The H19 long noncoding RNA is repressed after birth, except in skeletal muscle. Dey et al. discover that H19 exon1 encodes miR-675-3p and miR-675-5p. The inhibition of myogenesis by H19 depletion is rescued by expression of miR-675-3p and miR-675-5p. The abnormal skeletal muscle regeneration in H19-deficient mice is rectified by reintroducing miR-675-3p and miR-675-5p. These miRs target the anti-differentiation Smad transcription factors and the DNA replication initiation factor Cdc6. In summary, H19 gives rise to microRNAs to promote skeletal muscle differentiation and regeneration.
Keywords: H19, miR-675, long noncoding RNA, skeletal muscle, differentiation, regeneration
Abstract
Regulated expression of the H19 long noncoding RNA gene has been well characterized as a paradigm for genomic imprinting, but the H19 RNA's biological function remains largely unclear. H19 is abundantly expressed maternally in embryonic tissues but is strongly repressed after birth, and significant transcription persists only in skeletal muscle. Thus, we examined the role of the H19 RNA in skeletal muscle differentiation and regeneration. Knockdown of H19 RNA in myoblast cells and H19 knockout mouse satellite cells decreases differentiation. H19 exon1 encodes two conserved microRNAs, miR-675-3p and miR-675-5p, both of which are induced during skeletal muscle differentiation. The inhibition of myogenesis by H19 depletion during myoblast differentiation is rescued by exogenous expression of miR-675-3p and miR-675-5p. H19-deficient mice display abnormal skeletal muscle regeneration after injury, which is rectified by reintroduction of miR-675-3p and miR-675-5p. miR-675-3p and miR-675-5p function by directly targeting and down-regulating the anti-differentiation Smad transcription factors critical for the bone morphogenetic protein (BMP) pathway and the DNA replication initiation factor Cdc6. Therefore, the H19 long noncoding RNA has a critical trans-regulatory function in skeletal muscle differentiation and regeneration that is mediated by the microRNAs encoded within H19.
H19 is one of the best known imprinted genes that was discovered from several genetic screens (Pachnis et al. 1984; Davis et al. 1987; Poirier et al. 1991). It was first isolated in a screen for genes that were up-regulated by α-fetoprotein in the liver (Pachnis et al. 1984). Concurrently, H19 was identified in the same genetic screen for myogenic differentiation that identified MyoD and was called MyoH (Davis et al. 1987). H19 was also found to be up-regulated in a screen during embryonic stem cell differentiation (Poirier et al. 1991). These findings indicate that H19 may have a role in cellular differentiation.
The H19 gene is located on chromosome 7 in mice and chromosome 11 in humans and is expressed only from the maternal allele in both species (Bartolomei et al. 1991; Zhang and Tycko 1992). Although the H19 gene is imprinted paternally, the H19 RNA itself does not participate in the imprinting mechanism (Brannan et al. 1990). The locus has been intensively analyzed as a model system for genomic imprinting; however, the biological functions of the H19 gene product are only now being elucidated. The H19 RNA does not contain any conserved ORFs between mice and humans, and evolutionarily conserved structure prediction studies suggest that H19 is a noncoding RNA (Brannan et al. 1990; Juan et al. 2000). It has recently been established that H19 exon1 encodes two conserved microRNAs: miR-675-3p and miR-675-5p (Cai and Cullen 2007).
H19 is dysregulated in many cancers, and various studies have suggested both tumorigenic and anti-tumorigenic roles for the H19 RNA (Moulton et al. 1994; Adriaenssens et al. 1998; Yoshimizu et al. 2008). Especially, loss of H19 expression is associated with Wilms' tumor and rhabdosarcoma (Chung et al. 1996; Lynch et al. 2002; Rump et al. 2005; Ecke et al. 2009), and, in fact, restoration of H19 expression can mitigate the tumor phenotypes (Hao et al. 1993). More recently, mouse genetic studies indicate that H19 RNA represses embryonic placental growth (Keniry et al. 2012) and trans-regulates a network of imprinted genes (Gabory et al. 2009) during fetal development.
H19 is a highly abundant transcript in almost all embryonic and neonatal tissues, especially in skeletal muscle, where expression is mediated by a downstream skeletal muscle-specific enhancer, and represents ∼1% of all cellular mRNA (Eun et al. 2013a,b). Shortly after birth, H19 RNA is dramatically down-regulated in all tissues. However, skeletal muscle is unusual in that H19 repression is only partial so that significant amounts of H19 RNA are detected in adult animals. Interestingly, H19 expression is induced by MyoD (Borensztein et al. 2013). However, we do not know whether H19 is important for skeletal muscle differentiation and, if so, what its mechanism of action is.
Here we establish a definitive role of H19 in skeletal muscle differentiation and regeneration. H19 is essential and required for proper muscle differentiation in vitro and muscle regeneration in vivo. Two conserved microRNAs, miR-675-3p and miR-675-5p, encoded by the exon1 of H19 are responsible for this biological function of H19. miR-675-3p represses the bone morphogenetic protein (BMP) pathway by targeting anti-differentiation Smad transcription factors, smad1 and Smad5, and miR-675-5p represses Cdc6, a DNA replication initiation factor. Thus, H19 long noncoding RNA has an essential function in skeletal muscle differentiation and regeneration that is mediated by the microRNAs embedded within it.
Results
H19 long noncoding RNA and its encoded microRNAs, miR-675-3p and miR-675-5p, are expressed in the skeletal muscles and up-regulated during myoblast differentiation and muscle regeneration
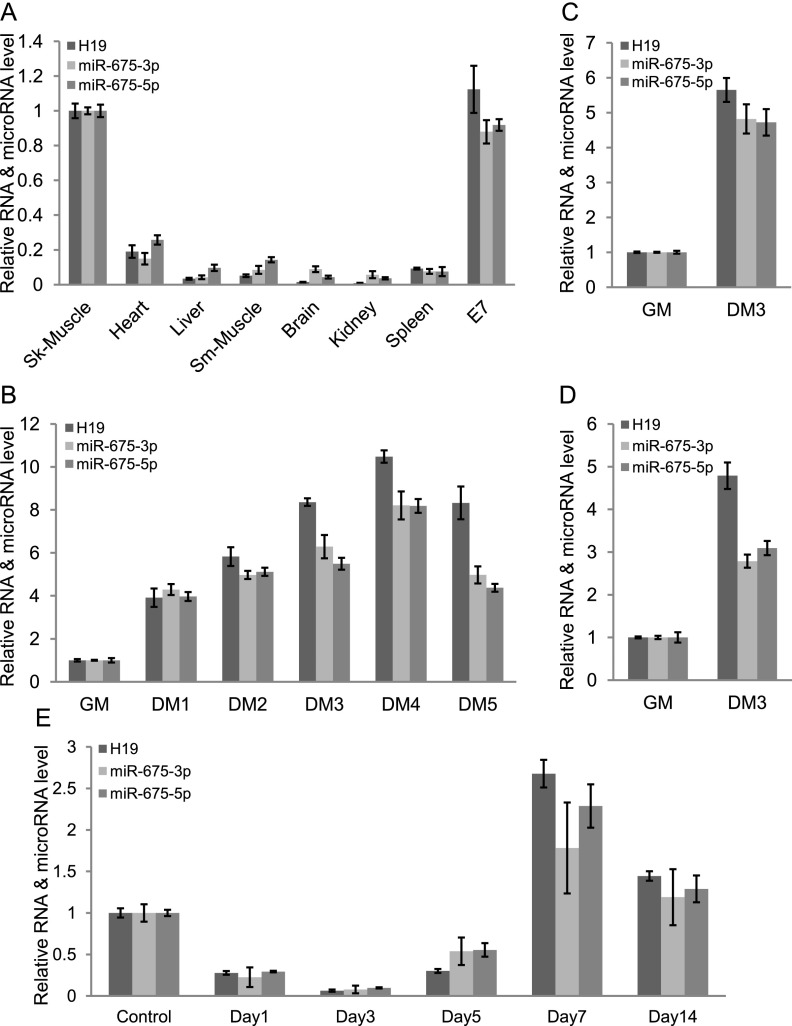
To gain an understanding of H19 function, we first looked at the expression pattern of H19 in a panel of adult mouse tissues and embryonic samples using quantitative RT–PCR (qRT–PCR). After expression in embryos, H19 RNA was dramatically down-regulated in most tissues except skeletal muscle (Fig. 1A; Supplemental Fig. 1A). The C2C12 myoblast cells serve as an excellent model system for studying muscle cell differentiation in vitro. Differentiation of myoblast cells into myocytes or myotubes can be accomplished by reducing serum supplements. H19 was gradually up-regulated during differentiation of C2C12 myoblast cells, with a slight decrease after differentiation day 4 (DM4), when myotubes were completely formed (Fig. 1B; Supplemental Fig. 1B). Similarly, H19 was also up-regulated during both mouse and human skeletal muscle satellite cell differentiation (Fig. 1C,D; Supplemental Fig. 1C).
