ABSTRACT
A novel avian-origin H7N9 influenza A virus (IAV) emerged in China in 2013, causing mild to lethal human respiratory infections. H7N9 originated with multiple reassortment events between avian viruses and carries genetic markers of human adaptation. Determining whether H7N9 induces a host response closer to that with human or avian IAV is important in order to better characterize this emerging virus. Here we compared the human lung epithelial cell response to infection with A/Anhui/01/13 (H7N9) or highly pathogenic avian-origin H5N1, H7N7, or human seasonal H3N2 IAV. The transcriptomic response to H7N9 was highly specific to this strain but was more similar to the response to human H3N2 than to that to other avian IAVs. H7N9 and H3N2 both elicited responses related to eicosanoid signaling and chromatin modification, whereas H7N9 specifically induced genes regulating the cell cycle and transcription. Among avian IAVs, the response to H7N9 was closest to that elicited by H5N1 virus. Host responses common to H7N9 and the other avian viruses included the lack of induction of the antigen presentation pathway and reduced proinflammatory cytokine induction compared to that with H3N2. Repression of these responses could have an important impact on the immunogenicity and virulence of H7N9 in humans. Finally, using a genome-based drug repurposing approach, we identified several drugs predicted to regulate the host response to H7N9 that may act as potential antivirals, including several kinase inhibitors, as well as FDA-approved drugs, such as troglitazone and minocycline. Importantly, we validated that minocycline inhibited H7N9 replication in vitro, suggesting that our computational approach holds promise for identifying novel antivirals.
IMPORTANCE
Whether H7N9 will be the next pandemic influenza virus or will persist and sporadically infect humans from its avian reservoir, similar to H5N1, is not known yet. High-throughput profiling of the host response to infection allows rapid characterization of virus-host interactions and generates many hypotheses that will accelerate understanding and responsiveness to this potential threat. We show that the cellular response to H7N9 virus is closer to that induced by H3N2 than to that induced by H5N1, reflecting the potential of this new virus for adaptation to humans. Importantly, dissecting the host response to H7N9 may guide host-directed antiviral development.
INTRODUCTION
Between February and May 2013, the novel avian influenza A (H7N9) virus was associated with 132 human cases, including 37 deaths, occurring mainly in Eastern China (1). Most reports of infection have presented a severe clinical picture characterized by bilateral pneumonia progressing to acute respiratory distress syndrome associated with a cytokine storm and multiorgan failure (2, 3). This clinical presentation is similar to that with H5N1 infection (2, 3) but is unusual for H7 viruses, which cause mostly mild respiratory illness or conjunctivitis (4). The hospitalized case fatality rate for H7N9 is estimated at 36% (5), compared to 60 to 70% for H5N1 (6, 7) and 6.5 to 8% for seasonal influenza infection (8, 9). However, mild cases of H7N9 infections were recently detected through sentinel influenza-like illness surveillance, suggesting that H7N9 could have caused a substantial number of asymptomatic to moderate infections (5, 10). This is in line with infections by most influenza virus strains, with the exception of H5N1, for which there is no report of a laboratory-confirmed mild case of infection. Other epidemiological differences between H7N9 and H5N1 include the more rapid accumulation of human infections with H7N9 than with H5N1 and differences in the age distribution of infected individuals, with H5N1 infections occurring mostly in children and young adults while 55% of H7N9 cases have been in individuals older than 60 years (7).
Sequence analysis indicates that H7N9 originated from multiple reassortment events between avian influenza viruses (11, 12). However, the H7N9 sequence bears several genetic markers of human adaptation, including PB2 E627K and hemagglutinin (HA) Q226L (11, 13). The consequence of these mutations for the host response has not been assessed. Previous studies of H7N9 virus-host interaction have focused on transmissibility and HA binding properties. The H7N9 strain A/Anhui/01/13 (Anhui01) has a mixed α2-3/α2-6 receptor preference, associated with a capacity to efficiently transmit by direct contact but poorly by respiratory droplets in the ferret model (14). These properties are intermediary between human seasonal influenza viruses that have an α2-6 preference and efficiently transmit by respiratory droplets and H5N1 viruses with an α2-3 preference that do not transmit efficiently in either respiratory droplet or direct-contact transmission models (14, 15). Crystallographic structures of HA from H7N9 strains isolated from human patients show that increased avidity for human receptors compared to that of avian H7 virus is due to the Q226L and G186V mutations (16). Assays of HA binding to human tissue sections revealed an intermediate binding pattern between human IAV (preferentially binding nonciliated cells of apical regions of the trachea) and H5N1 (binding alveolar sections). In addition, a single mutation, G228S, substantially increased HA binding to human receptors in the human respiratory tract (17).
There is great concern over potential further adaptation of H7N9 to humans and acquisition of sustainable person-to-person transmissibility. In addition, a recent study pointed out that H7N9 acquired resistance to neuraminidase (NA) inhibitors in 2 out of 14 hospitalized patients that had been treated with oseltamivir or peramivir (18). Emergence of H7N9 virus resistant to neuraminidase inhibitors is concerning, since this virus is also naturally resistant to M2 ion channel blockers and no vaccine is yet available.
In this study, we applied global transcriptomic profiling to human cells infected with H7N9 Anhui01 or with other avian or human IAV to broadly characterize the host response to infection and quickly identify FDA-approved drugs that may act as antivirals. We show that H7N9 induces both a specific response and responses intermediate between those to H3N2 and those to avian H5N1 and H7N7. Notably, H7N9 induced a downregulation of the antigen presentation pathway and delayed proinflammatory cytokine induction to a greater extent than H5N1 and H7N7, which could have an important impact on in vivo immune responses. Similar to H3N2, H7N9 induced minor changes in eicosanoid signaling and genes implicated in chromatin modification. Finally, we computationally predicted that several FDA-approved drugs were able to reverse the host response to H7N9 and may be antivirals against this emerging virus. One of these drugs, minocycline, exhibited anti-H7N9 activity in vitro.
RESULTS
H7N9 replicates to levels similar to those of other avian or human IAVs in human bronchial epithelial cells.
