Abstract
Increased expression of HBEGF in ER negative breast tumors is correlated with enhanced metastasis to distant organ sites and more rapid disease recurrence upon removal of the primary tumor. Our previous work has demonstrated a paracrine loop between breast cancer cells and macrophages in which the tumor cells are capable of stimulating macrophages through the secretion of CSF-1 while the tumor associated macrophages (TAMs) in turn aid in tumor cell invasion by secreting EGF. To determine how the autocrine expression of EGFR ligands by carcinoma cells would affect this paracrine loop mechanism, and in particular whether tumor cell invasion depends on spatial ligand gradients generated by TAMs, we generated cell lines with increased HBEGF expression. We find that autocrine HBEGF expression enhanced in vivo intravasation and metastasis, and resulted in a novel phenomenon in which macrophages were no longer required for in vivo invasion of breast cancer cells. In vitro studies revealed that expression of HBEGF enhanced invadopodium formation, thus providing a mechanism for cell autonomous invasion. The increased invadopodium formation was directly dependent on EGFR signaling, as demonstrated by a rapid decrease in invadopodia upon inhibition of autocrine HBEGF/EGFR signaling as well as inhibition of signaling downstream of EGFR activation. HBEGF expression also resulted in enhanced invadopodium function via upregulation of MMP2 and MMP9 expression. We conclude that high levels of HBEGF expression can short-circuit the tumor cell/macrophage paracrine invasion loop, resulting in enhanced tumor invasion that is independent of macrophage signaling.
Keywords: HBEGF, breast cancer invasion, EGFR, metastasis
Introduction
Despite advances in screening and prevention, breast cancer is still associated with a high mortality rate as a result of the development of metastatic disease (1). In order to metastasize, tumor cells must invade the local tissue parenchyma and blood vessels to enter the blood stream and establish distant metastases (2). Breast cancer cell migration in primary tumors and metastatic potential depend not only on tumor cell characteristics but also on interactions with cells in the tumor microenvironment, such as tumor associated macrophages (TAMs) (3, 4). TAMs can contribute to angiogenesis, remodeling of the matrix, and secretion of chemotactic factors to stimulate tumor cell motility and intravasation (5, 6). We have previously shown a paracrine interaction between tumor cells and macrophages that involves epidermal growth factor (EGF) and colony-stimulating factor-1 (CSF-1) that drives macrophage-mediated invasion of tumor cells in transgenic mouse, rat and human mammary tumors (7-9). Inhibition of either EGF or CSF-1 signaling resulted in decreased invasion and intravasation, thus suggesting that the EGF/CSF-1 paracrine signaling loop plays a key role in the in vivo invasion response to either EGF or CSF-1 in this model (7).
An important question regarding this paracrine loop mechanism involves whether the enhanced invasion induced by epidermal growth factor receptor (EGFR) ligands secreted by macrophages involves oriented migration induced by gradients of the ligands. If invasion critically depends on cell migration directed by a spatial gradient, then autocrine expression of EGFR ligands by tumor cells themselves would be expected to reduce chemotactic response to gradients of ligands from other cells, resulting in reduced invasion, intravasation, and metastasis. Paradoxically, however, the EGFR ligand HBEGF (10-12) is preferentially expressed in tumors coexpressing the EGFR (13), and elevated HBEGF expression is correlated with higher histoprognostic grading, especially in triple negative tumors (13-15) as well as worse patient prognosis and lower overall survival rate (14, 15). Therefore, we have evaluated the effect of increased expression of HBEGF on invasion, intravasation, and metastasis of ER negative breast cancer cells. As predicted, increased expression of HBEGF resulted in reduced sensitivity to gradients of EGF as reflected by reduced chemotaxis and invasion in response to EGF. However, in spite of the reduced in vitro chemotactic sensitivity to applied gradients of EGF, in vivo we found that HBEGF expression increased invasion, intravasation, and metastasis. Remarkably, inhibition of CSF-1 receptor (CSF-1R) signaling and macrophage function did not inhibit in vivo invasion of HBEGF expressing cells, indicating that autocrine expression of HBEGF results in paracrine loop independent invasion.
Results
HBEGF expression increases EGFR activation but not proliferation
MDA-MB 231 cells and MTLn3 mammary adenocarcinoma cells expressing either human ErbB1 (MTLn3 ErbB1) or its corresponding empty vector control (MTLn3 pLXSN) were used as independent breast cancer cell lines to evaluate the effects of HBEGF expression (5, 9, 16). All lines expressed GFP to enable intravital imaging of cell motility. These cell lines were transfected with either the pBM IRES retroviral expression vector containing full-length HBEGF or with the empty vector as control. The HBEGF expressing transductants were designated as 231 HBEGF, MTLn3 pLXSN HBEGF and MTLn3 ErbB1 HBEGF, and the empty vector control transductants were designated 231 control, MTLn3 pLXSN control, and MTLn3 ErbB1 control. Evaluation of supernatants collected from the HBEGF transductants using an HBEGF ELISA showed a significant increase in HBEGF secreted into the medium compared to the empty vector control transductants (Fig. 1A; Supplemental Fig. S1A). We did not find induction of expression of other EGFR ligands at the mRNA level for MTLn3-ErbB1 cells (data not shown). For the MDA-MB 231 cells, although AREG and EREG mRNA levels were increased, levels of AREG and EREG secretion were not (Supplemental Fig. S2). Initial studies indicated that HBEGF expression in the MTLn3 pLXSN line had limited effect on in vivo metastasis or in vitro properties (Supplemental Fig. S1B – F). This finding was consistent with the clinical data indicating that HBEGF's impact was greatest in tumors with high levels of EGFR expression (13). Therefore, we focused on the MDA-MB 231 and MTLn3 ErbB1 transductant cell lines, which have higher levels of EGFR.
Figure 1. Validation of HBEGF expression and EGFR activation.
A, HBEGF concentrations in supernatants collected from empty vector control transductants (control) or HBEGF transductants (HBEGF) as measured by an HBEGF ELISA. N = 3, data are means and SEM; *** p < 0.001 by t-test. B, representative cropped bands from western blots of total EGFR, phospho-Tyr, and total and phospho-ERK expression. Beta-tubulin was used as a loading control. C, quantitation of relative EGFR and ERK phosphorylation normalized to total EGFR or ERK. D, representative cropped bands from western blots of total and phospho-ERK expression in HBEGF expressing transductants treated with 2ug/mL CRM197 for 15 minutes. Beta-tubulin was used as a loading control. E, quantitation of relative ERK phosphorylation normalized to total ERK. Data are means and SEM; *p < 0.05, ***p < 0.001 by z-test.
To determine the effects of HBEGF expression on EGFR activation, Western blots of whole cell lysates was performed and showed an increase in tyrosine phosphorylation of the EGFR and ERK in the HBEGF transductants (Figs. 1B-C). Inhibition of autocrine HBEGF/EGFR signaling was achieved using CRM197, a specific HBEGF inhibitor (17), which resulted in a significant decrease in ERK phosphorylation in the HBEGF expressing transductants (Figs. 1D-E). To evaluate the effect of HBEGF expression on in vitro growth rate, MTLn3 and 231 transductant cell lines were grown in low serum (0.5% FBS) and cell numbers were determined by direct cell counting. No significant changes in growth rate in vitro were seen for the HBEGF transductants relative to the empty vector controls (doubling times were 15.7 ± 0.7 vs 15.6 ± 0.7 hours for the MTLn3 lines and 23.2 ± 1.3 vs 22.5 ± 0.3 hours for MDA-MB 231 lines). Thus expression of HBEGF leads to increased activation of the EGFR with no change in proliferation rate.