Figure 1.
H19 long noncoding RNA and its encoded microRNAs, miR-675-3p and miR-675-5p, are expressed in the adult skeletal muscles and up-regulated during myoblast differentiation and muscle regeneration. (A) H19, miR-675-3p, and miR-675-5p are abundantly expressed in the adult skeletal muscles and whole embryos. (E7) Embryonic day 7. H19, miR-675-3p, and miR-675-5p are up-regulated during differentiation of C2C12 myoblast cells (B), mouse satellite cells (C), and human satellite cells (D). qRT–PCR of H19, miR-675-3p, and miR-675-5p after the indicated days in differentiation medium (DM) is shown. (GM) Growth medium. Each value is normalized to GAPDH (for H19) or U6sn RNA (for microRNAs) and expressed relative to skeletal muscle (A) or cells in GM (B–D). (E) Expression of H19, miR-675-3p, and miR-675-5p is down-regulated on days 1–3, is up-regulated on days 5–7, and decreases slightly on day 14 after TA muscle injury induced by injection of CTX. Mean ± standard error of the mean (SEM) of three biological replicates.
As differentiation is an important event during adult skeletal muscle regeneration, we investigated H19 RNA levels after cardiotoxin (CTX)-mediated injury of mouse skeletal muscle, tibialis anterior (TA). The injury activates quiescent muscle satellite cells, which re-enter the cell cycle and, after proliferation, differentiate to replenish the degenerated muscle fibers (Yan et al. 2003; Dey et al. 2012). H19 RNA decreased rapidly on days 1 and 3 after injury, during the phase of muscle degeneration, and then increased through days 5–7, during muscle regeneration (Fig. 1E). Similar to what we observed after terminal myoblast differentiation in vitro, H19 levels were slightly decreased at day 14, a stage when new myofibers were already formed (Fig. 1E). We then sought to determine the expression pattern of two conserved microRNAs, miR-675-3p and miR-675-5p, encoded by exon1 of H19. These microRNAs displayed an expression pattern similar to H19 in mouse tissues and during both myoblast differentiation in vitro and skeletal muscle regeneration in vivo (Fig. 1A–E; Supplemental Figs. 1A–C, 2A,B).
H19 promotes skeletal muscle differentiation, and its trans-regulatory function is mediated by miR-675-3p and miR-675-5p
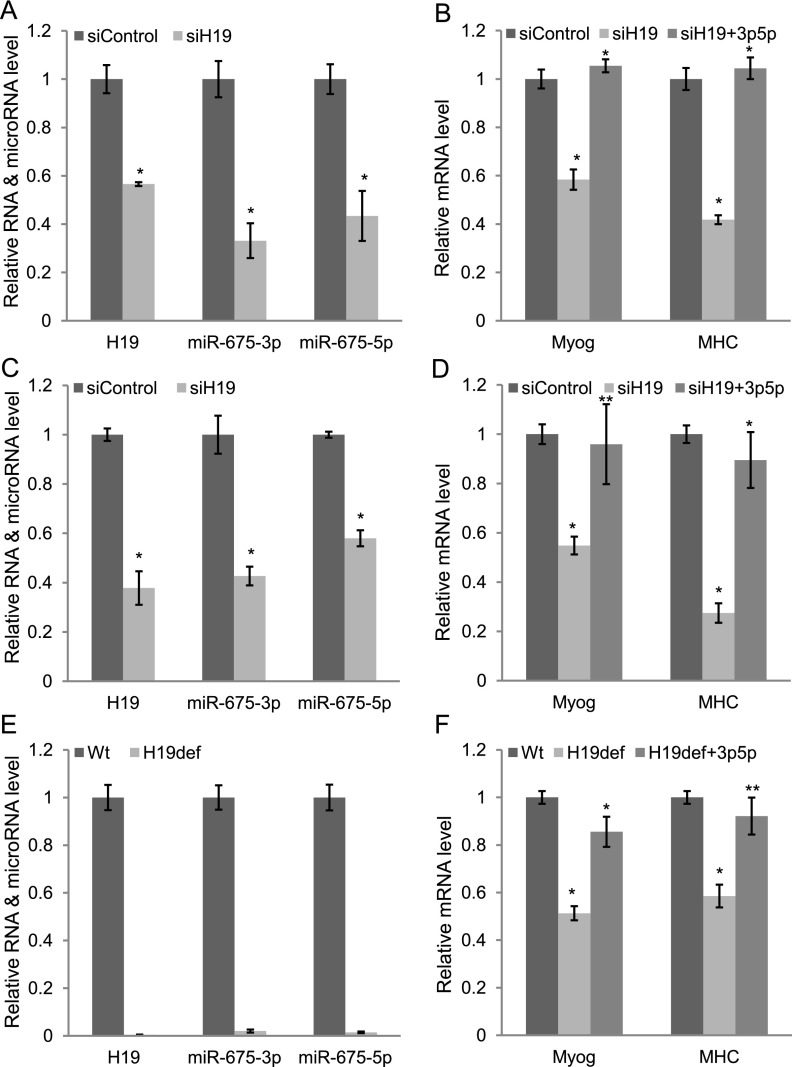
To determine the role of H19 long noncoding RNA in skeletal muscle differentiation, we knocked down H19 in C2C12 myoblast cells and human skeletal muscle satellite cells. Although the conventional view is that siRNA cannot target nuclear RNA, several nuclear long noncoding RNAs have been knocked down by many groups using siRNAs (Clemson et al. 2009; Matoba et al. 2011; Tripathi et al. 2013). Therefore, we transfected siRNAs specific to mouse H19 in C2C12 myoblast cells in growth medium (GM) and changed to differentiation medium (DM). The next day (DM1), siRNAs were again transfected, and the cells were harvested on DM3. H19 siRNAs reduced H19 RNA level by 43% (Fig. 2A; Supplemental Fig. 3) and decreased myogenesis, as shown by significant (41% and 58%, respectively) reduction of two established myogenic markers, myogenin and MHC (Dey et al. 2011), and morphology (Fig. 2B; Supplemental Fig. 4). Genome-wide microarray analyses revealed that a large number of differentiation-specific genes, including MyoD, myogenin, myosin family, Mef2c, IGF2, troponin, titin, and creatin kinase, were significantly down-regulated, whereas cell cycle and replication factors such as Cyclin E1, Cyclin E2, Cyclin D1, Cyclin D2, Brca1, Mcm4, Mcm6, Mcm7, and Cdc6 were up-regulated after knockdown of H19 (Supplemental Table 1). We performed similar experiments in human skeletal muscle satellite cells using siRNAs against human H19. The siRNAs against human H19 reduced H19 RNA level by 62% (Fig. 2C), and this knockdown was associated with decrease in differentiation, as measured by reduction of myogenin and MHC mRNA level by 41% and 72%, respectively (Fig. 2D). A similar result was obtained from mouse primary satellite cells (Supplemental Fig. 5A,B). Finally, a similar decrease in differentiation was seen in primary myoblast cells obtained from H19-deficient mice (ΔMuscle Enhancer/+, where the + allele is paternal and so is repressed by imprinting) (Fig. 2E,F). Altogether, these results strongly demonstrate that H19 is a critical factor for skeletal muscle differentiation.