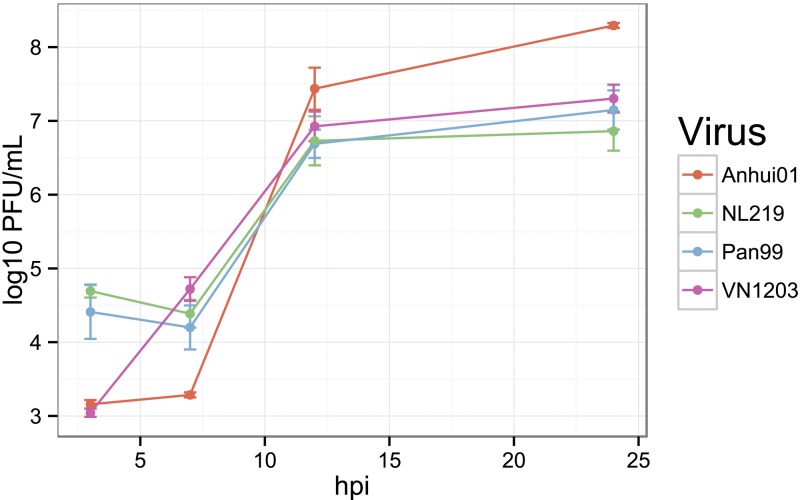
Polarized human bronchial epithelial cells (Calu-3) possess multiple features of in vivo airway epithelium and have been used previously as an in vitro model for multiple studies of influenza virus-host interaction (19–21) or for studying other respiratory viruses (22, 23). Here, polarized confluent monolayers of Calu-3 cells were infected apically with the avian-origin IAVs A/Anhui/01/2013 (H7N9) (Anhui01), A/Netherland/219/2003 (H7N7) (NL219), A/Vietnam/1203/2004 (H5N1) (VN1203), or a human seasonal virus, A/Panama/2007/1999 (H3N2) (Pan99) at a multiplicity of infection (MOI) of 1. Viral replication was assessed at 3, 7, 12, and 24 h postinoculation [hpi] by standard plaque assays. As shown in Fig. 1, H7N9 replicated to levels similar to those of H5N1 at 3 hpi, and to those of other IAVs at 12 hpi. Titers of H7N9 were slightly lower than those of other IAVs at 7 hpi but reached a slightly higher peak at 24 hpi, with an average number of PFU/ml of 8.3 (±0.04) for H7N9 and 7 (±0.3) for other IAVs. Cytopathic effects and destruction of the cell monolayer were observed at 24 hpi for human H3N2 and avian H7N7 IAVs and to a more limited extent after infection by H7N9 and H5N1 IAVs.
FIG 1 .
H7N9 replicates to a level similar to that of other influenza A viruses (IAVs) in polarized human bronchial epithelial Calu-3 cells. Polarized Calu-3 cells were infected apically with Anhui01 (H7N9), NL219 (H7N7), Pan99 (H3N2), or VN1203 (H5N1) at an MOI of 1 for an hour. After washing, medium was added and supernatants were collected at 3, 7, 12, and 24 h for determination of viral titers by standard plaque assay. Cells were harvested at the same time postinfection for transcriptomic profiling. Values represent means of titers in PFU/ml from quadruplicate wells ± SD.
The global transcriptomic response to H7N9 is specific and is intermediate between the cell responses to avian and human IAVs.
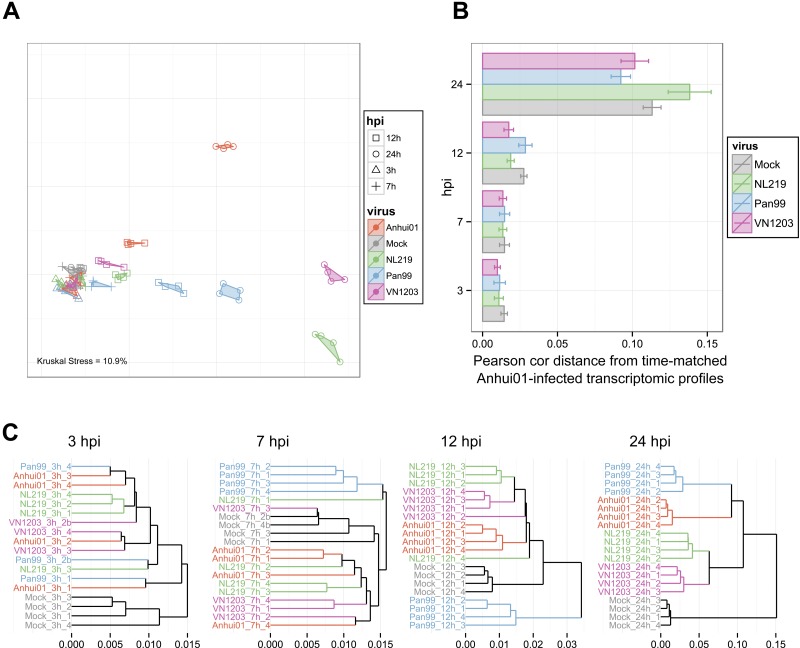
To assess the whole transcriptomic cellular response to IAV infection, we used several metrics (Fig. 2). First, multidimensional scaling (MDS) was used to visualize Euclidian distances between each transcriptomic profile in two dimensions (Fig. 2A). In this plot, each sample is represented by a dot; samples with similar transcriptomic profiles (i.e., a small Euclidian distance) are close together, while increasing distance is relative to increasing transcriptomic dissimilarity. We observed that samples clustered by time point and viral treatment, indicating a good agreement between biological replicates. At 3 and 7 hpi, IAV-infected samples were very similar to mock-infected ones (mocks), suggesting few transcriptomic changes early after infection, while the distance from mocks increased at 12 and 24 hpi. At 24 hpi, each viral condition was in a distinct location, indicating a specific response to each IAV. However, while avian H5N1 and H7N7 samples were relatively close together, H7N9 samples were located relatively far from each of the other IAVs, indicating a specific host response to this virus. To determine whether the host response to H7N9 infection was closer to the avian or human IAV response, we measured the transcriptomic distance between Anhui01-infected and other IAV-infected samples at each time point (Fig. 2B). At 12 hpi, H7N9 was significantly closer to both H7N7 and H5N1 than to H3N2. Surprisingly, at 24 hpi, the shortest distance was between H7N9 and H3N2, while the largest distance was between H7N9 and H7N7, indicating that responses to H7N7 and H7N9 were the most distinct. Of note, the response to H7N9 infection was as different from that of mock samples as that to avian IAV at 24 hpi, suggesting very specific response to H7N9 infection. Finally, to determine transcriptomic distances between all IAV- and mock-infected samples at each time point, we used hierarchical clustering (Fig. 2C). This analysis confirmed that H3N2 induced the largest host response early after infection and that at 24 hpi, responses to H7N7 and H5N1 were the most similar, while the response to H7N9 was closer to that to H3N2.