HBEGF expression increases intravasation and metastasis with no effect on primary tumor growth
Empty vector control and HBEGF transductants were orthotopically injected into the mammary fat pads of SCID/NCr mice, and in vivo tumor properties were monitored. No significant differences in the average growth rate or tumor volume were observed (Fig. 2A). Thus, consistent with the in vitro data, increased HBEGF expression does not affect tumor growth rate in MTLn3 or MDA-MB 231 cells.
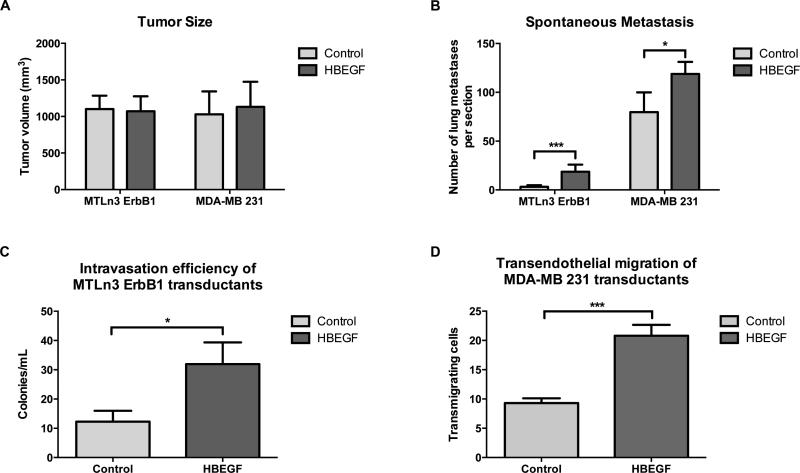
Figure 2. HBEGF expression enhances intravasation and metastasis with no effect on primary tumor growth.
HBEGF transductants (HBEGF) or empty vector controls (Control) were injected into the mammary fat pads of SCID/NCr mice, and at 4 weeks (for MTLn3) and 14 weeks (for MDA-MB 231) primary tumor size, metastasis and intravasation were measured. A, primary tumor size B, spontaneous lung metastasis. C, intravasation efficiency of MTLn3 tumor cells in vivo. D, in vitro transendothelial migration (iTEM) of MDA-MB 231 transductant cell lines. Data are means and SEM; *p < 0.05, ***p < 0.001 by t-test.
We then evaluated the effect of HBEGF expression on spontaneous metastatic potential by quantifying metastatic foci in H&E stained lung sections from tumor-bearing animals. Surprisingly, mice bearing HBEGF transductant tumors generated significantly more lung metastases than mice carrying empty vector control tumors (Fig. 2B). To test whether the increased spontaneous metastasis could be due to increased intravasation, we evaluated circulating tumor cells in the MTLn3 transductants (Fig. 2C). We found a significant increase in intravasation in the HBEGF transductants. Because the intravasation frequency of the 231 transductants was extremely low in this assay, we utilized an in vitro intravasation transendothelial migration assay to evaluate intravasation efficiency in the 231 transductants (Fig. 2D) and found enhanced intravasation efficiency of the 231 transductants as well.
HBEGF expression enhances in vitro and in vivo invasion in a cell autonomous fashion
The increased intravasation with HBEGF transductants suggested that HBEGF could increase invasion capability or motility. To investigate the effect of increased expression of HBEGF on invasion in vivo, the in vivo invasion assay was performed (18). Microneedles containing Matrigel were placed in primary mammary tumors to collect invasive cells. In the absence of an added chemoattractant, HBEGF transductants displayed a significantly greater basal in vivo invasion response compared to the empty vector control transductants, indicating that overexpression of HBEGF increases the basal invasiveness of carcinoma cells (Fig. 3A). In addition, the presence of EGF in the needle did not induce a significant increase over basal invasion in the HBEGF transductants. Using multiphoton microscopy and intravital imaging (IVI) of primary tumors, in vivo tumor cell motility was also significantly enhanced in the HBEGF transductants compared to the empty vector control transductants (Fig. 3B; Supplemental movies 1-4).
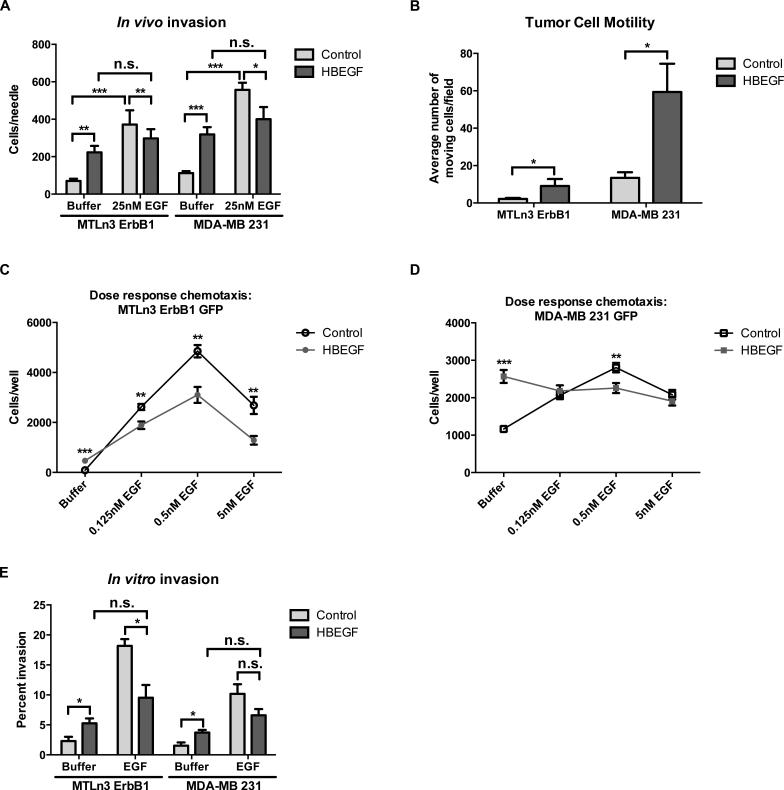
Figure 3. HBEGF expression increases basal motility and invasion.
A, in vivo invasion of MTLn3 ErbB1 or MDA-MB 231 tranductant primary tumors in response to buffer or 25nM EGF. B, intravital motility (IVI) using multiphoton imaging of GFP-labeled tumor cells. Total cell motility was quantified per 100um z-stack using 5um steps for 30 minute intervals. N = 5 – 10 mice per transductant cell line, data are means and SEM; *p < 0.05 by Mann-Whitney analysis. C,D, dose-response for EGF-stimulated chemotaxis for MTLn3 ErbB1 (C) and MDA-MB 231 (D) transductant cell lines. E, invasion in vitro in response to buffer or 25nM EGF. Samples were fixed and stained as described in the Materials and Methods and invasion was quantitated as the percent area of the filter covered by invading cells. Data are means and SEM; n.s., not significant; *p < 0.05, **p < 0.01, ***p < 0.001 by t-test.