Figure 2.
H19 promotes muscle differentiation in C2C12 cells, primary human satellite cells, and primary mouse myoblasts; its trans-regulatory function is mediated by miR-675-3p and miR-675-5p. We transfected siRNAs specific to mouse or human H19 in C2C12 myoblast cells or primary human satellite cells in GM and changed to DM. The next day (DM1), siRNAs were again transfected, and the cells were harvested for RNA analysis on DM3. Alternatively, we transfected cells with GL2 (22 bases from luciferase gene) as negative controls. Knockdown of H19 decreases H19, miR-675-3p, and miR-675-5p levels in C2C12 myoblast cells (A) and human satellite cells (C) and results in decreased expression of the differentiation markers myogenin and MHC mRNAs (B,D). (B,D) Exogenous miR-675-3p and miR-675-5p rescue the differentiation in both cell types. (E,F) Satellite cells derived from H19-deficient mice (H19ΔME/+) lack H19, miR-675-3p, and miR-675-5p (E) and show decreased expression of myogenin and MHC mRNAs at DM1 (F). (F) Exogenous miR-675-3p and miR-675-5p restore expression of the differentiation markers. The normalization for H19, myogenin, MHC mRNAs, and microRNAs was as described in Figure 1. Mean ± SEM of three biological replicates are shown. (*) P < 0.001; (**) P < 0.005.
H19 exon1 encodes two conserved microRNAs (miR-675-3p and miR-675-5p) (Cai and Cullen 2007), and knockdown in H19 RNA by siRNA treatment or genetic ablation results in the expected concomitant reduction in these microRNAs (Fig. 2A,C,E; Supplemental Fig. 3). To examine whether the prodifferentiation function of the H19 noncoding RNA is mediated by these microRNAs, we cotransfected mature miR-675-3p and miR-675-5p when H19 was down-regulated in the myoblast cells. Exogenous miR-675-3p and miR-675-5p together rescued the reduction in differentiation almost entirely in all three cell types examined (Fig. 2B,D,F). Thus, miR-675-3p and miR-675-5p appear to be the major contributors to the trans-regulatory prodifferentiation function of H19.
miR-675-3p and miR-675-5p promote muscle differentiation
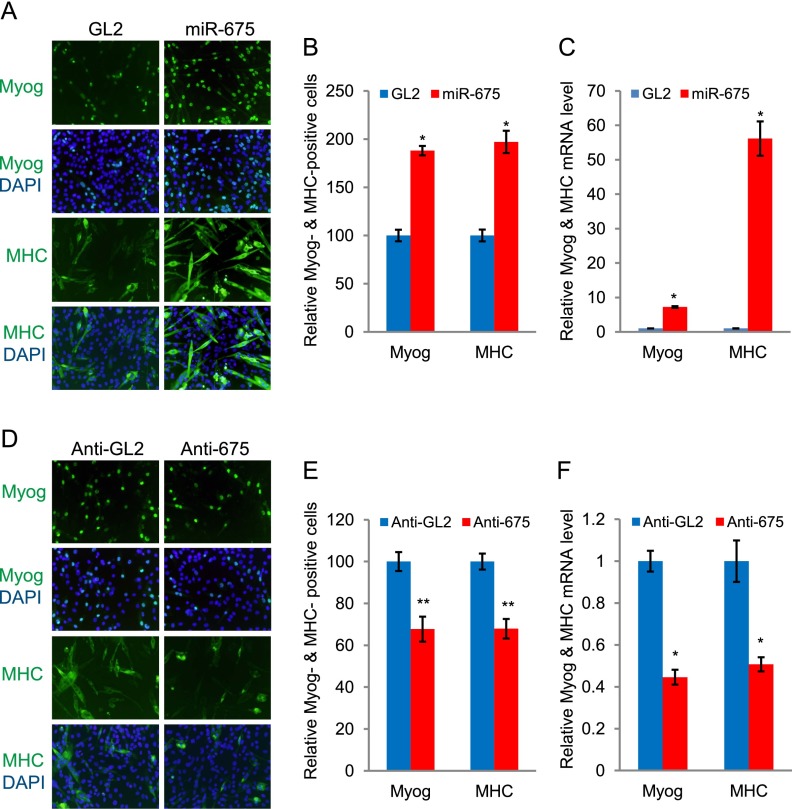
We next determined whether miR-675-3p and miR-675-5p can promote myogenic differentiation in C2C12 myoblast cells. C2C12 myoblast cells were transfected with RNA duplexes encoding miR-675-3p and miR-675-5p (miR-675) or GL2 control microRNA (22 bases from luciferase gene) (Dey et al. 2011) in GM and then transferred to DM. The exogenous miR-675-3p and miR-675-5p increased differentiation, as seen by the morphology and the number of myogenin- and MHC-positive cells (Fig. 3A,B; Supplemental Table 2). Myogenin and MHC mRNAs were up-regulated in miR-675-3p- and miR-675-5p-transfected myoblasts even when the cells were transfected and continuously grown in GM (Fig. 3C), suggesting that miR-675-3p and miR-675-5p are sufficient to induce myogenic differentiation. Consistent with this, ectopic expression of the fragment of H19 (mid) containing the pre-microRNA specifically increased myogenic differentiation, and this ability was lost when this fragment was mutated in the sequences of both microRNAs (mid-Mut) (Supplemental Fig. 6A,B).
Figure 3.
miR-675-3p and miR-675-5p promote muscle differentiation. (A) C2C12 myoblast cells in GM were transfected twice at 24-h intervals with GL2 control microRNA or miR-675-3p and miR-675-5p (miR-675). The cells were then transferred to DM and stained for myogenin at 32 h or MHC at 60 h. (Green) Myogenin or MHC; (blue) nuclei stained by DAPI. (B) Numbers of myogenin- and MHC-positive cells are presented relative to the GL2 control, which is set as 100%. Mean ± SEM of 10 random fields (see Supplemental Table 2 for details). (*) P < 0.001. (C) C2C12 myoblast cells were transfected as in A and kept in GM for an extra 24 h before harvesting to measure myogenin and MHC mRNA by qRT–PCR. Each value was normalized to GAPDH in the same sample and then again to the value in GL2-transfected cells. Mean ± SEM of three biological replicates. (*) P < 0.001. (D,E) 2′O-methyl antisense oligonucleotides against GL2 (anti-GL2) or miR-675-3p and miR-675-5p (anti-675) were transfected as in A, and the cells were stained for myogenin at 32 h or MHC at 60 h in DM. Data are presented as in B. (F) Measurement of myogenin and MHC mRNAs by qRT–PCR as in C. Mean ± SEM of three biological replicates are shown. (*) P < 0.001; (**) P < 0.005.
In a reciprocal experiment, we knocked down miR-675-3p and miR-675-5p levels in the C2C12 cells by transfecting 2′O-methyl antisense oligonucleotides against both of these microRNAs (anti-675) or a control oligonucleotide (anti-GL2) in GM and then transferring the cells to DM. We show later that both antisense oligonucleotides effectively neutralize the actions of the microRNAs on two specific targets (Supplemental Fig. 7A,B). Anti-675-transfected cells reduced differentiation, as measured by myogenin and MHC staining and their respective RNA levels (Fig. 3D–F). Therefore, miR-675-3p and miR-675-5p are prodifferentiation factors for C2C12 differentiation.