FIG 2 .
The host response to H7N9 infection is specific but closer to that to H3N2 than to the response to other avian-origin IAVs at late time points. (A) Similarities in transcriptomic responses are depicted using nonparametric multidimensional scaling (MDS). Each RNA sample is represented as a single point colored by viral treatment and with a different shape according to the time point. Euclidian distance was calculated using the whole normalized transcriptomic data, such that proximity indicates similarity, whereas distance indicates dissimilarity, of gene expression profiles. Kruskal’s stress quantifies the quality of the representations as a fraction of the information lost during the dimensionality reduction procedure. (B) Average distances between Anhui01-infected samples and time-matched infected or mock samples quantifying whole-transcriptome diversity after infection. Pearson correlation distance (Pearson cor distance) is defined as 1 − Pearson correlation coefficient calculated using normalized transcriptomic data. Error bars indicate SD. Similar results were obtained with Euclidian, Manhattan, or Spearman correlation distance (data not shown). (C) Hierarchical clustering by average linkage of IAV- and mock-infected samples at each time hpi based on their gene expression profiles. Distances were calculated using Pearson correlation distance, but similar clustering was observed with other distance metrics.
Altogether, these results show that the host response to H7N9 is highly specific to this virus but has globally more similarities with the response to H3N2 than to those to other IAVs at 24 hpi. Interestingly, although H7N9 and H7N7 have similar HA sequences, the host responses to these two viruses were very different.
Functional characterization of host responses specific to H7N9 and to human and avian IAVs.
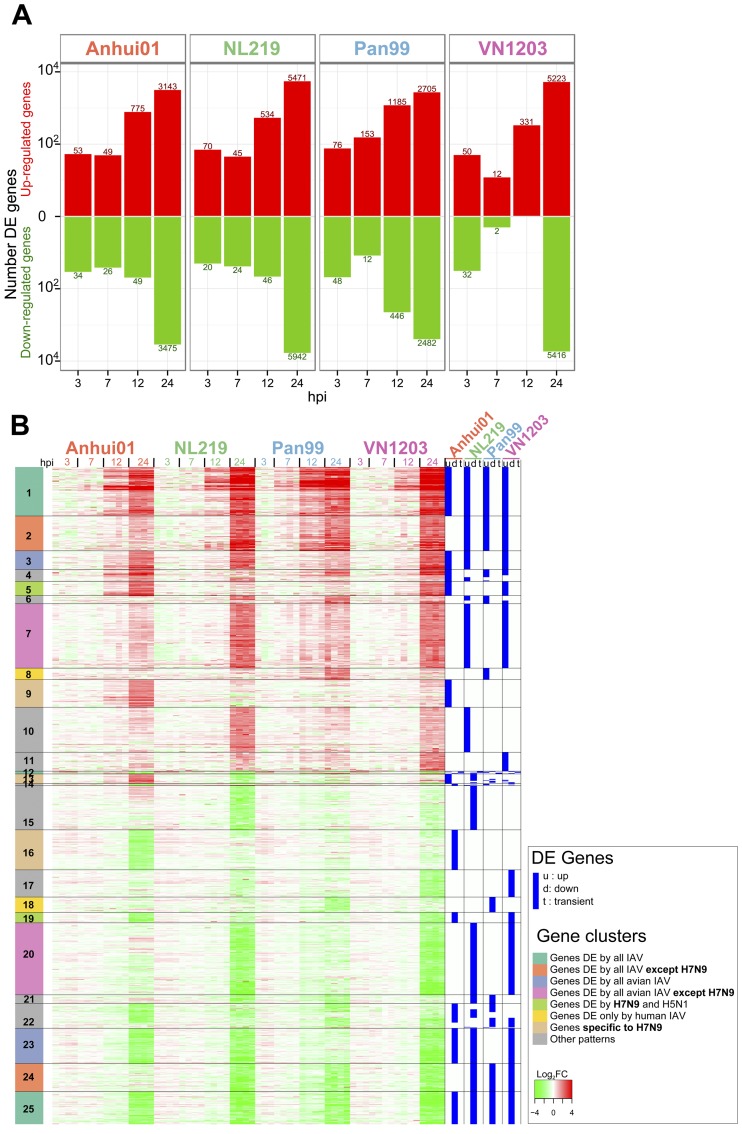
To further study specificities in the host response to each IAV, we identified genes differentially expressed (DE) between IAV-infected samples and mock-infected samples for each time point using the following criteria: a q value of <0.01, as determined by Limma’s empirical Bayes moderated t test, and a log2 fold change (log2FC) value of >1.5 (Fig. 3). Consistent with transcriptomic distance analysis, IAVs induced few transcriptomic changes at 3 and 7 hpi (Fig. 3A). At 12 hpi, the response to H3N2 infection was quite robust, with 1,631 DE genes, while there were only 824 DE genes after H7N9 infection and 580 and 331 for H7N7 and H5N1, respectively. At 24 hpi, all IAVs induced a drastic host response, with the number of DE genes ranging from 5,187 genes for H3N2 to 10,637 genes for H5N1 and 11,413 genes for H7N7. With 6,618 DE genes for H7N9 at 24 hpi, the amplitude of the host response to H7N9 at 24 hpi was closer to that for human than that for avian IAV (Fig. 3A). Therefore, we observed a more progressive host response to human H3N2 infection, whereas avian IAVs delayed the host response until 24 hpi, when a massive dysregulation of the host response was observed. The response to H7N9 was characterized by a number of DE genes intermediate between those of avian and human IAVs.
FIG 3 .
H7N9 induces a robust host response at 24 hpi that contains unique signatures and signatures in common with those induced by other avian and human IAVs. (A) Numbers of upregulated (red) and downregulated (green) differentially expressed (DE) genes after infection with IAV compared to time-matched mocks. Criteria used for differential expression analysis are a q value of <0.01 as determined by Limma’s empirical Bayes moderated t test and |log 2FC| > 1.5. (B) Heat map depicting the expression values of 16,327 genes DE after infection of at least one virus at one time point. For each viral condition, DE genes were classified as up- or downregulated across infection or with a transient expression for genes both up- and downregulated during the time course of infection. These DE profiles are shown in blue on the right of the heat map. Genes were clustered in 25 clusters by a constant height cut of the hierarchical clustering dendrogram of Euclidean distance of DE profiles. Functional enrichment for each cluster is presented in Table 1 and in Fig. S1 to S3 in the supplemental material.