We then tested whether these in vivo changes were reflected in the in vitro motility and invasion properties of the HBEGF transductants. A microchemotaxis chamber migration assay was used to measure basal motility and chemotaxis. The HBEGF transductants demonstrated stronger basal motility, but their chemotactic responses at all concentrations of EGF were diminished when compared with the empty vector control transductants (Figs. 3C and 3D). To examine the effects of HBEGF on invasive capability, we used an in vitro invasion assay in which cells were monitored for their ability to cross a Matrigel-coated transwell in response to EGF. As in the chemotaxis assays, the HBEGF transductants displayed enhanced basal invasion, while EGF-induced invasion was reduced or similar in comparison to the empty vector control lines (Fig. 3E). Thus both in vitro and in vivo, HBEGF expression enhances basal motility and invasion while reducing the relative enhancement induced by EGF.
Previous studies have shown that the metastatic potential of carcinoma cells is affected by their interactions with other cell types present in the tumor microenvironment, such as TAMs (8, 9). In particular, an EGF/CSF-1 paracrine loop between carcinoma cells and TAMs has been shown to drive invasion and metastasis in rat, mouse, and human breast cancer models (7-9). However, our in vitro motility and invasion studies described above were performed in the absence of macrophages, and suggested that the HBEGF induced invasive properties might be macrophage independent. We therefore tested the dependence of the in vivo invasion properties of the HBEGF transductants on the paracrine loop and macrophages. To evaluate the role of the CSF-1R, the in vivo invasion assay was conducted in the presence and absence of a CSF-1R inhibitor, JnJ. In vivo invasion of the MTLn3 control transductants relied upon CSF-1/CSF-1R signaling as expected, with the presence of 1uM JnJ in the microneedles inhibiting invasion (Fig. 4A). Remarkably, in vivo invasion of the MTLn3 HBEGF transductants was independent of CSF-1/CSF-1R signaling, since the presence of 1uM JnJ in the microneedles did not suppress invasion (Fig. 4B). Because basal invasion was enhanced in the HBEGF transductants and was not affected by inhibition of the CSF-1R, this suggested that in vivo invasion was no longer reliant on macrophage function. To test this hypothesis, in vivo macrophage function was inhibited using clodronate liposome pre-treatment. Effective inhibition of macrophage function in vivo was confirmed by demonstrating the loss of Texas-Red dextran phagocytosis (Fig. 4C) (19-24). In vivo invasion of the MTLn3 HBEGF transductants was found to be independent of macrophage function (Fig. 4D). Next we tested whether these in vivo changes were recapitulated in vitro by utilizing the in vitro 3D invasion assay. The HBEGF expressing transductants demonstrated enhanced basal tumor cell invasion, but the presence of macrophages did not further stimulate invasion when compared with the empty vector control transductants (Fig. 4E). Thus, we conclude that the enhanced invasion in vivo and in vitro is not dependent upon macrophage function.
Figure 4. Invasion in vivo no longer requires the CSF-1/CSF-1R paracrine loop upon HBEGF expression.
A,B, MTLn3 ErbB1 control (A) and HBEGF (B) primary tumors were tested for their ability to invade in vivo upon inhibition of the paracrine loop of invasion by using 1uM JnJ, a CSF-1R inhibitor. C, PBS-containing control or clodronate-containing liposomes were injected i.v. 48 h and 24 h prior to analysis into the tail veins of animals bearing MTLn3 ErbB1 HBEGF tumors. Four hours prior to analysis, 70kDa Texas Red dextran was injected intravenously, and the uptake of dextran by macrophages in the spleen and tumor was assessed using multiphoton imaging. Representative images of the spleen (left columns) and primary tumor (right columns) are shown. Magenta, collagen fibers; red, macrophages; green, tumor cells. Scale bar, 50 um. D, Needles containing buffer or 25nM EGF were inserted into the primary tumors of animals that were treated with PBS-containing control liposomes (vehicle) or clodronate-containing liposomes (clodronate) and the number of cells invading into the needle was determined. E, 3D in vitro invasion of MTLn3 ErbB1 and MDA-MB 231 transductant cell lines in the absence and presence of BAC macrophages measured as the fraction of carcinoma cells invading ≥ 20um into collagen gel. Data are means and SEM; **p < 0.01, ***p < 0.001 by t- test.
HBEGF expression stimulates invadopodium formation through activation of EGFR
One mechanism by which increased HBEGF expression could enhance invasion is via the formation of invadopodia, actin-rich structures capable of degrading ECM barriers (25). To evaluate the effect of HBEGF expression on invadopodium formation and matrix degradation, an invadopodium degradation assay was performed (26, 27). Cells were plated on Alexa Fluor 405-coupled gelatin, fixed and stained for total cortactin and Tks5. Invadopodia were identified as punctate structures showing cortactin and Tks5 co-localization. Expression of HBEGF resulted in increases in the total number of invadopodia (Figs. 5A, B, C), degradation area (Figs. 5A, B, D), and activation of cortactin (measured as relative level of cortactin phosphorylation in the entire cell) (Fig. 5E) (26). Induction of invasion, invadopodia and matrix degradation by autocrine HBEGF was confirmed using a second human triple negative cell line, BT549 (Supplemental Fig. S3).
Figure 5. HBEGF expression increases basal invadopodium formation and matrix degradation.
Representative images of invadopodia and matrix degradation for MTLn3 ErbB1 (A) and MDA-MB 231 (B) transductants. Invadopodia are defined as areas of cortactin and Tks5 co-localized puncta, examples of which are indicated by the red arrows. Scale bar, 25um. C, quantitation of the total number of invadopodia per cell for MTLn3 and MDA-MB 231 transductants. D, quantitation of total degradation area per field of view for MTLn3 (16 hours) and MDA-MB 231 (4 hours) transductants. All values were normalized to the corresponding control. E, cortactin activation measured as relative level of phosphorylation as described in Materials and Methods. F, quantitation of total number of invadopodia per cell after treatment with DMSO or 1uM Iressa for 15 minutes. G, quantitation of total number of invadopodia per cell after treatment with vehicle or 2ug/mL CRM197 for 15 minutes. H, quantitation of total number of invadopodia per cell after treatment with DMSO or 20uM SRCI1 for the duration of the invadopodium formation and matrix degradation assay (16 hours for MTLn3s and 4 hours for MDA-MB 231s). I, quantitation of total degradation area per field of view for HBEGF expressing transductants after treatment with DMSO or 20uM SrcI1. Three independent experiments were performed for each condition (N = 3), and for each independent experiment, eight to ten random fields of view were imaged for further data analysis. Data are means and SEM; *p < 0.05, **p < 0.01, ***p < 0.001 by t- test.