H19 mutant mice impair skeletal muscle regeneration that can be rescued by reintroduction of miR-675-3p and miR-675-5p
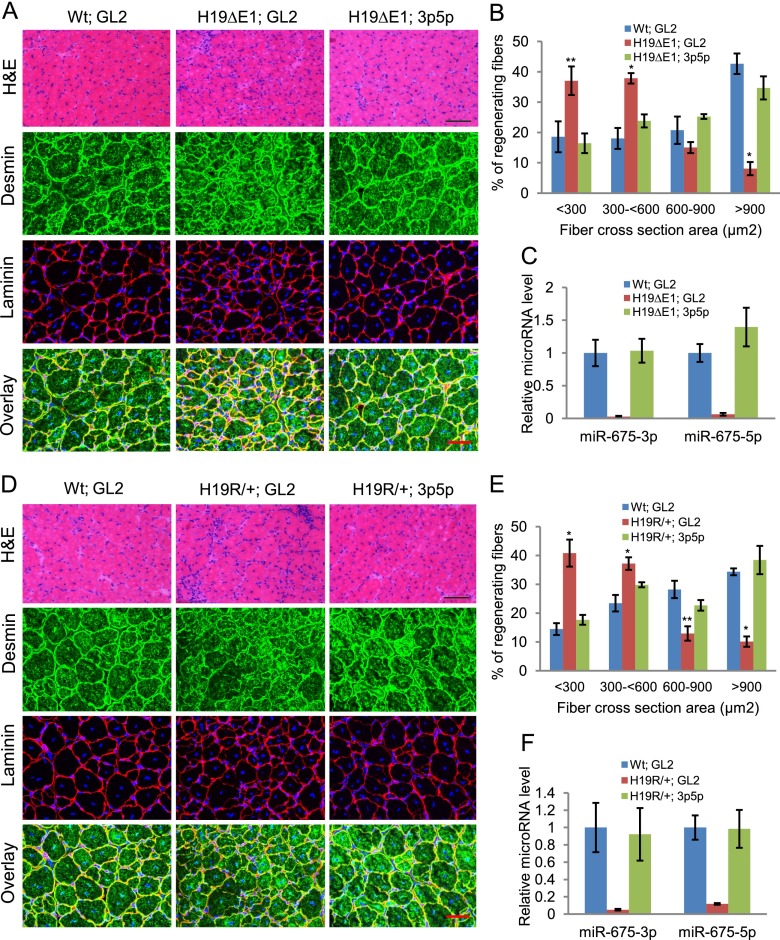
To test whether H19 plays a role in skeletal muscle regeneration in vivo, we employed a mouse model for skeletal muscle regeneration (Yan et al. 2003; Dey et al. 2012). Specifically, we examined mice carrying a 1-kb deletion of H19 (H19ΔExon1/+) that removes almost the entire exon1, including the region encoding the microRNAs, and reduces levels of even the partial H19 transcript by >100-fold (Srivastava et al. 2003; C Gebert and K Pfeifer, unpubl.). During the first 3 d following CTX injury, wild-type mouse TA muscles display extensive myofiber degeneration, but muscle regeneration is nearly complete on day 14 after injury (Dey et al. 2012). H19ΔExon1/+ mice showed impaired regeneration on day 14 after CTX injury, as marked by the persistence of inflammatory cells and smaller myofibers (Fig. 4A, first two panels, cf. the GL2 control duplex-injected wild type and H19ΔExon1/+). We immunostained the muscle sections for desmin and laminin. Desmin is an intermediate filament protein that is abundantly expressed in newly made muscle fibers during muscle regeneration (Kuisk et al. 1996), whereas laminin demarcates the muscle fiber boundary. The newly made muscle fibers were significantly smaller in H19ΔExon1/+ muscle as compared with the wild-type muscle (Fig. 4B). Thus, we conclude that lack of H19 significantly impairs the regeneration of adult skeletal muscle.
Figure 4.
H19 mutant mice show defects in muscle regeneration that can be rescued by exogenous miR-675-3p and miR-675-5p. (A) TA muscles in wild-type (Wt) or H19-deficient (H19ΔE1) littermates were injured by CTX injection and, after 3 d, were injected with miR-675-3p and miR-675-5p or with GL2 control microRNA in the contralateral leg. (Top panel) Representative H&E-stained images are from TA muscles harvested 14 d after CTX injury. Bar, 100 μm. (Bottom three panels) TA muscles from the same specimens were stained with desmin and laminin. Bar, 50 μm. Five mice were used in each group. (B) Cross-section areas of regenerating fibers of these samples were quantitated using Image J software. More than 200 individual fibers were counted in each group. (C) miR-675-3p and miR-675-5p levels were measured in these samples by qRT–PCR. (D–F) Data were obtained as in A–C from H19R/+ and control littermates. Bars: H&E, 100 μm; desmin and laminin, 50 μm. Mean ± SEM (n = 5). (*) P < 0.001; (**) P < 0.005. Note that the H19 mutations used in this study were specifically chosen because they do not disrupt imprinting of the adjoining Igf2 gene.
We next examined whether introduction of mature miR-675-3p and miR-675-5p can rescue the defects in regeneration in the H19-deficient muscles. Mature miR-675-3p and miR-675-5p were injected in the TA muscle of one leg, and GL2 control microRNA was injected in the contralateral leg as described (Ge et al. 2011; Eulalio et al. 2012). Injection of mature miR-675-3p and miR-675-5p in the TA muscle deficient in H19 restored miR-675-3p and miR-675-5p levels (Fig. 4C) and improved the regeneration at day 14, whereas control GL2-injected samples failed to regenerate completely (Fig. 4A,B).
We repeated these analyses in mice with an alternative mutation called H19R. The H19R chromosome carries an insertion of a transcriptional insulator that prevents interactions between the H19 promoter and the downstream skeletal muscle enhancer (ME) and thus results in a dramatic reduction of H19 specifically in muscle cells while leaving expression in endodermal cells unaffected (Yoon et al. 2007). As with H19ΔExon1/+ mice, H19R/+ mice showed defects in regeneration that was rescued by injection of miR-675-3p and miR-675-5p (Fig. 4D–F). These results confirm that H19 is required for normal muscle regeneration in vivo. Moreover, the cell type specificity of the mutation confirms that H19-dependent regeneration is cell-autonomous: The muscle cells themselves require H19.