In total, 16,327 genes were DE in at least one viral treatment and at one time point (Fig. 3B). For each gene, we defined whether the gene was up- or downregulated during the course of infection or, for a minority of genes, had a transient expression pattern. Using this DE profile, the 16,327 DE genes were grouped in 25 clusters and functional enrichment was performed for each cluster (Table 1; see also Fig. S1 to S3 in the supplemental material).
TABLE 1 .
DE gene cluster characterizationa
| Cluster | No. of genes | Description of regulation | Top GO BP | GO ES | Top IPA canonical pathway | Path. ES | Top regulator | Reg. ES |
|---|---|---|---|---|---|---|---|---|
| 1 | 1,218 | Up by all viruses | Type I interferon-mediated signaling pathway | 21.8 | Role of hypercytokinemia/ hyperchemokinemia in pathogenesis of influenza |
13.0 | IRF7 | 54.0 |
| 2 | 864 | Up by all but not significantly by H7N9 | Response to stimulus | 2.4 | Role of cytokines in mediating communication between immune cells | 4.1 | IRF7 | 6.1 |
| 3 | 465 | Up by all but not significantly by H3N2 | Negative regulation of T cell activation | 3.0 | cAMP-mediated signaling | 1.3 | EGOT | 1.8 |
| 4 | 297 | Up by H7N9 and 1 or 2 other viruses | Inflammatory response | 5.6 | IL-10 signaling | 6.4 | Poly(rI ⋅ rC) RNA | 15.1 |
| 5 | 345 | Up by H7N9 and H5N1 | Cellular response to granulocyte-macrophage colony-stimulating factor stimulus | 3.1 | HMGB1 signaling | 2.0 | Lipopolysac- charide |
5.6 |
| 6 | 205 | Up by H3N2 and H5N1 or H7N7 | Positive regulation of nitric oxide bio-synthetic process | 5.3 | Cross talk between dendritic cells and natural killer cells | 5.8 | Poly(rI ⋅ rC) RNA | 7.8 |
| 7 | 1,603 | Up by H5N1 and H7N7 | Positive regulation of transcription from RNA polymerase I promoter | 2.9 | Eicosanoid signaling | 2.3 | POU3F3 | 3.6 |
| 8 | 287 | Up by H3N2 only | Gamma interferon-mediated signaling pathway | 8.2 | Antigen presentation pathway | 7.7 | IFNG | 19.9 |
| 9 | 686 | Up by H7N9 only | Regulation of transcription, DNA dependent | 6.9 | Cyclins and cell cycle regulation | 3.0 | MLL | 5.5 |
| 10 | 1,105 | Up by H7N7 only | Genes | 4.3 | Nucleotide excision repair pathway | 2.3 | INPP1 | 2.9 |
| 11 | 474 | Up by H5N1 only | Arginine metabolic process | 2.6 | NFKB activation by virus | 1.6 | FRS2 | 4.4 |
| 12 | 73 | Genes up and then down | Regulation of immune response | 4.2 | Granulocyte adhesion and diapedesis | 3.1 | IL1 | 4.7 |
| 13 | 241 | Up by H7N9 but down by H7N7 or H5N1 | Regulation of transcription, DNA dependent | 5.8 | 1D-myoinositol hexakisphosphate biosynthesis V [from Ins(1,3,4)P3] | 1.4 | LTBP1 | 4.0 |
| 14 | 52 | Down by H7N9 but up by one other virus | Antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 10.9 | Antigen presentation pathway | 9.3 | CIITA | 4.8 |
| 15 | 1,091 | Down only by H7N7 | Regulation of transcription, DNA dependent | 12.0 | Role of NFAT in cardiac hypertrophy | 3.6 | Beta-estradiol | 4.6 |
| 16 | 1,002 | Down only by H7N9 | Proteolysis | 9.6 | Serotonin degradation | 5.6 | MYC | 7.6 |
| 17 | 676 | Down only by H5N1 | tRNA processing | 3.7 | Role of BRCA1 in DNA damage response | 3.0 | I kappa B kinase | 3.1 |
| 18 | 380 | Down only by H3N2 | Negative regulation of RNA polymerase II transcriptional pre-initiation complex assembly | 2.8 | TR/RXR activation | 2.8 | TRIM63 | 3.5 |
| 19 | 259 | Down by H7N9 and H5N1 | Intracellular protein transport | 3.6 | Role of BRCA1 in DNA damage response | 3.7 | CDH1 | 2.9 |
| 20 | 1,785 | Down by H7N7 and H5N1 | Regulation of transcription, DNA dependent | 16.9 | Glioma signaling | 2.0 | miR-143-3p (or w/ seed GAGAUGA) | 5.7 |
| 21 | 217 | Down by H7N7 and H3N2 | Sodium ion transport | 2.9 | Triacylglycerol biosynthesis | 2.3 | Cisplatin | 2.5 |
| 22 | 617 | Down by at least 2 viruses | Ethanol oxidation | 4.3 | Ethanol degradation IV | 5.0 | Meloxicam | 5.7 |
| 23 | 871 | Down by all but H3N2 | Transmembrane transport | 8.2 | GDP-mannose biosynthesis | 2.6 | mir-210 | 4.1 |
| 24 | 701 | Down by all but H7N9 | Regulation of transcription, DNA dependent | 12.8 | Factors promoting cardiogenesis in vertebrates | 3.7 | ASXL1 | 2.8 |
| 25 | 813 | Down by all viruses | Xenobiotic metabolic process | 5.2 | Alpha-tocopherol degradation | 3.5 | HNF1A | 6.3 |
Most-enriched gene ontology (GO) biological process (BP), IPA canonical pathway (path.), and top regulator (reg.) were reported for each cluster identified in Fig. 3. Enrichment scores (ES) were calculated as –log10 P values, using a right-tailed Fisher exact test.