To test whether EGFR activity was directly regulating invadopodium formation, MTLn3 ErbB1 and MDA-MB 231 transductants were treated with 1uM Iressa for 15 minutes and fixed and stained immediately after treatment. This relatively brief inhibition of EGFR signaling resulted in a significant reduction in the total number of invadopodia (Fig. 5F). In addition, a significant decrease in the total number of invadopodia in the HBEGF expressing transductants was observed upon the inhibition of autocrine HBEGF/EGFR signaling by treating the cell lines with 2ug/mL CRM197 for 15 minutes. (Fig. 5G). Such a rapid response to EGFR or HBEGF inhibition indicates that autocrine HBEGF/EGFR signaling is directly leading to the increased invadopodium numbers observed in the HBEGF expressors (28).
A number of kinases are capable of phosphorylating cortactin, including Src (29). More recently, Mader and colleagues established that Src activation, which can occur downstream of EGFR activation, is essential for the activation and functional maturation of invadopodia in breast carcinoma cell invasion (28). To investigate whether Src activity was important for HBEGF-induced invadopodium function, Src activity was inhibited in HBEGF expressing MTLn3 ErbB1 and MDA-MB 231 transductants by treatment with 20uM SrcI1, a Src inhibitor. The significant decreases observed in invadopodium formation and matrix degradation in the MTLn3 ErbB1 and MDA-MB 231 cells expressing HBEGF demonstrate a requirement for activation of Src in the stimulation of invadopodia by HBEGF expression (Figs. 5H, I).
Expression of HBEGF enhances invadopodium function and tumor cell invasion by promoting expression of MMP2 and MMP9
In addition to stimulating invadopodium formation and activation, autocrine HBEGF could be enhancing invadopodium function and breast carcinoma cell invasion through the upregulation of matrix metalloprotease (MMP) production, especially MMP2, MMP9 and MMP14 (also known as membrane type-1 MMP [MT1-MMP]) (25, 30, 31). Previous studies conducted in models of ovarian and prostate carcinoma have shown that enhanced HBEGF expression can induce MMP2 and MMP9 expression (32, 33). Quantitative PCR (qPCR) was performed to evaluate the effect of HBEGF expression on MMP2, MMP9 and MMP14 expression, and a significant increase in MMP2 and MMP9 expression at the mRNA level was present in the HBEGF expressing transductants; MMP14 expression was unchanged with HBEGF expression (Fig. 6A). The increase in MMP2 and MMP9 expression was dependent upon the ERK signaling pathway; treatment with a MEK inhibitor resulted in significant decreases in both MMP2 and MMP9 expression (Fig. 6B).
Figure 6. HBEGF expression enhances invadopodium function and carcinoma cell invasion via upregulation of MMP2 and MMP9 expression.
A, relative fold change in MMP2, MMP9 and MMP14 mRNA expression in MTLn3 ErbB1 (left) and MDA-MB 231 (right) transductants as determined by qPCR. B, relative fold change in MMP2 and MMP9 mRNA expression in HBEGF expressing MTLn3 ErbB1 (left) and MDA-MB 231 (right) transductants after treatment with 10uM MEK inhibitor (MEKi) overnight as determined by qPCR. GAPDH was used as a housekeeping gene, and all values were normalized to the corresponding control. Data are means and SEM; **p < 0.01, ***p < 0.001 by z-test. C, invasion in vitro in response to DMSO or 0.5uM BiPS. D, quantitation of total degradation area per field of view for HBEGF expressing transductants after treatment with 0.5uM BiPS. All values were normalized to the corresponding control. Data are means and SEM; *p < 0.05, ***p < 0.001 by t-test.
To test if MMP2 and MMP9 were important in invadopodium function, HBEGF expressing MTLn3 ErbB1 and MDA-MB 231 transductants were treated with 0.5uM BiPS, a dual MMP2/MMP9 inhibitor. Inhibition of MMP2 and MMP9 activity in the HBEGF expressing transductants resulted in a significant reduction in the amount of invasion in vitro (Fig. 6C). Similarly, a significant decrease in matrix degradation resulted when MTLn3 ErbB1 and MDA-MB 231 cells expressing HBEGF were treated with 0.5uM BiPS (Fig. 6D). These results indicate that HBEGF stimulation of invadopodium function and tumor cell invasion is dependent upon increased MMP2 and MMP9 expression.
Discussion
These results provide novel insights into the mechanisms by which autocrine HBEGF expression can contribute to breast cancer malignancy using in vivo models. We originally hypothesized that autocrine expression would reduce chemotactic responses and therefore inhibit paracrine loop-induced invasion and metastasis. We generated cell lines with increased HBEGF expression and confirmed that autocrine expression of HBEGF by carcinoma cells inhibited their chemotactic responses to EGF. However, in vivo invasion, intravasation and metastasis were enhanced in the HBEGF expressing lines. This led to the insight that HBEGF expression in tumor cells abrogated the need for macrophages and the EGF/CSF-1 paracrine loop for invasion in vivo. In vitro studies revealed that HBEGF expression increased both basal cell motility and invadopodium production, providing a mechanism for cell autonomous invasion. The enhanced invasion and invadopodium production were dependent upon Src and MMP2/MMP9 activity.
Previous studies have demonstrated that the autocrine expression of EGF in HMEC can lead to constant activation of the EGFR, which can drive in vitro cellular motility but have no effect of proliferation rates even though the EGFR gets downregulated as a consequence of receptor activation (34). In this work we have extended these studies to evaluate the effect of autocrine HBEGF expression on tumor cell invasion, intravasation, and metastasis. We find persistent EGFR activation as indicated by increased EGFR tyrosine phosphorylation and ERK activation. The basal motility and invasion of cells is increased but EGF-induced chemotaxis and invasion in vitro are reduced, consistent with responses to spatial gradients of EGFR ligands being reduced due to high local basal levels of receptor occupancy.
Surprisingly, although primary tumor growth was not affected and EGF induced chemotaxis and invasion were reduced, intravasation and metastasis were enhanced by increased HBEGF expression. This raises the possibility that under some conditions spatial gradients of EGFR ligands are not critical for tumor cell invasion and metastasis. Indeed our in vivo invasion measurements revealed an increased basal level of invasion for HBEGF expressing lines compared to empty vector controls but reduced invasion in response to EGF gradients. In addition, we found that the paracrine loop between macrophages and tumor cells was not enhancing invasion under these conditions: inhibition of the CSF-1R or overall macrophage function had no effect on invasion. Similarly, in the in vitro 3D invasion assay, the basal level of invasion of the HBEGF expressing lines was increased and addition of macrophages had no effect. We interpret this to indicate that autocrine expression of HBEGF can substitute for macrophage production of EGFR ligands.
Our in vitro invasion studies indicate that the increased invasion that we observe upon the expression of HBEGF is due to increased production of invadopodia, which are specialized actin-containing structures used by carcinoma cells to degrade and invade through ECM during metastasis (25). However, work by Hayes et al. showed that the exogenous addition of recombinant HBEGF reduced invadopodium formation and matrix degradation in MDA-MB 231 cells (35). Autocrine stimulation can differ from exogenous stimulation in a number of ways. EGFR ligands are initially generated as type I transmembrane proteins, with an N terminal extracellular ligand attached to a C terminal membrane anchor. Release of the ligand from the transmembrane domain via proteases such as ADAMs can be regulated, potentially resulting in enhanced activation of EGFR compared to exogenous addition (34, 36, 37). In addition, after cleavage and release of the N-terminal ligand domain, the C-terminal domain can have additional functions including gene regulation (38).