Smad1, Smad5, and Cdc6 are the important targets for miR-675-3p and miR-675-5p
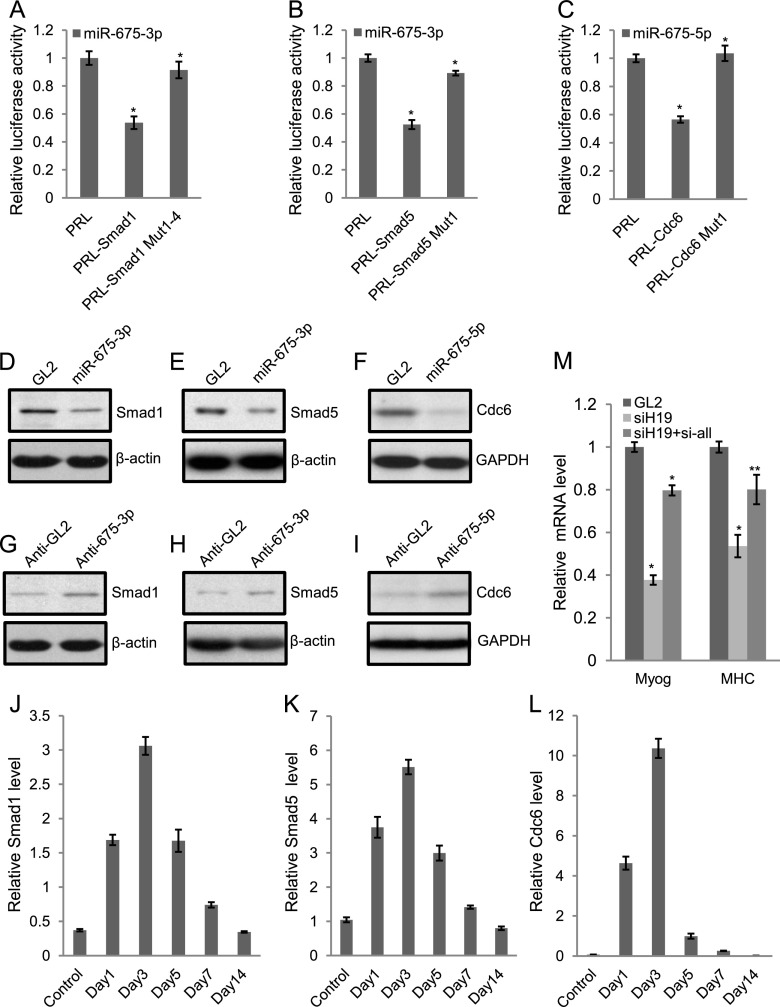
Because miR-675-3p and miR-675-5p promote skeletal muscle differentiation and regeneration, we anticipated that their direct targets would include anti-differentiation or proproliferation genes. Target prediction algorithms predicted a large number (>100) of targets for miR-675-3p and miR-675-5p (Supplemental Fig. 8; Supplemental Table 3; data not shown). Of the predicted targets, the transcription factors that mediate anti-differentiation effects of the BMP pathway, Smad1 and Smad5, and a DNA replication factor, Cdc6, attracted our attention because BMP pathway and DNA replication factors need to be down-regulated during myoblast differentiation (Dey et al. 2012; data not shown). To demonstrate whether these are the direct targets, the 3′ untranslated regions (UTRs) of Smad1, Smad5, and Cdc6 were separately fused to a luciferase reporter gene. Smad1 and Smad5 reporters were repressed by miR-675-3p by 47% and 48%, respectively (Fig. 5A,B). Similarly miR-675-5p repressed a luciferase reporter fused to the 3′ UTR of Cdc6 by 46% (Fig. 5C). Mutation of the microRNA target sites relieved the repression in all three cases (Fig. 5A–C). Not only were the luciferase reporters containing 3′ UTRs of Smad1 or Cdc6 repressed by miR-675-3p or miR-675-5p, respectively, but the repression was relieved by the 2′O-methyl inhibitor of miR-675-3p or miR-675-5p (Supplemental Fig. 7A,B). Consistent with Smad1, Smad5, and Cdc6 being direct targets of these microRNAs, all three proteins were down-regulated by transfection of C2C12 cells in GM with the cognate RNAs (Fig. 5D–F). In a reciprocal experiment, we inhibited the endogenous miR-675-3p or miR-675-5p using 2′O-methyl or LNA antisense inhibitor and observed increased levels of endogenous Smad1, Smad5, and Cdc6 proteins during C2C12 differentiation (Fig. 5G–I). These results are consistent with the hypothesis that miR-675-3p and miR-675-5p can repress Smad1, Smad5, and Cdc6 proteins and thus promote myogenic differentiation.
Figure 5.
Smad1, Smad5, and Cdc6 are important targets for miR-675-3p and miR-675-5p. (A–C) Cotransfection of miR-675-3p or miR-675-5p microRNA into U2OS osteosarcoma cells repressed Renilla (rr) luciferase activity when the rr reporter was fused to the target 3′ UTR. Mutations in the target sites in the 3′ UTR (Supplemental Fig. 7) relieved the repression. A firefly (pp) luciferase plasmid was cotransfected as a transfection control. The rr/pp was normalized to that for a control Renilla luciferase plasmid with a vector 3′ UTR (PRL) and expressed relative to the normalized rr/pp in cells transfected with the GL2 RNA control. Mean ± SEM of three individual experiments. (*) P < 0.001. (D–F) Transfection of miR-675-3p or miR-675-5p in C2C12 myoblast cells in GM decreased Smad1, Smad5, and Cdc6 protein levels as detected by Western blotting 2 d after the second transfection. GL2 served as a control oligonucleotide. β-Actin or GAPDH was used as a loading control. (G–I) 2′O-methyl oligonucleotide against miR-675-3p or LNA antisense oligonucleotide against miR-675-5p (anti-675-3p or anti--675-5p) increased endogenous Smad1, Smad5, and Cdc6 protein levels in C2C12 cells cultured in DM for 2 d. 2′O-methyl or LNA antisense GL2 (anti-GL2) served as a control oligonucleotide. β-Actin or GAPDH is used as a loading control. (J–L) As measured by qRT–PCR, miR-675-3p or miR-675-5p target genes display anti-correlation in their temporal expression pattern during regeneration of skeletal muscle after CTX injury. (M) C2C12 myoblast cells were transfected with GL2 control siRNA, H19 siRNA, or H19 siRNA combined with siRNAs specific to Smad1, Smad5, and Cdc6 once in GM and then on DM1, and cells were harvested on DM3. qRT–PCR was performed for myogenin and MHC. The normalization for myogenin and MHC mRNAs was as described in Figure 1. Mean ± SEM of three biological replicates are shown. (*) P < 0.001; (**) P < 0.005.
As shown in Figure 1, H19 or its encoded miR-675-3p and miR-675-5p are down-regulated on days 1–3 after CTX-induced injury but then increases steadily on days 5–14 as new myofibers are formed (Fig. 1E). Thus, we tested the mRNA level of targets of miR-675-3p and miR-675-5p, Smad1, Smad5, and Cdc6, in these samples during regeneration. The levels of these targets were anti-correlated to H19 and miR-675-3p and miR-675-5p, increasing on days 1–3 after injury and then decreasing steadily on days 5–14 after injury (Fig. 5J–L). This result is consistent with the notion that miR-675-3p and miR-675-5p repress Smad1, Smad5, and Cdc6 during regeneration.
We showed that H19 generates miR-675-3p and miR-675-5p to down-regulate Smad1, Smad5, and Cdc6 during differentiation. To test whether misregulation of these proteins is sufficient to explain the reduced differentiation due to H19 deficiency, we performed coknockdown of Smad1, Smad5, and Cdc6 with siH19 in C2C12 cells. The depletion of Smad1, Smad5, and Cdc6 in H19-depleted cells was sufficient to overcome the decrease in differentiation as measured by the levels of myogenin and MHC mRNAs (Fig. 5M). Thus, Smad1, Smad5, and Cdc6 are important targets for the prodifferentiation action of H19.
Discussions
Our findings reveal a new function of H19 in promoting skeletal muscle differentiation and regeneration. Interestingly, H19, named MyoH, was identified in the same genetic screen for myogenic differentiation that identified MyoD (Davis et al. 1987). H19 was also found to be up-regulated in a screen during embryonic stem cell differentiation (Poirier et al. 1991). Together, these findings suggest that H19 plays a role in cellular differentiation. Our result is consistent with the earlier findings that inactivation of H19 due to loss of heterozygosity or hypermethylation of maternal alleles is associated with Wilms' tumor and rhabdomyosarcoma (Chung et al. 1996; Lynch et al. 2002; Ecke et al. 2009). Rhabdomyosarcoma arises from defects in skeletal muscle differentiation, and treatment of DNA methylation inhibitor 5-AzaC-2′-deoxycytidine reactivated H19 in rhabdomyosarcoma cell lines and prevented rhabdomyosarcoma formation in mice (Chung et al. 1996; Lynch et al. 2002; Ecke et al. 2009). As miR-675-3p and miR-675-5p can induce muscle cell differentiation, we are curious whether introduction of these microRNAs is sufficient to differentiate rhabdomyosarcoma cell lines or prevent rhabdomyosarcoma formation in vivo. Conversely, ectopic expression of H19 transgene causes prenatal lethality (Brunkow and Tilghman 1991). However, other groups found that the H19 transgenic mice are viable, and the transgene regulates the imprinted gene network (Pfeifer et al. 1996; Gabory et al. 2009). Thus, it will be interesting to test whether ectopic expression of miR-675-5p and miR-675-3p produces phenotypes similar to H19 transgenic mice.
