Abstract
We have identified and functionally characterized a new Escherichia coli gene, dsbC, whose product is involved in disulfide bond formation in the periplasmic space. It corresponds to a previously sequenced open reading frame mapping upstream of recJ with no previously assigned function. Null mutations in dsbC were obtained using a screen for dithiothreitol (DTT)-sensitive mutants and were shown to result in the accumulation of reduced forms of a variety of disulfide bond-containing periplasmic proteins. This defect could be rescued by the addition of either oxidized DTT or cystine or by multicopy expression of dsbA, a known periplasmic disulfide oxidase. The DsbC protein is synthesized as a precursor form of 25.5 kDa which is processed to a 23.3 kDa mature species located in the periplasmic space. The DsbC protein was overexpressed, purified to homogeneity and shown to catalyse the reduction of insulin in a DTT-dependent manner at levels comparable with those of purified DsbA. The replacement of either cysteine residue of the predicted active site, F-(X4)-C-G-Y-C, completely inactivates DsbC protein function. We have further shown that in vivo overexpression of DsbC can functionally substitute for a loss of DsbA function. Taken together, all of our results demonstrate that DsbC acts in vivo as a disulfide oxidase.
Full text
PDF



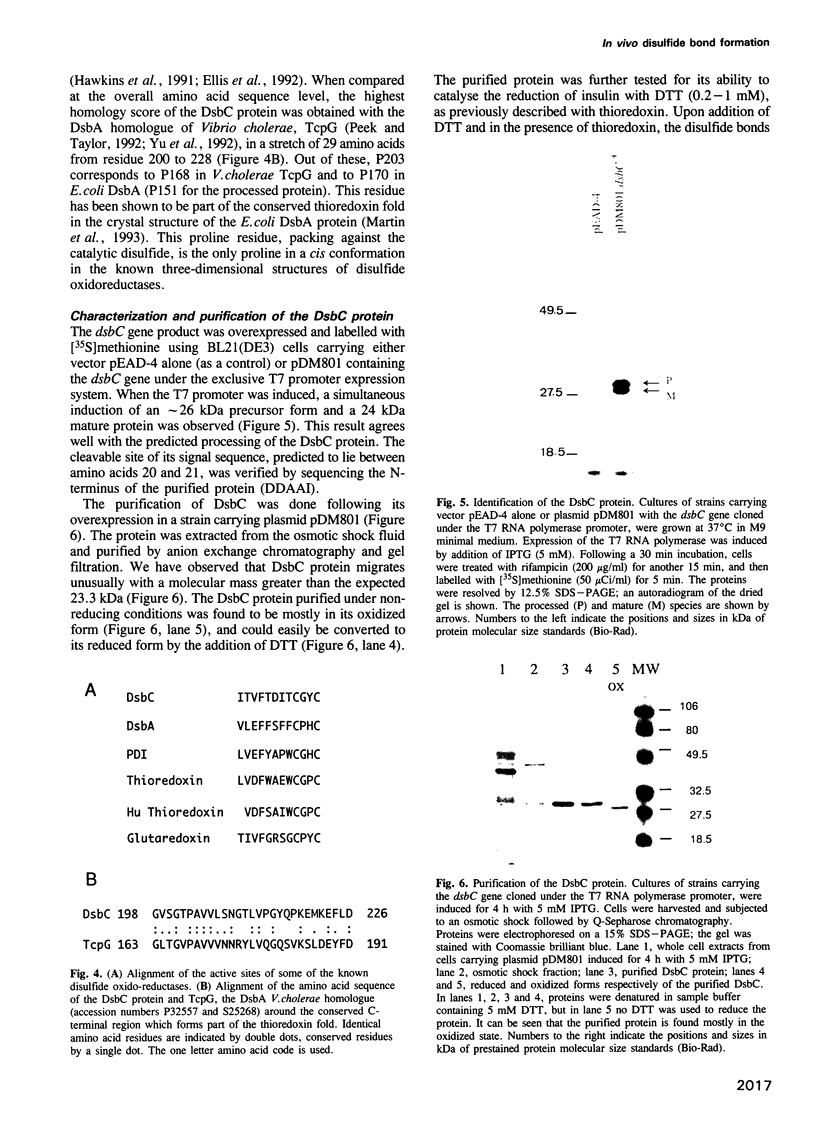
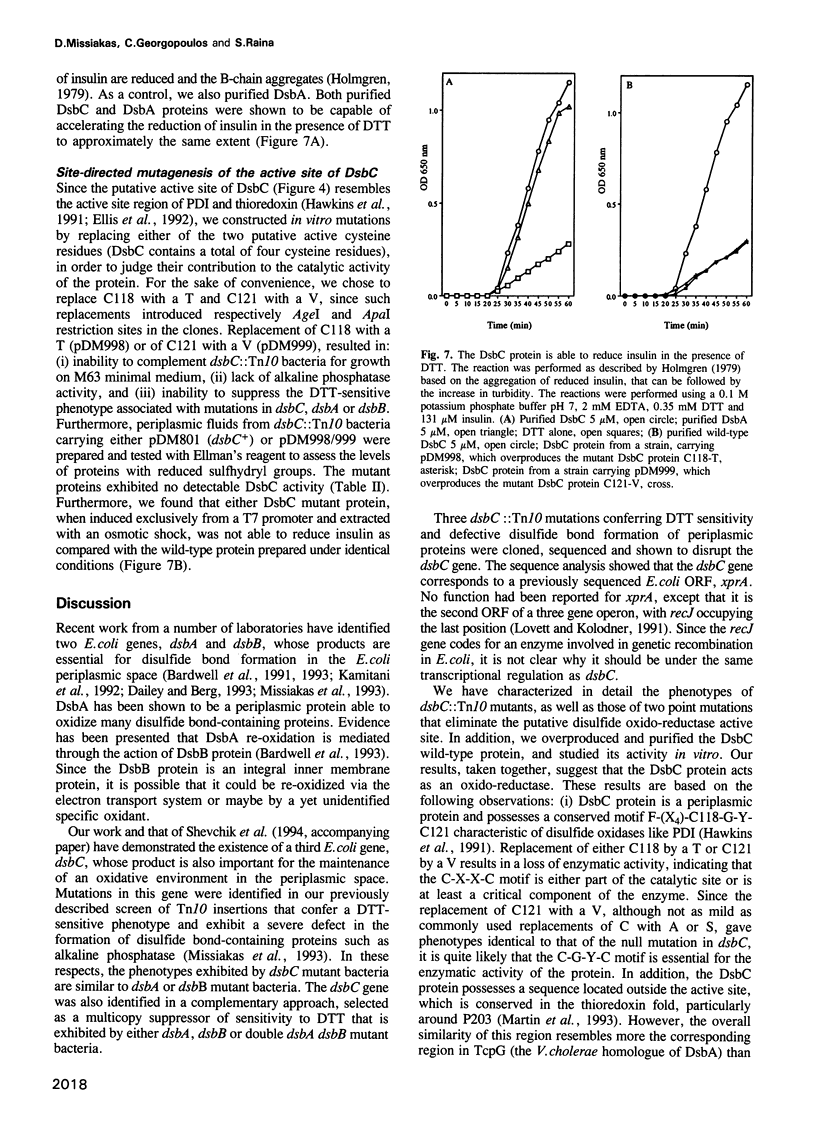


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bardwell J. C., Lee J. O., Jander G., Martin N., Belin D., Beckwith J. A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):1038–1042. doi: 10.1073/pnas.90.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell J. C., McGovern K., Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991 Nov 1;67(3):581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- Creighton T. E., Freedman R. B. A model catalyst of protein disulphide bond formation. Curr Biol. 1993 Nov 1;3(11):790–793. doi: 10.1016/0960-9822(93)90034-l. [DOI] [PubMed] [Google Scholar]
- Creighton T. E., Hillson D. A., Freedman R. B. Catalysis by protein-disulphide isomerase of the unfolding and refolding of proteins with disulphide bonds. J Mol Biol. 1980 Sep 5;142(1):43–62. doi: 10.1016/0022-2836(80)90205-3. [DOI] [PubMed] [Google Scholar]