More than 20% of the total DE genes were specific to H7N9 infection, with genes changing only in response to H7N9 (clusters 9, 13, and 16: 1,929 genes) or genes dysregulated by all IAVs except H7N9 (clusters 2 and 24: 1,565 genes). Genes specifically induced by H7N9 were broadly enriched among genes implicated in global regulation of gene transcription and the cell cycle (Table 1; see also Fig. S1 to S3 in the supplemental material). Interestingly, the top predicted regulator of cluster 9 was MLL, a histone methyltransferase involved in epigenetic regulation of diverse gene types associated with cell cycle regulation and development (Table 1). Regulation of transcription and the cell cycle by H7N9 could be related to specific interactions between H7N9 proteins and nuclear factors. Among the genes significantly upregulated by all IAVs except H7N9 (cluster 2) were several interferon (IFN) type 1 genes (IFN-A10, IFN-A16, IFN-A21, and IFNw1), IL-27, IL-32, and IL-12A (see Fig. S4A). Most other cytokines were induced by all IAVs and belonged to cluster 1. However, closer examination of the cytokine induction level revealed an earlier and stronger induction after H3N2 infection than with avian IAV, especially for CCL5, CXCL10, IFN-B1, IL-12A, IL-1A, and IL-6 (see Fig. S4A). Among avian IAVs, levels for some cytokines, including CCL5, IFN-A7, and IFN-B1, were significantly lower in response to H7N9 infection than to infection with H7N7 or H5N1 (see Fig. S4A).
More than 12% of the whole DE gene list was changed similarly in response to the H7N9, H7N7, and H5N1 viruses, including genes up- and downregulated similarly by each of these viruses (clusters 3 and 23: 1,336 genes) and genes similarly unchanged after avian IAV infection and dysregulated only after H3N2 infection (clusters 8 and 18: 667 genes). Cluster 8 was particularly interesting, since it included several genes from the antigen presentation pathway upregulated only after H3N2 infection, including those encoding class I major histocompatibility complex (MHC) molecules (HLA-A, HLA-B, and HLA-F) and transcription factors (CIITA and NLRC5). Some genes belonging to the antigen presentation pathway were also found in cluster 14 and were downregulated after H7N9 infection but upregulated in H3N2-infected cells, and these included genes encoding class II MHC molecules (HLA-DPA1, HLA-DPB1, HLA-DRA, HLA-DRB1, HLA-DRB4, HLA-DRB5, and CD74) (see Fig. S4B in the supplemental material).
Finally, more than 20% of the DE genes had similar profiles in H7N9 and H3N2 IAV versus H5N1 and H7N7 IAVs: these genes were significantly DE only after H5N1 and H7N7 infection (clusters 7 and 20: 3,388 genes). These genes could reflect adaptation of H7N9 to the human host, similar to findings for the seasonal H3N2 virus. Cluster 7 genes were significantly upregulated only in H5N1- and H7N7-infected samples and included genes from the eicosanoid signaling pathway (see Fig. S4C in the supplemental material). Upstream of this pathway, gene expression levels for PLA2G5 were 2 to 4 times more increased after H7N7 and H5N1 infection than after that with H3N2 and H7N9 at 24 hpi; PLA2G5 encodes a phospholipase A2 enzyme that catalyzes arachidonic acid production, a key inflammatory intermediate that is further catalyzed in different lipid mediators, regulating many biological processes, including inflammation and immune function. Several receptors of eicosanoids (LTB4R, TBXA2R, and PTGER3) were also more increased by H5N1 and H7N7 than by H7N9 or H3N2 IAVs. Forty-seven genes from cluster 20 were involved in chromatin modification, with the majority of genes involved in this biological process having similar profiles between H7N9 and H3N2 versus H5N1 and H7N7, but some genes were specifically upregulated only in H7N9-infected samples (cluster 9 and 13 genes) (see Fig. S5). This could reflect both H7N9-specific and human-IAV-like ability to modify expression of genes involved in chromatin and epigenetic regulation.
In conclusion, cluster analysis revealed an extensive list of functions dysregulated specifically by H7N9 or similar to findings for other IAVs, including proinflammatory cytokines, the antigen presentation pathway, and several nuclear pathways. Confirming transcriptomic distance analysis, there were more similarities between H5N1 and H7N9 responses, with 2 clusters specific to these viruses (clusters 5 and 19), than between H7N7 and H7N9, since there was no gene DE specifically in response to H7N9 and H7N7 only.
Transcriptome-based antiviral prediction.
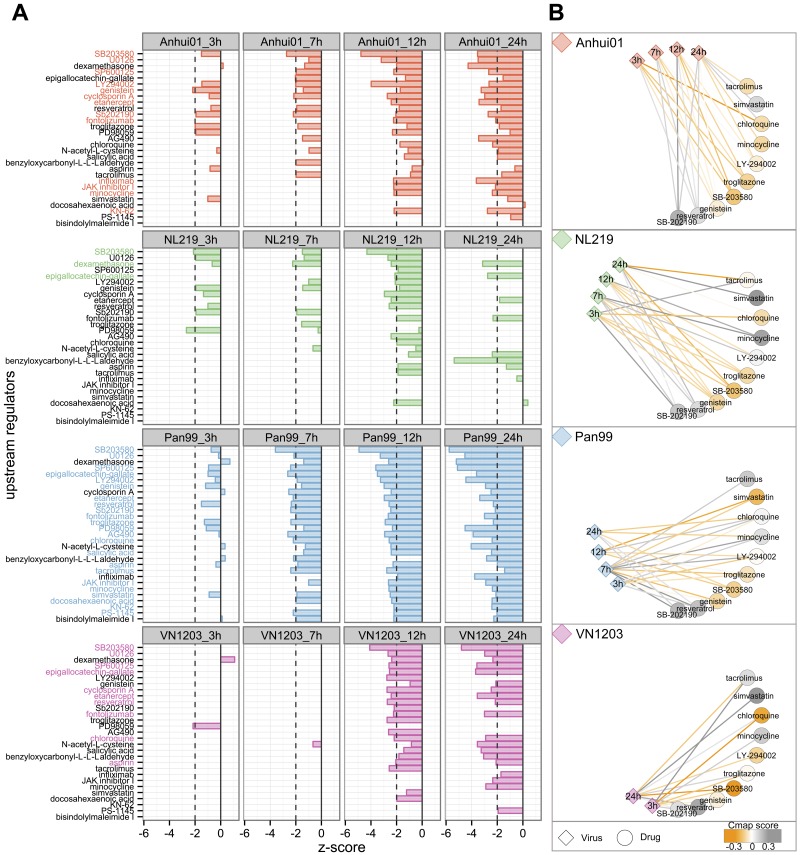
We have previously shown that drugs reversing the host response to IAV or coronavirus infection may inhibit viral replication (24, 25). In an effort to identify such potential antivirals that target the cellular response to these IAVs, we used two independent and complementary methods. The first method is a literature-based prediction using the Ingenuity Knowledge Database that identifies molecules known to regulate DE genes in an opposite direction (z score < −2) from that with IAV infection (Fig. 4A). For each IAV, potential antivirals were identified as upstream regulators having significant z scores on at least 2 time points and no positive z scores at any other time point. Only a few drugs were identified as potential anti-H7N7 therapies while 23 compounds were identified as potential regulators of the response to H3N2 IAV. Thirteen compounds were identified as negative regulators of the response to H7N9, including several kinases inhibitors (SB203580, U0126, SP600125, LY294002, genistein, SB202190, JAK inhibitor I and KN-32), immunosuppressive drugs (cyclosporine A, fontolizumab, infliximab, etanercept) and one antibiotic (minocycline). Other drugs that were predicted antivirals for H5N1 and H3N2, but with lower z score for H7N9, included chloroquine, aspirin and resveratrol.