A genome-wide survey predicted that ∼100 long noncoding RNAs encode microRNAs (He et al. 2008). Our findings demonstrate how one could experimentally validate whether the encoded microRNAs mediate specific functions of specific long noncoding RNAs.
miR-675-3p and miR-675-5p promote muscle differentiation and regeneration by negatively regulating two important classes of targets: the BMP pathway transcription factors Smad1 and Smad5 and DNA replication initiation factor Cdc6. We showed earlier that miR-26a, another microRNA induced during muscle differentiation, promotes myogenesis by repressing signal transduction by other members of the BMP/TGF-β family (Dey et al. 2012). Recent studies identified Igf2 and/or Igf1r as targets of miR-675-3p/miR-675-5p in embryonic placental growth and maintenance of hemopoietic stem cell quiescence (Keniry et al. 2012; Venkatraman et al. 2013). However, Igf2 levels increase during differentiation (Ge et al. 2011), when the miR-675-3p and miR-675-5p increase (Fig. 1B), and we did not see any significant difference in the expression of Igf2 and Igf1r in the muscles from mice that were wild type, H19 mutant, or H19 mutant restored with miR-675-3p and miR-675-5p (data not shown). Interestingly, Igf2 is inhibited by miR-125b, whose expression decreases during muscle differentiation and regeneration (Ge et al. 2011). Thus, it is likely that during skeletal muscle differentiation, Igf2/Igf1r genes are regulated by miR-125b or some other unknown mechanisms, while the proproliferative Smad1, Smad5, and Cdc6 are the major targets of miR-675-3p and miR-675-5p during skeletal muscle differentiation and regeneration.
Other muscle differentiation-induced microRNAs inhibit other anti-differentiation factors: miR-206 inhibits Pax7 (Dey et al. 2011), HDAC4 (Chen et al. 2006), and Notch3 (Gagan et al. 2012), while miR-378 targets MyoR (Gagan et al. 2011). Similarly, we showed that three other microRNAs induced during muscle differentiation with promyogenic activity target cell cycle factors: miR-206 down-regulates DNA polymerase α (Kim et al. 2006), and miR-322 and miR-503 repress Cdc25a, the phosphatase that removes an inhibitory phosphate from CDK2 (Sarkar et al. 2010). These results indicate that the concerted actions of multiple microRNAs are important for muscle development by collectively down-regulating anti-differentiation and cell cycle factors. The redundancy of multiple prodifferentiation microRNAs is similar to the redundancy of myogenic transcription factors like MyoD and explains the normal skeletal muscle development following the knockout of miR-206 (Williams et al. 2009) or H19 (Leighton et al. 1995). This bears a striking parallel with the normal development seen after knockout of one myogenic transcription factor like MyoD (Rudnicki et al. 1992). The requirement of these factors during differentiation becomes more evident under stressed conditions such as skeletal muscle regeneration; e.g., miR-206 (Liu et al. 2012), MyoD (White et al. 2000), and now H19 in our study. As with the myogenic transcription factors, simultaneous knockout of multiple muscle differentiation-induced microRNAs will need to be tested to uncover the essential functions of microRNAs during skeletal muscle development. Consistent with the notion that the compensatory mechanisms are activated after chronic knockout of microRNA genes is the observation in Figure 2 that acute knockdown of H19 by siRNA impairs differentiation to almost the same extent as in the H19 knockout cells even though the former have more residual microRNAs.
A recent study reports that H19 is a molecular sponge for let-7 in a human embryonic kidney (HEK293) cell line (Kallen et al. 2013). The investigators suggest that H19 sponges let-7 in C2C12 cells to inhibit muscle differentiation. Our results are in stark contrast because we provide evidence from knockdown and genetic knockout of H19 that it is a prodifferentiation factor: in vitro in C2C12 cells and primary myoblasts in culture and in vivo in regenerating skeletal muscle. Moreover, we show that the H19-encoded microRNAs miR-675-5p and miR-675-3p can execute the prodifferentiation function of H19 in skeletal muscle lacking endogenous H19. A huge difference between the 90-fold induction of let-7 during C2C12 differentiation reported by Kallen et al. (2013) and the lack of any such induction of let-7 in our cells (Supplemental Table 4; Supplemental Fig. 9A–C) suggests that there is a significant difference between the C2C12 cell lines and the in vitro differentiation conditions, which may account for the contradictory roles attributed to H19. In this context, it is particularly important that the prodifferentiation function of H19 is confirmed in our experiments in primary myoblasts in vitro and in regenerating skeletal muscle in vivo. In addition, our studies provide important insight into how this critical trans-regulatory function of H19 in skeletal muscle differentiation and regeneration is mediated by the microRNAs embedded within.
Materials and methods
Cell culture
The C2C12 mouse myoblast cell line was supplied by American Type Culture Collection (ATCC) (Yaffe and Saxel 1977). Cells were cultured at subconfluent densities in GM, made up of DMEM supplemented with 10% heat-inactivated FCS and 1% penicillin/streptomycin. C2C12 myoblast cells were differentiated into myocytes or myotubes in DM (Andres and Walsh 1996), consisting of DMEM containing 2% heat-inactivated horse serum and 1% penicillin/streptomycin. U2OS cells were cultured in DMEM supplemented with 10% heat-inactivated FCS and 1% penicillin/streptomycin. Mouse primary myoblasts and differentiated myotubes were used as described (Dey et al. 2011). H19-deficient primary myoblast cells were obtained from a ME-deleted (H19ΔME/+) mouse line (Kaffer et al. 2001). ΔME removed a 10-kb region far downstream from H19 that is essential for H19 expression in skeletal muscle. Human satellite cells and H19-deficient primary myoblast cells were cultured as described (Bois and Grosveld 2003; Zhu et al. 2007).
Generation of constructs
Mouse Smad1 3′ UTR of 1.4 kb, Smad5 3′ UTR of 1.5, and Cdc6 3′ UTR of 1.4 kb were amplified by PCR from C2C12 myoblast genomic DNA or cDNA and cloned into pRL-CMV vector. Specific point mutations in Smad1, Smad5, and Cdc6 3′ UTRs cloned into pRL-CMV vector were created using a site-directed mutagenesis kit (Stratagene). Fragments of H19 (5′, bases 1–956; mid, bases 741–1407; 3′, bases 1296–2068) were subcloned to the pMSCV retroviral vector. Mid-Mut carried mutations in the sequences of miR-675-5p and miR-675-3p as follows: TGGTGCGGAAAGGGCCCACAGT was changed to CCACATTTCAAGGGCCCACAGT, and CTGTATGCCCTAACCGCTCAGT was changed to CCACATCGCCATACCGCTCAGT. Retrovirus was made in HEK293T cells cotransfected with virus packaging plasmids using a standard protocol.
Transfection of plasmids, siRNAs, mature microRNAs, and 2′O-methyl or LNA antisense microRNAs
We used Lipofectamine 2000 (Invitrogen) to transfect plasmid DNA and RNAiMAX (Invitrogen) to transfect siRNAs, microRNA mimics, and 2′O-methyl or LNA antisense microRNAs following the manufacturer's instructions. The following siRNA sequences were used: mouse siH19, GGACUGGAGACUAGGGUAAdTdT; human siH19, UAAGUCAUUUGCACUGG UUdTdT; GCUAGAGGAACCAGACCUdTdT; si-Smad1, GGGCGAUGAAGAAGAGAAAdTdT; si-Smad5, CCUGGGAUUGUUGUCAAAUGUUAAUdTdT. A pool of three siRNAs against mouse Cdc6 was purchased from Santa Cruz Biotechnology. We transfected 50 nM siRNAs specific to mouse or human H19 (and control siRNA) in C2C12 myoblast or human satellite cells in GM and changed to DM. The next day (DM1), siRNAs were again transfected, and the cells were harvested on DM3.