- Dailey F. E., Berg H. C. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):1043–1047. doi: 10.1073/pnas.90.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis L. B., Saurugger P., Woodward C. Identification of the three-dimensional thioredoxin motif: related structure in the ORF3 protein of the Staphylococcus aureus mer operon. Biochemistry. 1992 May 26;31(20):4882–4891. doi: 10.1021/bi00135a020. [DOI] [PubMed] [Google Scholar]
- Fellay R., Frey J., Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52(2-3):147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- Franklin N. C. A plasmid to visualize and assay termination and antitermination of transcription in Escherichia coli. Plasmid. 1989 Jan;21(1):31–42. doi: 10.1016/0147-619x(89)90084-x. [DOI] [PubMed] [Google Scholar]
- GOLDBERGER R. F., EPSTEIN C. J., ANFINSEN C. B. Acceleration of reactivation of reduced bovine pancreatic ribonuclease by a microsomal system from rat liver. J Biol Chem. 1963 Feb;238:628–635. [PubMed] [Google Scholar]
- Groisman E. A., Casadaban M. J. Mini-mu bacteriophage with plasmid replicons for in vivo cloning and lac gene fusing. J Bacteriol. 1986 Oct;168(1):357–364. doi: 10.1128/jb.168.1.357-364.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins H. C., Blackburn E. C., Freedman R. B. Comparison of the activities of protein disulphide-isomerase and thioredoxin in catalysing disulphide isomerization in a protein substrate. Biochem J. 1991 Apr 15;275(Pt 2):349–353. doi: 10.1042/bj2750349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem. 1979 Oct 10;254(19):9627–9632. [PubMed] [Google Scholar]
- Kamitani S., Akiyama Y., Ito K. Identification and characterization of an Escherichia coli gene required for the formation of correctly folded alkaline phosphatase, a periplasmic enzyme. EMBO J. 1992 Jan;11(1):57–62. doi: 10.1002/j.1460-2075.1992.tb05027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lovett S. T., Kolodner R. D. Nucleotide sequence of the Escherichia coli recJ chromosomal region and construction of recJ-overexpression plasmids. J Bacteriol. 1991 Jan;173(1):353–364. doi: 10.1128/jb.173.1.353-364.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. L., Bardwell J. C., Kuriyan J. Crystal structure of the DsbA protein required for disulphide bond formation in vivo. Nature. 1993 Sep 30;365(6445):464–468. doi: 10.1038/365464a0. [DOI] [PubMed] [Google Scholar]