FIG 4 .
Potential antiviral prediction based on transcriptomic profiles after IAV infection. (A) Prediction based on expression of known targets for molecules within the IPA database. A negative z score indicates that the regulator is known to downregulate the same genes that were significantly upregulated after infection and/or to upregulate genes that were downregulated after infection. Dashed lines depict the limit of significance (|z score| > 2). For each virus, z scores were calculated using log2FC expression of DE genes at each time point, and molecules with significant negative z scores for at least 2 time points were selected for this representation (29 drugs). Potential antivirals were defined as drugs with a z score < −2 for at least 2 time points and no positive z score (molecules highlighted in color). Twenty-six regulators were predicted to have an effect opposite to that of infection for at least one IAV. Molecules were ranked from most- to least-negative mean z score across all conditions. (B) The Connectivity Map (Cmap) was used to confirm potential anti-IAV effects of regulators predicted in IPA. Gene expression profiles for 10 molecules (out of the 26) were found in the Cmap database and compared with IAV-infected profiles. Cmap scores go from −1 to 1, with positive scores for drugs inducing changes similar to the viral signature and negative scores for opposite changes. Relationships between viral signatures and drugs were depicted in a network with a circular layout. Edges between virus and drugs are colored based on the Cmap scores comparing drug and viral profiles at each time point. Drugs are colored based on the mean of the Cmap scores for all time points. Drugs colored in orange induced gene expression changes that are the reverse of those for IAVs after cell treatment. Note that the Cmap query requires a list of up- and downregulated genes and was therefore not performed for VN1203 at 7 and 12 hpi, for which there were too few downregulated genes.
To confirm these predictions, we used a second independent method which is a data-driven prediction based on the Connectivity Map (Cmap). Cmap is a database of more than 7,000 gene expression profiles, representing 1,309 compounds, and we used it to identify drugs reverting IAV signatures (Cmap score close to −1) or inducing changes similar to those induced by IAVs (Cmap score close to 1) (Fig. 4B). Because Cmap profiles were generated using cell lines and array platforms different from those in this experiment, we used these results as a second line of validation, considering only drugs predicted in the Ingenuity Pathways Knowledge Base (IPA). Among the 26 compounds predicted to be upstream regulators of at least 1 IAV, 10 were found to be significant in Cmap. Relationships between these 10 drugs and each viral condition are depicted as a drug-virus network (Fig. 4B). These results confirmed that SB-203580 and genistein reverted the host response to all 4 IAVs. Conversely, SB-202190 and resveratrol were found to induce gene expression changes similar to those induced by IAV. Troglitazone and LY-294002 were predicted to reverse H7N9 signatures across infection but had positive Cmap scores for some viral treatment: H5N1 at 24 hpi for troglitazone and H3N2 and H7N7 at 7 hpi for LY-294002. Finally, some drugs had strain-specific effects, such as minocycline, predicted in both IPA and Cmap to negatively regulate H7N9 signatures, or simvastatin, which induced effects opposite to those of H3N2 only.
Altogether, a combination of two different approaches identified the kinase inhibitor SB-203580 and genistein as capable of reversing the host response to all tested IAVs, including H7N9. Additional molecules identified by both methods as reverting H7N9 signatures were troglitazone, minocycline, and LY-294002.
In vitro evaluation of minocycline antiviral activity.
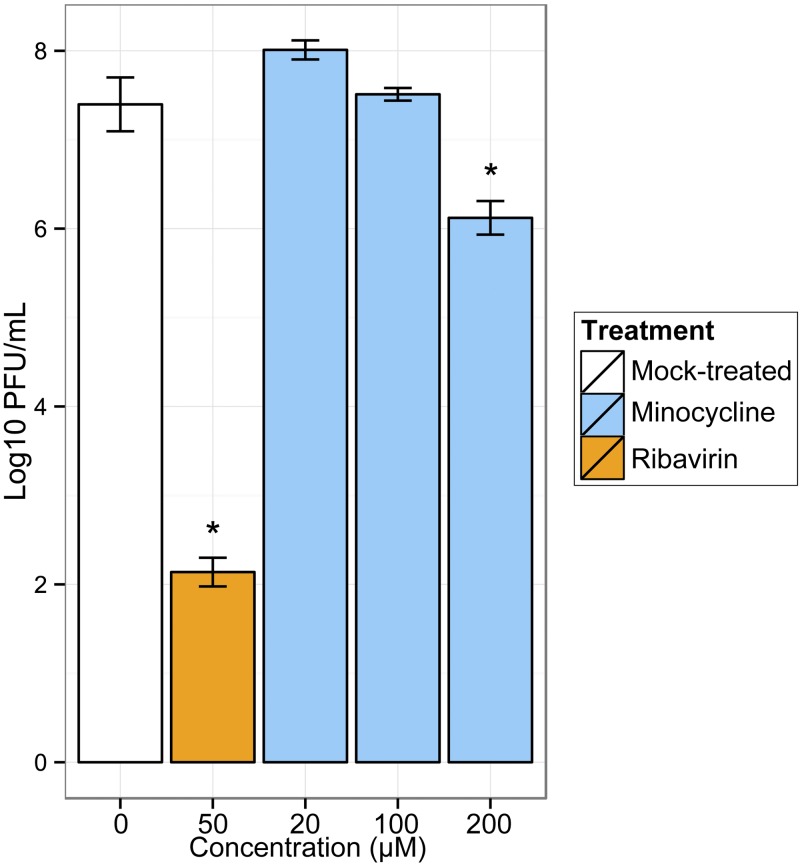
To validate our drug prediction, we evaluated the ability of minocycline, a tetracycline, to inhibit H7N9 viral replication in vitro. This molecule was chosen because it was predicted to reverse H7N9 signatures by both computational methods but had not been tested previously against influenza virus. Using a colorimetric viability assay, the 50% cytotoxicity concentration of minocycline on Calu-3 cells was evaluated to be around 1,000 µM, and we observed no significant cytotoxicity up to 266 µM (see Fig. S6 in the supplemental material). To evaluate potential antiviral effect of minocycline, polarized Calu-3 cells were pretreated with increasing concentrations of minocycline for 3 h and infected with Anhui01 at an MOI of 0.01 (Fig. 5). At 24 hpi, there was a significant decrease of 1.3 log10 (17%) of Anhui01 viral titers in cells treated with 200 µM minocycline (P < 0.01). Lower concentrations of minocycline did not inhibit Anhui01 viral replication. Ribavirin, an antiviral drug with in vitro activity against both DNA and RNA viruses, was used as a positive control and induced a more robust decrease of 5.3 log10 (71%) of the log10 viral titer (Fig. 5). Overall, these data show that minocycline exerts a modest but significant antiviral effect against H7N9, validating our in silico drug screening.