The different properties of individual EGFR ligands may also account for differences in outcome for patients with tumors overexpressing specific ligands. As noted in the previous paragraph, the differing C-termini could affect both processing efficiency as well as intracellular targets (38). In addition, HBEGF has an N terminal heparin binding extension which enables interactions with cell surface heparin sulfate proteoglycans or extracellular matrix, which can lead to enhanced EGFR activation (39, 40). Conversely EGF has an extremely large N terminal extension whose cleavage may regulate the efficiency of EGF activation of EGFR (41). Finally, the efficiency of receptor activation and processing of the ligand-receptor complex, including recycling of the receptor, varies between ligands (37, 42).
Previous studies have shown that the phosphorylation of cortactin, which plays a pivotal role in the induction and maturation of invadopodia, can occur as a result of EGFR activation (28). The addition of 1uM Iressa or 2ug/mL CRM197 to HBEGF transductants for 15 minutes, rapidly blocking signaling downstream of EGFR activation, resulted in a significant reduction in invadopodium formation. Our results are consistent with a mechanism in which autocrine HBEGF expression results in sustained EGFR activation as well as concomitant increased invadopodium formation through increased phosphorylation of cortactin and enhanced invadopodium function and matrix degradation due increases in Src activation. Furthermore, we find a MEK-dependent upregulation of MMP2 and MMP9 expression, which results in enhanced invadopodium function and tumor cell invasion.
Based on these results, we propose the following model for ER negative tumors. For tumors with low levels of autocrine expression of EGFR ligands, local EGFR/CSF-1R paracrine loop interaction fields will stimulate a moderate degree of tumor cell invasion, with increased tumor cell invasion occurring near macrophages (which secrete EGFR ligands). For tumors with high levels of HBEGF autocrine expression and high EGFR expression levels, there is sustained EGFR activation, resulting in increased spontaneous motility as well as enhanced invadopodium formation and matrix degradation. Because of the higher basal motility and invasion, tumor cells are more autonomous and no longer rely on macrophages and the EGF/CSF1 paracrine loop for invasion. We have previously reported that head and neck cancers showed macrophage independent in vivo invasion (19). Head and neck cancers have relatively high expression of EGFR and HBEGF, and thus a mechanism similar to that described here may explain the macrophage-independent invasion observed in head and neck cancers. Similarly, other tumors with high levels of EGFR and HBEGF expression may utilize macrophage independent invasion. Because the EGF/CSF-1 paracrine loop invasion mechanism may not be operating in these tumors, the effectiveness of prognostic tests based on macrophage function, as well as therapies that target TAMs using CSF-1R inhibitors, may be most effective with tumors that express low levels of HBEGF.
Materials and Methods
Cell culture and generation of stable cell lines
Rat mammary adenocarcinoma MTLn3 cells expressing green fluorescent protein (GFP) and human ErbB1 (MTLn3 ErbB1) or corresponding empty vector control pLXSN (MTLn3 pLXSN), human mammary carcinoma MDA-MB 231 cells expressing GFP (MDA-MB 231), and BAC1.2F51.2F5 cells (BAC macrophages) were propagated as described previously (5, 8, 9). BT549 cells were cultured in DMEM (Cellgro, Manassas, VA, USA) containing 10% fetal bovine serum (FBS), 0.5% penicillin/streptomycin solution (Life Technologies, Rockville, MD, USA) and 0.023IU/mL insulin (EMD Millipore, Billerica, MA, USA). Human umbilical vein endothelial cells (HUVECs) (Lonza Inc., Allendale, NJ, USA) were propagated and maintained according to the manufacturer's instructions, and not used beyond passage number 4 for all assays.
The HBEGF precursor in the pJMU2-1 vector was originally obtained from Dr. Robert Coffey at Vanderbilt University. PCR primers were designed to add NotI (5’) and BamHI (3’) sites to the flanking ends of the ligand, which were used for ligation into a pBM-IRES vector modified to include puromycin resistance (43). The empty pBM-IRES retroviral vector or retroviral vector containing full-length human HBEGF was transfected into the 293GP cells, kindly provided by Jonathan Backer, with packaging vector VSV-G using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. To generate stable transductants, MTLn3 pLXSN, MTLn3 ErbB1, MDA-MB 231, and BT549 cells were transduced with supernatants containing virus in the presence of 8ug/mL polybrene and placed under puromycin selection (1ug/mL) for one week.
Animal models
All experiments conducted in mice were in accordance with the National Institutes of Health regulations on the use and care of experimental animals and approved by the Albert Einstein College of Medicine (AECOM) Animal Use Committee. MTLn3 and MDA-MB 231 orthotopic tumor xenografts were generated by injecting tumor cells in the lower right mammary fat pad of 6- to 8-week-old female severe combined immunodeficiency (SCID)/NCr mice (National Cancer Institute, Frederick, MD, USA) as described previously (9, 44, 45). Tumor volumes, spontaneous metastasis, and intravasation efficiency in vivo were measured as previously described (5). All experiments were performed on tumor bearing mice 4 weeks and 14 weeks post-injection for the MTLn3 and MDA-MB 231 transductant cell lines, respectively.
In vivo invasion assay
Invasive tumor cell collection into 33-gauge microneedles placed into live anesthetized tumor bearing SCID/NCr mice was performed as described previously (9, 18). To inhibit the mouse CSF-1 receptor (CSF-1R), the small molecule 4-cyano-1H-pyrrole-2-carboxylic acid [4-(4-methyl-piperazin-1-yl)-2-(4-methyl-piperidin-1-yl)-phenyl]-amide, referred to as “JnJ” hereafter, (JNJ-28312141; Johnson & Johnson Pharmaceutical Research & Development, New Brunswick, NJ, USA) was used at a final concentration of 1uM; DMSO at the same quantity was used as a control vehicle (46).
Intravital imaging of tumor cell motility and uptake of Texas Red dextran by macrophages in primary tumors and spleens
A detailed protocol for the intravital imaging of primary tumors has been described elsewhere (47). In brief, orthotopic tumor xenografts were generated in SCID/NCr mice as described above. A skin flap surgery was performed on anesthetized tumor bearing animals to expose the primary tumor. The mouse was placed on an inverted Olympus FV1000-MPE multiphoton microscope with a 25X 1.05 NA water objective, and the collagen matrix and GFP-positive tumor cells were imaged at an excitation wavelength of 880nm. Multiple fields were imaged per tumor. For each field, a 30 min z-stack time-lapse series was collected and the number of moving cells per field was quantified.
Intravital imaging of Texas Red dextran uptake by macrophages in primary tumors and spleens was carried out as described previously (19, 48). Texas Red-labeled and GFP cells were imaged using a 25X 1.05 NA water objective. Multiple z-series were taken for the tumor and spleen using 5um steps. All subsequent imaging was performed using the Olympus FV1000-MPE multiphoton microscope at an 880nm excitation wavelength with a 25X 1.05 NA water objective unless otherwise stated.