Isolation of total RNA and performance of qRT–PCR
Trizol reagent (Invitrogen) or RNeasy minikit (Qiagen) were used to extract total RNA from various cell lines and tissue samples following the manufacturer's instructions. Mouse tissue panels were purchased from Clontech Laboratories, Inc. cDNA synthesis for mRNA detection was carried out using SuperScript III first strand synthesis system for RT–PCR (Invitrogen). MicroRNA was detected using TaqMan microRNA assays (Applied Biosystems) or Ncode microRNA first strand cDNA synthesis kit (Invitrogen). qPCR for mRNA and microRNA detection was carried out in an ABI thermal cycler using SYBR Green according to the manufacturer's instructions. Quantification of amplicons was done using ABI 7300 software (Applied Biosystem).
Microarray analysis
si-H19 or si-control was transfected into C2C12 cells twice (once in GM and then on DM1), and cells were harvested on DM3. Total RNA was isolated, converted to cDNA probe, and hybridized to a mouse gene 1.0 ST array (Affymetrix). The signals for specific probes and fold change (si-H19/si-Control) were determined.
Luciferase assays
We transfected U2OS osteosarcoma cells with the desired microRNAs twice at 24-h intervals. We then transfected luciferase plasmids 6 h after the last transfection. pGL3 (Promega) was cotransfected as an internal control. Luciferase assays were performed with dual-luciferase reporter assay system (Promega) using a luminometer (Monolight 3020, BD Biosciences) or GlomaX microplate reader luminometer (Promega) at 32–48 h after plasmid transfection. We normalized Renilla luciferase values (rr) first to the cotransfected pGL3 control firefly Photinus pyralis luciferase values (pp), and then each rr/pp value in the microRNA-transfected samples was normalized with the rr/pp values obtained in control GL2-transfected samples.
Western blotting and antibodies
Cells were harvested, washed with 1× PBS, and lysed in NP40 lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 0.1% NP-40, 5 mM EDTA, 10% glycerol) with protease inhibitors cocktail (Sigma). Proteins were separated in SDS-PAGE, transferred, and immunoblotted with various antibodies. The antibodies used were anti-Smad1 (dilution 1:1500; Invitrogen), anti-Smad5 (dilution 1:500; Cell Signaling), anti-Cdc6 (dilution 1:500; Cell Signaling), anti- β-actin (dilution 1:3000; Santa Cruz Biotechnology), and anti-GAPDH (dilution 1:10000; Sigma).
Mouse strains
The following mouse strains were used : H19R/+ (Yoon et al. 2007), H19ΔE1 (Srivastava et al. 2003), H19ΔME/+ (Kaffer et al. 2001), and C57BL/6 (Harlan). The use of animals in all of the studies was done following protocols approved by the Animal Care and Use Committee (ACUC) of University of Virginia.
Skeletal muscle regeneration model and TA muscle injection
We injured mouse skeletal muscle by injecting CTX from Naja nigricollis (EMD Millipore) essentially following the procedure described earlier (Yan et al. 2003). Briefly, ∼10-wk-old male mice of the desired genotype were injected on TA muscles with 100 μL of 10 μM CTX. A high volume (100 μL) and pressure of injection and post-injection massage spread the injected material throughout the TA compartment. We did all of our analysis on the middle two-thirds of the TA muscle that was closest to the injection site. We optimized the concentration and amount of microRNA injections to obtain “restored” microRNA levels that were close to wild-type levels. Many other groups have successfully injected microRNA and various forms of microRNA inhibitors in the TA muscle (Ge et al. 2011; Yin et al. 2013) and cardiac muscles (Eulalio et al. 2012). Finally, we made stock microRNA and lipid complexes with modifications as described earlier (Ge et al. 2011; Eulalio et al. 2012). One-hundred microliters each of 100 μM miR-675-3p and miR-675-5p (Invitrogen) together with 100 μL of RNase-free water were mixed with 300 μL of RNAiMAX transfection reagent (Invitrogen) and incubated for 30 min at room temperature. For control, 200 μL of 100 μM GL2 (Invitrogen) and 100 μL of RNase-free water was similarly mixed with RNAiMAX transfection reagent and incubated for 30 min. Finally, 100 μL of microRNA complex was injected into the TA muscle of one leg, and the control microRNA complex was injected into the contralateral leg 3 d after the CTX injection. Mice were anesthetized and sacrificed by cervical dislocation to harvest muscle samples.
Immunocytochemistry and immunohistochemistry
Immunocytochemistry was carried out as described previously (Kim et al. 2006). Cells were grown on sterile glass coverslips and fixed with 2% formaldehyde in PBS for 15 min. Next, the cells were permeabilized with 0.2% Triton X-100 and 1% normal goat serum in ice-cold PBS for 5 min and blocked with 1% NGS in PBS twice for 15 min. Cells were incubated with primary antibody (myogenin [1:50; Santa Cruz Biotechnology] and MHC [1:400; Sigma], in 1% NGS) for 1 h. After washing twice with 1× PBS, FITC-conjugated anti-mouse IgG (dilution 1:500; Dako Cytomation) was incubated for another 1 h. Again, after two washes, nuclei were counterstained with DAPI, and the coverslips were mounted on a glass slide (H-1200, Vector Laboratories). Images were captured with an Olympus Hi-Mag microscope. Immunostaining of mouse tissue sections was carried out with slight modifications of the protocols as described by others: anti-laminin and anti-desmin (Liu et al. 2012). The primary antibodies used were rat anti-laminin (1:100; Millipore) and mouse anti-desmin (1:200; Dako). The secondary antibodies used were Alexa Fluor 594 goat anti-rat IgG1 (1:400; Invitrogen) and Alexa Fluor 488 goat anti-mouse IgG (1:400; Invitrogen). Images were taken using a Zeiss LSM-700 confocal microscope. H&E staining was carried out using a standard protocol of the University of Virginia histology core facility. Bright-field images were captured using an Olympus microscope.
Statistical analyses
Data are presented as the mean ± standard error of mean (SEM) of three or more biological replicates. Two-tailed Student's t-test was employed to determine P-values.
Acknowledgments
We thank Megan Sampley and Claudia Gebert for primary myoblast culture and genotyping, and members of the Dutta laboratory for many helpful discussions. This work was supported by R01 AR053948 and P01CA104106 to A.D., and partially supported by a post-doctoral fellowship from Heart and Stroke Foundation of Canada (HSFC) to B.K.D.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.234419.113.