- Missiakas D., Georgopoulos C., Raina S. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):7084–7088. doi: 10.1073/pnas.90.15.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu H. C., Heppel L. A. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965 Sep;240(9):3685–3692. [PubMed] [Google Scholar]
- Peek J. A., Taylor R. K. Characterization of a periplasmic thiol:disulfide interchange protein required for the functional maturation of secreted virulence factors of Vibrio cholerae. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):6210–6214. doi: 10.1073/pnas.89.13.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitt S., Zalkin H. Role of primary structure and disulfide bond formation in beta-lactamase secretion. J Bacteriol. 1983 Jan;153(1):27–32. doi: 10.1128/jb.153.1.27-32.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley A. P. Translocation of a folded protein across the outer membrane in Escherichia coli. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):12058–12062. doi: 10.1073/pnas.89.24.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchik V. E., Condemine G., Robert-Baudouy J. Characterization of DsbC, a periplasmic protein of Erwinia chrysanthemi and Escherichia coli with disulfide isomerase activity. EMBO J. 1994 Apr 15;13(8):2007–2012. doi: 10.1002/j.1460-2075.1994.tb06470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., Dove W., Jaacks K. J., Grossman A. D., Erickson J. W., Gross C. A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989 Mar;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene. 1991 Apr;100:189–194. doi: 10.1016/0378-1119(91)90365-i. [DOI] [PubMed] [Google Scholar]
- Wang R. F., Kushner S. R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991 Apr;100:195–199. [PubMed] [Google Scholar]
- Wunderlich M., Glockshuber R. In vivo control of redox potential during protein folding catalyzed by bacterial protein disulfide-isomerase (DsbA). J Biol Chem. 1993 Nov 25;268(33):24547–24550. [PubMed] [Google Scholar]
- Wunderlich M., Otto A., Seckler R., Glockshuber R. Bacterial protein disulfide isomerase: efficient catalysis of oxidative protein folding at acidic pH. Biochemistry. 1993 Nov 16;32(45):12251–12256. doi: 10.1021/bi00096a039. [DOI] [PubMed] [Google Scholar]
- Yu J., Webb H., Hirst T. R. A homologue of the Escherichia coli DsbA protein involved in disulphide bond formation is required for enterotoxin biogenesis in Vibrio cholerae. Mol Microbiol. 1992 Jul;6(14):1949–1958. doi: 10.1111/j.1365-2958.1992.tb01368.x. [DOI] [PubMed] [Google Scholar]