In vitro transendothelial migration (iTEM) assay
To prepare the endothelial monolayer, the underside of each transwell (EMD Millipore) was coated with 50uL of 2.5ug/mL Matrigel (BD Biosciences). Approximately 100,000 HUVECs were plated on inverted transwells in 50uL of EGM-2 (Lonza Inc.) and allowed to adhere for 4 hours at 37°C. Transwells were then placed in a 24-well plate containing 200ul or 1mL of EGM-2 in the lower and upper chambers, respectively, and cultured for 48 hours to allow a monolayer to form. Carcinoma cells were labeled with cell tracker green dye (Invitrogen), resuspended in M199 medium (Invitrogen), plated at 10,000 cells/transwell, and allowed to transmigrate towards EGM-2 for 18 hours. Samples were then fixed in 4% paraformaldehyde (PFA), permeabilized in 0.1% triton-X100 and stained with rhodamine-phalloidin (Invitrogen). Z-series were taken in 8 random fields per transwell using 2um step sizes.
Invadopodium formation, matrix degradation assay and immunofluorescence
Invadopodium formation and thin gelatin matrix degradation assays were performed as described (26, 49-51). For Tks5 or phospho-cortactin and total cortactin colocalization and gelatin matrix degradation, 100,000 MTLn3 ErbB1 or MDA-MB 231 transductants were plated on Alexa Fluor-405 thin gelatin matrix for 16 hours (for MTLn3 ErbB1 cells), 4 hours (for MDA-MB 231 cells), or 12 hours (for BT549 cells) prior to fixation in 3.7% PFA, and immunofluorescence for Tks5, total cortactin, and phospho-cortactin was performed as described previously (27). Matrix degradation area was quantified as described previously (52). Cortactin activation was quantitated by measuring the signal intensity of total and phospho-cortactin in ImageJ, and a ratio of phospho-cortactin/total cortactin was determined.
Additional reagents and methods are described in the Supplemental Materials and Methods.
Statistical analysis
All of the results shown are representative of at least three independent experiments for the in vitro experiments and at least five different animals per point for the in vivo experiments. Statistical analyses were assessed using a two-tailed Student's t test, Mann Whitney u-test, or z-test as indicated.
Supplementary Material
Acknowledgements
We would like to thank the Condeelis, Cox, Hodgson, and Segall labs for their comments and suggestions. We thank Carl Manthey and Johnson & Johnson for providing the JnJ compound.
Grant Support
J.E.S. is the Betty and Sheldon Feinberg Senior Faculty Scholar in Cancer Research. Funding was provided by CA100324 (ERS, ARB, DC, JWP, JC, and JES), CA77522 (JES), T32-GM007288 (ZNZ), a postdoctoral fellowship from Susan G. Komen for the Cure ®, KG111405 (VPS), a NIH F32 postdoctoral fellowship F32-CA159663-01 (MRJ), and NIH 1K12GM102779-01 (EAP).
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Conflict of Interest
JC has received compensation as a member of the scientific advisory board of MetaStat and owns stock in the company. The remaining authors declare no conflict of interest.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA: a cancer journal for clinicians. 2010 Sep-Oct;60(5):277–300. doi: 10.3322/caac.20073. PubMed PMID: 20610543. [DOI] [PubMed] [Google Scholar]
- 2.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nature reviews Cancer. 2003 Jun;3(6):453–8. doi: 10.1038/nrc1098. PubMed PMID: 12778135. [DOI] [PubMed] [Google Scholar]
- 3.Place AE, Jin Huh S, Polyak K. The microenvironment in breast cancer progression: biology and implications for treatment. Breast cancer research : BCR. 2011;13(6):227. doi: 10.1186/bcr2912. PubMed PMID: 22078026. Pubmed Central PMCID: 3326543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Artacho-Cordon A, Artacho-Cordon F, Rios-Arrabal S, Calvente I, Nunez MI. Tumor microenvironment and breast cancer progression: a complex scenario. Cancer biology & therapy. 2012 Jan 1;13(1):14–24. doi: 10.4161/cbt.13.1.18869. PubMed PMID: 22336584. [DOI] [PubMed] [Google Scholar]
- 5.Xue C, Wyckoff J, Liang F, Sidani M, Violini S, Tsai KL, et al. Epidermal growth factor receptor overexpression results in increased tumor cell motility in vivo coordinately with enhanced intravasation and metastasis. Cancer research. 2006 Jan 1;66(1):192–7. doi: 10.1158/0008-5472.CAN-05-1242. PubMed PMID: 16397232. [DOI] [PubMed] [Google Scholar]
- 6.Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer research. 2007 Mar 15;67(6):2649–56. doi: 10.1158/0008-5472.CAN-06-1823. PubMed PMID: 17363585. [DOI] [PubMed] [Google Scholar]
- 7.Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, et al. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer research. 2004 Oct 1;64(19):7022–9. doi: 10.1158/0008-5472.CAN-04-1449. PubMed PMID: 15466195. [DOI] [PubMed] [Google Scholar]
- 8.Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, et al. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer research. 2005 Jun 15;65(12):5278–83. doi: 10.1158/0008-5472.CAN-04-1853. PubMed PMID: 15958574. [DOI] [PubMed] [Google Scholar]
- 9.Patsialou A, Wyckoff J, Wang Y, Goswami S, Stanley ER, Condeelis JS. Invasion of human breast cancer cells in vivo requires both paracrine and autocrine loops involving the colony-stimulating factor-1 receptor. Cancer research. 2009 Dec 15;69(24):9498–506. doi: 10.1158/0008-5472.CAN-09-1868. PubMed PMID: 19934330. Pubmed Central PMCID: 2794986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris RC, Chung E, Coffey RJ. EGF receptor ligands. Experimental cell research. 2003 Mar 10;284(1):2–13. doi: 10.1016/s0014-4827(02)00105-2. PubMed PMID: 12648462. [DOI] [PubMed] [Google Scholar]
- 11.Singh AB, Harris RC. Autocrine, paracrine and juxtacrine signaling by EGFR ligands. Cellular signalling. 2005 Oct;17(10):1183–93. doi: 10.1016/j.cellsig.2005.03.026. PubMed PMID: 15982853. [DOI] [PubMed] [Google Scholar]
- 12.Ongusaha PP, Kwak JC, Zwible AJ, Macip S, Higashiyama S, Taniguchi N, et al. HB-EGF is a potent inducer of tumor growth and angiogenesis. Cancer research. 2004 Aug 1;64(15):5283–90. doi: 10.1158/0008-5472.CAN-04-0925. PubMed PMID: 15289334. [DOI] [PubMed] [Google Scholar]
- 13.Revillion F, Lhotellier V, Hornez L, Bonneterre J, Peyrat JP. ErbB/HER ligands in human breast cancer, and relationships with their receptors, the bio-pathological features and prognosis. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2008 Jan;19(1):73–80. doi: 10.1093/annonc/mdm431. PubMed PMID: 17962208. [DOI] [PubMed] [Google Scholar]
