009. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science 324: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]