References
- Adriaenssens E, Dumont L, Lottin S, Bolle D, Lepretre A, Delobelle A, Bouali F, Dugimont T, Coll J, Curgy JJ 1998. H19 overexpression in breast adenocarcinoma stromal cells is associated with tumor values and steroid receptor status but independent of p53 and Ki-67 expression. Am J Pathol 153: 1597–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres V, Walsh K 1996. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J Cell Biol 132: 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS, Zemel S, Tilghman SM 1991. Parental imprinting of the mouse H19 gene. Nature 351: 153–155 [DOI] [PubMed] [Google Scholar]
- Bois PR, Grosveld GC 2003. FKHR (FOXO1a) is required for myotube fusion of primary mouse myoblasts. EMBO J 22: 1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borensztein M, Monnier P, Court F, Louault Y, Ripoche MA, Tiret L, Yao Z, Tapscott SJ, Forne T, Montarras D, et al. 2013. Myod and H19-Igf2 locus interactions are required for diaphragm formation in the mouse. Development 140: 1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan CI, Dees EC, Ingram RS, Tilghman SM 1990. The product of the H19 gene may function as an RNA. Mol Cell Biol 10: 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkow ME, Tilghman SM 1991. Ectopic expression of the H19 gene in mice causes prenatal lethality. Genes Dev 5: 1092–1101 [DOI] [PubMed] [Google Scholar]
- Cai X, Cullen BR 2007. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13: 313–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ 2006. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38: 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WY, Yuan L, Feng L, Hensle T, Tycko B 1996. Chromosome 11p15.5 regional imprinting: Comparative analysis of KIP2 and H19 in human tissues and Wilms' tumors. Hum Mol Genet 5: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51: 987–1000 [DOI] [PubMed] [Google Scholar]
- Dey BK, Gagan J, Dutta A 2011. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol 31: 203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey BK, Gagan J, Yan Z, Dutta A 2012. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev 26: 2180–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecke I, Petry F, Rosenberger A, Tauber S, Monkemeyer S, Hess I, Dullin C, Kimmina S, Pirngruber J, Johnsen SA, et al. 2009. Antitumor effects of a combined 5-aza-2'deoxycytidine and valproic acid treatment on rhabdomyosarcoma and medulloblastoma in Ptch mutant mice. Cancer Res 69: 887–895 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, Giacca M 2012. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 492: 376–381 [DOI] [PubMed] [Google Scholar]
- Eun B, Sampley ML, Good AL, Gebert CM, Pfeifer K 2013a. Promoter cross-talk via a shared enhancer explains paternally biased expression of Nctc1 at the Igf2/H19/Nctc1 imprinted locus. Nucleic Acids Res 41: 817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun B, Sampley ML, Van Winkle MT, Good AL, Kachman MM, Pfeifer K 2013b. The Igf2/H19 muscle enhancer is an active transcriptional complex. Nucleic Acids Res 41: 8126–8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, Jammes H, Ainscough JF, Surani MA, Journot L, et al. 2009. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development 136: 3413–3421 [DOI] [PubMed] [Google Scholar]
- Gagan J, Dey BK, Layer R, Yan Z, Dutta A 2011. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. J Biol Chem 286: 19431–19438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagan J, Dey BK, Layer R, Yan Z, Dutta A 2012. Notch3 and Mef2c proteins are mutually antagonistic via Mkp1 protein and miR-1/206 microRNAs in differentiating myoblasts. J Biol Chem 287: 40360–40370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Sun Y, Chen J 2011. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J Cell Biol 192: 69–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B 1993. Tumour-suppressor activity of H19 RNA. Nature 365: 764–767 [DOI] [PubMed] [Google Scholar]
- He S, Su H, Liu C, Skogerbo G, He H, He D, Zhu X, Liu T, Zhao Y, Chen R 2008. MicroRNA-encoding long non-coding RNAs. BMC Genomics 9: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan V, Crain C, Wilson C 2000. Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Res 28: 1221–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffer CR, Grinberg A, Pfeifer K 2001. Regulatory mechanisms at the mouse Igf2/H19 locus. Mol Cell Biol 21: 8189–8196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, et al. 2013. The imprinted H19 LncRNA antagonizes Let-7 microRNAs. Mol Cell 52: 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W 2012. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol 14: 659–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A 2006. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol 174: 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuisk IR, Li H, Tran D, Capetanaki Y 1996. A single MEF2 site governs desmin transcription in both heart and skeletal muscle during mouse embryogenesis. Dev Biol 174: 1–13 [DOI] [PubMed] [Google Scholar]
- Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM 1995. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375: 34–39 [DOI] [PubMed] [Google Scholar]
- Liu N, Williams AH, Maxeiner JM, Bezprozvannaya S, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN 2012. MicroRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest 122: 2054–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch CA, Tycko B, Bestor TH, Walsh CP 2002. Reactivation of a silenced H19 gene in human rhabdomyosarcoma by demethylation of DNA but not by histone hyperacetylation. Mol Cancer 1: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Inoue K, Kohda T, Sugimoto M, Mizutani E, Ogonuki N, Nakamura T, Abe K, Nakano T, Ishino F, et al. 2011. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc Natl Acad Sci 108: 20621–20626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulton T, Crenshaw T, Hao Y, Moosikasuwan J, Lin N, Dembitzer F, Hensle T, Weiss L, McMorrow L, Loew T, et al. 1994. Epigenetic lesions at the H19 locus in Wilms' tumour patients. Nat Genet 7: 440–447 [DOI] [PubMed] [Google Scholar]
- Pachnis V, Belayew A, Tilghman SM 1984. Locus unlinked to α-fetoprotein under the control of the murine raf and Rif genes. Proc Natl Acad Sci 81: 5523–5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer K, Leighton PA, Tilghman SM 1996. The structural H19 gene is required for transgene imprinting. Proc Natl Acad Sci 93: 13876–13883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier F, Chan CT, Timmons PM, Robertson EJ, Evans MJ, Rigby PW 1991. The murine H19 gene is activated during embryonic stem cell differentiation in vitro and at the time of implantation in the developing embryo. Development 113: 1105–1114 [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Braun T, Hinuma S, Jaenisch R 1992. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell 71: 383–390 [DOI] [PubMed] [Google Scholar]
- Rump P, Zeegers MP, van Essen AJ 2005. Tumor risk in Beckwith-Wiedemann syndrome: A review and meta-analysis. Am J Med Genet A 136: 95–104 [DOI] [PubMed] [Google Scholar]
- Sarkar S, Dey BK, Dutta A 2010. miR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol Biol Cell 21: 2138–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Frolova E, Rottinghaus B, Boe SP, Grinberg A, Lee E, Love PE, Pfeifer K 2003. Imprint control element-mediated secondary methylation imprints at the Igf2/H19 locus. J Biol Chem 278: 5977–5983 [DOI] [PubMed] [Google Scholar]
- Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, et al. 2013. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet 9: e1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman A, He XC, Thorvaldsen JL, Sugimura R, Perry JM, Tao F, Zhao M, Christenson MK, Sanchez R, Yu JY, et al. 2013. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature 500: 345–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JD, Scaffidi A, Davies M, McGeachie J, Rudnicki MA, Grounds MD 2000. Myotube formation is delayed but not prevented in MyoD-deficient skeletal muscle: Studies in regenerating whole muscle grafts of adult mice. J Histochem Cytochem 48: 1531–1544 [DOI] [PubMed] [Google Scholar]
- Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN 2009. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 326: 1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe D, Saxel O 1977. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270: 725–727 [DOI] [PubMed] [Google Scholar]
- Yan Z, Choi S, Liu X, Zhang M, Schageman JJ, Lee SY, Hart R, Lin L, Thurmond FA, Williams RS 2003. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J Biol Chem 278: 8826–8836 [DOI] [PubMed] [Google Scholar]
- Yin H, Pasut A, Soleimani VD, Bentzinger CF, Antoun G, Thorn S, Seale P, Fernando P, van Ijcken W, Grosveld F, et al. 2013. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab 17: 210–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon YS, Jeong S, Rong Q, Park KY, Chung JH, Pfeifer K 2007. Analysis of the H19ICR insulator. Mol Cell Biol 27: 3499–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimizu T, Miroglio A, Ripoche MA, Gabory A, Vernucci M, Riccio A, Colnot S, Godard C, Terris B, Jammes H, et al. 2008. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci 105: 12417–12422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Tycko B 1992. Monoallelic expression of the human H19 gene. Nat Genet 1: 40–44 [DOI] [PubMed] [Google Scholar]
- Zhu CH, Mouly V, Cooper RN, Mamchaoui K, Bigot A, Shay JW, Di Santo JP, Butler-Browne GS, Wright WE 2007. Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: Consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell 6: 515–523 [DOI] [PubMed] [Google Scholar]