- 14.Yotsumoto F, Oki E, Tokunaga E, Maehara Y, Kuroki M, Miyamoto S. HB-EGF orchestrates the complex signals involved in triple-negative and trastuzumab-resistant breast cancer. International journal of cancer Journal international du cancer. 2010 Dec 1;127(11):2707–17. doi: 10.1002/ijc.25472. PubMed PMID: 20499311. [DOI] [PubMed] [Google Scholar]
- 15.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast cancer research and treatment. 2010 Oct;123(3):725–31. doi: 10.1007/s10549-009-0674-9. PubMed PMID: 20020197. [DOI] [PubMed] [Google Scholar]
- 16.Kedrin D, Wyckoff J, Boimel PJ, Coniglio SJ, Hynes NE, Arteaga CL, et al. ERBB1 and ERBB2 have distinct functions in tumor cell invasion and intravasation. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009 Jun 1;15(11):3733–9. doi: 10.1158/1078-0432.CCR-08-2163. PubMed PMID: 19458057. Pubmed Central PMCID: 2859965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitamura T, Higashiyama S, Taniguchi N, Klagsbrun M, Mekada E. Diphtheria toxin binds to the epidermal growth factor (EGF)-like domain of human heparin-binding EGFlike growth factor/diphtheria toxin receptor and inhibits specifically its mitogenic activity. The Journal of biological chemistry. 1995 Jan 20;270(3):1015–9. doi: 10.1074/jbc.270.3.1015. PubMed PMID: 7836353. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez L, Smirnova T, Wyckoff J, Condeelis J, Segall JE. In vivo assay for tumor cell invasion. Methods in molecular biology. 2009;571:227–38. doi: 10.1007/978-1-60761-198-1_15. PubMed PMID: 19763970. Pubmed Central PMCID: 3156574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smirnova T, Adomako A, Locker J, Van Rooijen N, Prystowsky MB, Segall JE. In vivo invasion of head and neck squamous cell carcinoma cells does not require macrophages. The American journal of pathology. 2011 Jun;178(6):2857–65. doi: 10.1016/j.ajpath.2011.02.030. PubMed PMID: 21641405. Pubmed Central PMCID: 3124357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roussos ET, Balsamo M, Alford SK, Wyckoff JB, Gligorijevic B, Wang Y, et al. Mena invasive (MenaINV) promotes multicellular streaming motility and transendothelial migration in a mouse model of breast cancer. Journal of cell science. 2011 Jul 1;124(Pt 13):2120–31. doi: 10.1242/jcs.086231. PubMed PMID: 21670198. Pubmed Central PMCID: 3113666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian B, Deng Y, Im JH, Muschel RJ, Zou Y, Li J, et al. A distinct macrophage population mediates metastatic breast cancer cell extravasation, establishment and growth. PloS one. 2009;4(8):e6562. doi: 10.1371/journal.pone.0006562. PubMed PMID: 19668347. Pubmed Central PMCID: 2721818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonde AK, Tischler V, Kumar S, Soltermann A, Schwendener RA. Intratumoral macrophages contribute to epithelial-mesenchymal transition in solid tumors. BMC cancer. 2012;12:35. doi: 10.1186/1471-2407-12-35. PubMed PMID: 22273460. Pubmed Central PMCID: 3314544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaynagetdinov R, Sherrill TP, Polosukhin VV, Han W, Ausborn JA, McLoed AG, et al. A critical role for macrophages in promotion of urethane-induced lung carcinogenesis. Journal of immunology. 2011 Dec 1;187(11):5703–11. doi: 10.4049/jimmunol.1100558. PubMed PMID: 22048774. Pubmed Central PMCID: 3221921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeisberger SM, Odermatt B, Marty C, Zehnder-Fjallman AH, Ballmer-Hofer K, Schwendener RA. Clodronate-liposome-mediated depletion of tumour-associamacrophages: a new and highly effective antiangiogenic therapy approach. British journal of cancer. 2006 Aug 7;95(3):272–81. doi: 10.1038/sj.bjc.6603240. PubMed PMID: 16832418. Pubmed Central PMCID: 2360657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy DA, Courtneidge SA. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nature reviews Molecular cell biology. 2011 Jul;12(7):413–26. doi: 10.1038/nrm3141. PubMed PMID: 21697900. Pubmed Central PMCID: 3423958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oser M, Mader CC, Gil-Henn H, Magalhaes M, Bravo-Cordero JJ, Koleske AJ, et al. Specific tyrosine phosphorylation sites on cortactin regulate Nck1-dependent actin polymerization in invadopodia. Journal of cell science. 2010 Nov 1;123(Pt 21):3662–73. doi: 10.1242/jcs.068163. PubMed PMID: 20971703. Pubmed Central PMCID: 3037016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, et al. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. The Journal of cell biology. 2005 Jan 31;168(3):441–52. doi: 10.1083/jcb.200407076. PubMed PMID: 15684033. Pubmed Central PMCID: 2171731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mader CC, Oser M, Magalhaes MA, Bravo-Cordero JJ, Condeelis J, Koleske AJ, et al. An EGFR-Src-Arg-cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer research. 2011 Mar 1;71(5):1730–41. doi: 10.1158/0008-5472.CAN-10-1432. PubMed PMID: 21257711. Pubmed Central PMCID: 3057139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ammer AG, Weed SA. Cortactin branches out: roles in regulating protrusive actin dynamics. Cell motility and the cytoskeleton. 2008 Sep;65(9):687–707. doi: 10.1002/cm.20296. PubMed PMID: 18615630. Pubmed Central PMCID: 2561250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer metastasis reviews. 2006 Mar;25(1):9–34. doi: 10.1007/s10555-006-7886-9. PubMed PMID: 16680569. [DOI] [PubMed] [Google Scholar]
- 31.Artym VV, Zhang Y, Seillier-Moiseiwitsch F, Yamada KM, Mueller SC. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer research. 2006 Mar 15;66(6):3034–43. doi: 10.1158/0008-5472.CAN-05-2177. PubMed PMID: 16540652. [DOI] [PubMed] [Google Scholar]
- 32.Yagi H, Yotsumoto F, Miyamoto S. Heparin-binding epidermal growth factor-like growth factor promotes transcoelomic metastasis in ovarian cancer through epithelial-mesenchymal transition. Molecular cancer therapeutics. 2008 Oct;7(10):3441–51. doi: 10.1158/1535-7163.MCT-08-0417. PubMed PMID: 18852147. [DOI] [PubMed] [Google Scholar]
- 33.Lue HW, Yang X, Wang R, Qian W, Xu RZ, Lyles R, et al. LIV-1 promotes prostate cancer epithelial-to-mesenchymal transition and metastasis through HB-EGF shedding and EGFR-mediated ERK signaling. PloS one. 2011;6(11):e27720. doi: 10.1371/journal.pone.0027720. PubMed PMID: 22110740. Pubmed Central PMCID: 3218022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joslin EJ, Opresko LK, Wells A, Wiley HS, Lauffenburger DA. EGF-receptor-mediated mammary epithelial cell migration is driven by sustained ERK signaling from autocrine stimulation. Journal of cell science. 2007 Oct 15;120(Pt 20):3688–99. doi: 10.1242/jcs.010488. PubMed PMID: 17895366. [DOI] [PubMed] [Google Scholar]