FIG 5 .
Antiviral activity of minocycline. Polarized Calu-3 cells were treated with increasing concentrations of minocycline for 3 h and infected with Anhui01 at an MOI of 0.01. Viral titers were determined at 24 hpi. Ribavirin at 50 µM was used as a reference. Values represent means of titers in PFU/mL from triplicate wells ± SD. *, P < 0.01 (Student’s t test).
DISCUSSION
Although the H7N9 outbreak has entered a stationary state in China, with only a few cases reported since late May 2013, there are concerns that H7N9 may reappear in the winter. Other avian IAVs, such as H5N1, indeed have a seasonal pattern, with an increase in human infection during the winter. In addition, H7N9 is likely still circulating in poultry. Several studies have shown that H7N9 possess genetic markers associated with adaptation to humans, allowing higher replication in mammal cells (PB2 E627K) and increased binding of α2-6 human receptors (HA Q226L and G186V). Further adaptation of H7N9 to humans is of concern, since it could lead to efficient human-to-human transmission. In this study, we have shown that cell responses to H7N9 infection are more similar to those to H3N2 infection than to those to infection with avian H5N1 or H7N7 viruses, which implies that H7N9 has adapted to the human host. Some of the human IAV-like transcriptomic responses of H7N9 were related to similar lower induction of several genes involved in the eicosanoid pathway. Some lipid mediators from these pathways were recently identified as correlates of the pathogenic or resolution phase of IAV infection in mice (26) and had antiviral effect in cells and mice (27). Our analysis revealed that the eicosanoid pathway is differentially regulated by avian and human IAVs, with a potential impact on lung inflammation and viral replication.
Importantly, differences in the transcriptomic response to infection were not related to major differences in replication between viruses. We previously observed higher replication of the Anhui01 H7N9 strain than of the seasonal A/Texas/50/2012 (H3N2) virus in Calu-3 cells (14). However, at this higher MOI and using the more replicative A/Panama/2004/99 (H3N2), similar replication between avian and human IAVs was observed.
Surprisingly, the most distinct response was between H7N9 and H7N7, despite genetic similarities between HA segments. Therefore, analysis of viral sequences alone cannot predict the host response to infection. Analysis between HA and human receptors has revealed that H7N9 has a profile intermediate between those of human and avian IAVs. Here, we observed that the host response to H7N9 shared some similarities with the response to human and avian IAVs, but we also observed a large amount of specificity in the cell response to H7N9 infection. Genes significantly induced by H7N9 were involved in cell cycle regulation, transcription, and epigenetic modifications. Consistent with findings of a previous study in polarized Calu-3 cells (19), we noted that avian IAVs induced a weaker proinflammatory cytokine response than human IAV. For genes coding for type I IFN and some other cytokines, this reduced cytokine induction was even more striking in H7N9-infected than in H5N1- or H7N7-infected samples. The ability to delay the antiviral host response could also have a major impact on in vivo pathogenesis of H7N9 human infection. Finally, genes coding for antigen presentation pathway proteins were upregulated only in H3N2-infected cells, while the expression of these genes was either unchanged by avian IAVs or downregulated by H7N9 virus. Notably, downregulation of the antigen presentation pathway was also observed in nonpolarized Calu-3 cells infected with avian VN1203 H5N1 virus while upregulated following human A/California/04/2009 (H1N1) infection or alpha IFN (IFN-α) treatment (V. D. Menachery, A. J. Eisfeld, L. Josset, A. C. Sims, M. G. Katze, Y. Kawaoka, and R. S. Baric, unpublished data). Therefore, downregulation or absence of induction of antigen presentation in infected epithelial cells is a characteristic of avian versus human IAVs. Given concerns about the immunogenicity of avian H5N1 and H7N9 viruses, mechanisms of antigen presentation pathway control and its implication in vivo warrant further exploration.
Because no vaccine for the prevention of H7N9 infections is currently available, prophylaxis and treatment rely on antivirals. Studies in mice (28) and in a patient (29) have shown low efficacy of NA inhibitors for H7N9 morbidity. Moreover, emergence of H7N9 virus resistant to NA inhibitors has been observed (18), and one of the resistance mutations (NA-R292K) induced high-level oseltamivir resistance without impairing viral replication and in vivo virulence (30). Finally, ribavirin, which shows in vitro activity against influenza viruses, including H7N9, is not currently approved for the treatment of influenza and has controversial efficacy in vivo (31). Developing alternative antiviral therapeutics is therefore a priority. Genome-based drug repurposing strategies relying on the identification of drugs reversing cell responses to viral infection have successfully identified antivirals against IAVs (25) and coronavirus (24). These strategies have the advantage of being able to screen a large number of available drugs in silico and potentially accelerate their use in clinics. Here, we used a combination of two complementary methods, a knowledge-based approach (using IPA) prioritizing drugs with published effects that are opposite to IAV-induced gene expression changes, and a data-based approach using drug gene expression profiles present in Cmap to determine whether drug treatment induced transcriptomic changes opposite to viral infection. Twenty-six molecules were predicted by the first method to modulate the host response to at least one IAV, and out of the 10 present in Cmap, 8 were found to induce gene expression changes opposite to IAV signatures after multiple cell line treatment. Drugs that were predicted by both methods for H7N9 were the kinase inhibitors SB203580, genistein, LY-294002, and the antibiotic minocycline. In addition, the antidiabetic drug troglitazone, which was just below our threshold for IPA prediction, was found in Cmap to significantly revert H7N9 signatures at 3, 7, and 12 hpi.