- 35.Hayes KE, Walk EL, Ammer AG, Kelley LC, Martin KH, Weed SA. Ableson kinases negatively regulate invadopodia function and invasion in head and neck squamous cell carcinoma by inhibiting an HB-EGF autocrine loop. Oncogene. 2012 Nov 12; doi: 10.1038/onc.2012.513. PubMed PMID: 23146907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joslin EJ, Shankaran H, Opresko LK, Bollinger N, Lauffenburger DA, Wiley HS. Structure of the EGF receptor transactivation circuit integrates multiple signals with cell context. Molecular bioSystems. 2010 Jul;6(7):1293–306. doi: 10.1039/c003921g. PubMed PMID: 20458382. Pubmed Central PMCID: 3306786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson KJ, Mill C, Lambert S, Buchman J, Wilson TR, Hernandez-Gordillo V, et al. EGFR ligands exhibit functional differences in models of paracrine and autocrine signaling. Growth factors. 2012 Apr;30(2):107–16. doi: 10.3109/08977194.2011.649918. PubMed PMID: 22260327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimura T, Yoshida M, Fukuda S, Ebi M, Hirata Y, Mizoshita T, et al. Nuclear translocation of the cytoplasmic domain of HB-EGF induces gastric cancer invasion. BMC cancer. 2012;12:205. doi: 10.1186/1471-2407-12-205. PubMed PMID: 22646534. Pubmed Central PMCID: 3414754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishi E, Klagsbrun M. Heparin-binding epidermal growth factor-like growth factor (HB-EGF) is a mediator of multiple physiological and pathological pathways. Growth factors. 2004 Dec;22(4):253–60. doi: 10.1080/08977190400008448. PubMed PMID: 15621728. [DOI] [PubMed] [Google Scholar]
- 40.Tolino MA, Block ER, Klarlund JK. Brief treatment with heparin-binding EGF-like growth factor, but not with EGF, is sufficient to accelerate epithelial wound healing. Biochimica et biophysica acta. 2011 Sep;1810(9):875–8. doi: 10.1016/j.bbagen.2011.05.011. PubMed PMID: 21640162. Pubmed Central PMCID: 3143286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott J, Patterson S, Rall L, Bell GI, Crawford R, Penschow J, et al. The structure and biosynthesis of epidermal growth factor precursor. Journal of cell science Supplement. 1985;3:19–28. doi: 10.1242/jcs.1985.supplement_3.3. PubMed PMID: 3011823. [DOI] [PubMed] [Google Scholar]
- 42.Roepstorff K, Grandal MV, Henriksen L, Knudsen SL, Lerdrup M, Grovdal L, et al. Differential effects of EGFR ligands on endocytic sorting of the receptor. Traffic. 2009 Aug;10(8):1115–27. doi: 10.1111/j.1600-0854.2009.00943.x. PubMed PMID: 19531065. Pubmed Central PMCID: 2723868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garton KJ, Ferri N, Raines EW. Efficient expression of exogenous genes in primary vascular cells using IRES-based retroviral vectors. BioTechniques. 2002 Apr;32(4):830. doi: 10.2144/02324rr01. 2, 4 passim. PubMed PMID: 11962605. [DOI] [PubMed] [Google Scholar]
- 44.Roussos ET, Goswami S, Balsamo M, Wang Y, Stobezki R, Adler E, et al. Mena invasive (Mena(INV)) and Mena11a isoforms play distinct roles in breast cancer cell cohesion and association with TMEM. Clinical & experimental metastasis. 2011 Aug;28(6):515–27. doi: 10.1007/s10585-011-9388-6. PubMed PMID: 21484349. Pubmed Central PMCID: 3459587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smirnova T, Zhou ZN, Flinn RJ, Wyckoff J, Boimel PJ, Pozzuto M, et al. Phosphoinositide 3-kinase signaling is critical for ErbB3-driven breast cancer cell motility and metastasis. Oncogene. 2012 Feb 9;31(6):706–15. doi: 10.1038/onc.2011.275. PubMed PMID: 21725367. Pubmed Central PMCID: 3469325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coniglio SJ, Eugenin E, Dobrenis K, Stanley ER, West BL, Symons MH, et al. Microglial stimulation of glioblastoma invasion involves epidermal growth factor receptor (EGFR) and colony stimulating factor 1 receptor (CSF-1R) signaling. Molecular medicine. 2012;18:519–27. doi: 10.2119/molmed.2011.00217. PubMed PMID: 22294205. Pubmed Central PMCID: 3356419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kedrin D, Wyckoff J, Sahai E, Condeelis J, Segall JE. Imaging tumor cell movement in vivo. Current protocols in cell biology / editorial board. Juan S Bonifacino. 2007 Jun; doi: 10.1002/0471143030.cb1907s35. [et al] Chapter 19:Unit 19 7. PubMed PMID: 18228501. [DOI] [PubMed] [Google Scholar]
- 48.Van Rooijen N, Sanders A. Liposome mediated depletion of macrophages: mechanism of action, preparation of liposomes and applications. Journal of immunological methods. 1994 Sep 14;174(1-2):83–93. doi: 10.1016/0022-1759(94)90012-4. PubMed PMID: 8083541. [DOI] [PubMed] [Google Scholar]
- 49.Artym VV, Yamada KM, Mueller SC. ECM degradation assays for analyzing local cell invasion. Methods in molecular biology. 2009;522:211–9. doi: 10.1007/978-1-59745-413-1_15. PubMed PMID: 19247615. [DOI] [PubMed] [Google Scholar]
- 50.Artym VV, Matsumoto K, Mueller SC, Yamada KM. Dynamic membrane remodeling at invadopodia differentiates invadopodia from podosomes. European journal of cell biology. 2011 Feb-Mar;90(2-3):172–80. doi: 10.1016/j.ejcb.2010.06.006. PubMed PMID: 20656375. Pubmed Central PMCID: 3153956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma VP, Entenberg D, Condeelis J. High-resolution live-cell imaging and timelapse microscopy of invadopodium dynamics and tracking analysis. Methods in molecular biology. 2013 doi: 10.1007/978-1-62703-538-5_21. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oser M, Yamaguchi H, Mader CC, Bravo-Cordero JJ, Arias M, Chen X, et al. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. The Journal of cell biology. 2009 Aug 24;186(4):571–87. doi: 10.1083/jcb.200812176. PubMed PMID: 19704022. Pubmed Central PMCID: 2733743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hernandez L, Smirnova T, Kedrin D, Wyckoff J, Zhu L, Stanley ER, et al. The EGF/CSF- 1 paracrine invasion loop can be triggered by heregulin beta1 and CXCL12. Cancer research. 2009 Apr 1;69(7):3221–7. doi: 10.1158/0008-5472.CAN-08-2871. PubMed PMID: 19293185. Pubmed Central PMCID: 2820720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boimel PJ, Cruz C, Segall JE. A functional in vivo screen for regulators of tumor progression identifies HOXB2 as a regulator of tumor growth in breast cancer. Genomics. 2011 Sep;98(3):164–72. doi: 10.1016/j.ygeno.2011.05.011. PubMed PMID: 21672623. Pubmed Central PMCID: 3165059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.