Importantly, we evaluated the anti-H7N9 activity of minocycline, a tetracycline with antimicrobial action, and independent anti-inflammatory and neuroprotective properties. We observed that minocycline significantly inhibited H7N9 replication in vitro. Minocycline was previously shown to inhibit simian immunodeficiency virus and human immunodeficiency virus (SIV and HIV) replication in vitro and in vivo and to reduce SIV-induced encephalitis severity in macaques (32). Minocycline was also shown to inhibit West Nile virus replication in vitro, possibly by inhibiting Jun N-terminal protein kinase (JNK) induction (33), and to protect mice against Japanese encephalitis virus infection (34). This study is the first, to our knowledge, to show that minocycline also inhibits influenza virus replication in vitro. Moreover, this assay validates our in silico drug prediction. Minocycline is an interesting candidate to consider for in vivo testing, since it is FDA approved, readily available, and well tolerated. In addition to minocycline’s antiviral activity, its neuroprotective effect may potentially prevent influenza-associated neurological complications.
Our in silico drug prediction was further validated by literature mining, revealing that out of the 26 drugs predicted to have an effect on at least one IAV in the present study, 17 have already been tested against influenza in cell or mouse models and 14 had antiviral effects in those tests (see Table S1 in the supplemental material). Among the 3 drugs that did not show any effect on influenza virus replication or pathogenicity, dexamethasone was predicted only to be a potential anti-H7N7 agent, since it had a positive z score for H7N9, H3N2, and H5N1 infections at early time points and could therefore increase early viral replication. This could be linked to the controversial role of corticosteroids in patients with influenza acute respiratory distress syndrome (ARDS), for which very early corticosteroid therapy may be harmful (35). Most likely, several drugs predicted here that had previously known anti-IAV effects may have broad antiviral effects and could be active against H7N9. FDA-approved drugs that are already used in clinics for diverse indications but not influenza and that were identified in our in silico screening and with previously published anti-influenza effects, such as chloroquine (36), are particularly interesting to consider for therapeutic intervention against H7N9.
The in vivo pulmonary host response and outcome for influenza infection cannot be determined by directly transposing transcriptomic profiles from single cell types. The whole-lung response to infection depends on the interaction between infected cells, immune cells, and the lung microenvironment. In addition, the host response to influenza infection depends on the cell type and differentiation levels of infected cells (37). We have highlighted in a recent review that novel mathematical models are needed to model interactions between cells and to integrate in vitro and in vivo studies (38). By using a multivariate modeling approach, McDermott et al. were able to predict pulmonary expression of conserved regulatory modules in macaques and mice in response to H5N1 infection from a regulatory model built from Calu-3 cells infected with H5N1 (39). In addition, numerous studies have shown that cell cultures are extremely useful experimental models for extended influenza virus-host interaction studies and for the screening of potential antivirals. In particular, polarized Calu-3 cells were shown to be valuable models for the study of influenza virus and supported viral replication and type I IFN responses similarly to primary human bronchial epithelial (HBE) cells (19). Culture systems of differentiated primary epithelial cells from humans, like HBE cells, recapitulate more closely the morphological features of the human upper airway but provide a less convenient and reproducible model for the study of influenza virus because of experiment, passage, and donor variations, inducing a variable differentiated phenotype (40). In this study, we used Calu-3 cell cultures to rapidly characterize the emerging H7N9 virus by comparing its response to several avian and human influenza viruses and to perform in silico screening for potential antivirals. We believe that this data set is a valuable resource for further studies of H7N9 and other influenza viruses. In two ongoing studies, we have been profiling mouse and macaque pulmonary responses to H7N9 infection (J. Morrison, L. Josset, N. Tchitchek, J. Chang, J. Belser, T. Tumpey, and M. G. Katze, submitted for publication; E. de Wit, A. Rasmussen, F. Feldmann, T. Bushmaker, C. Martellaro, E. Haddock, A. Okumura, S. Proll, J. Chang, D. Gardner, M. G. Katze, and V. J. Munster, submitted for publication). Importantly, we found that a significant number of drugs predicted to reverse Calu-3 cell responses to H7N9, including minocycline, SB203580, KN-62, and Jak Inhibitor 1, were also predicted to reverse the mouse and macaque pulmonary response to H7N9 infection (Morrison et al., submitted). This demonstrates conserved functional responses between these models.
In conclusion, high-throughput profiling of the cellular response to H7N9 infection and comparison to those to other avian IAVs or a seasonal human H3N2 virus revealed that H7N9 induced both specific responses and responses in common with avian or human IAVs. Transcriptomic profiles similar to those of H7N9 and H3N2 indicate potential human adaptation of H7N9, and further evolution toward the response to human IAVs could be surveyed with novel H7N9 isolates. Our analysis pointed out several FDA-approved drugs, including minocycline, as potential anti-H7N9 drugs that would be important to explore in vivo.
MATERIALS AND METHODS
Viruses.
Three avian-origin IAVs isolated from fatal human cases, A/Anhui/01/2013 (H7N9) (Anhui01), A/Netherland/219/2003 (H7N7) (NL219), and A/Vietnam/1203/2004 (H5N1) (VN1203), and a seasonal human virus, A/Panama/2007/1999 (H3N2) (Pan99), were used in this study. Virus stocks were grown in the allantoic cavities of 10-day-old embryonated hen’s eggs for 24 to 28 h at 37°C for avian-origin IAVs or for 48 h at 34°C for the H3N2 virus. Allantoic fluids from multiple eggs were pooled, clarified by centrifugation, aliquoted, and stored at −70°C. Virus titers were determined by a plaque assay as previously described (19). All research with avian viruses was conducted under biosafety level 3 containment, including enhancements required by the U.S. Department of Agriculture and the Select Agent Program (http://www.cdc.gov/od/ohs/biosfty/bmbl5/bmbl5toc.htm).
Cell culture and viral infection.
The human bronchial epithelial cell line Calu-3 was originally derived from subbronchial epithelium of a lung adenocarcinoma (ATCC), and cells were grown as previously described (19). Confluent monolayers of polarized Calu-3 cells achieving stable transepithelial resistance were infected apically at an MOI of 1. After a 1-h incubation, monolayers were washed to remove nonadherent virus and 2 ml of minimal essential medium-bovine serum albumin (MEM-BSA) was added to both apical and basolateral reservoirs of cells and left for the duration of the experiment.
RNA isolation and microarray processing.
RNA extraction from IAV- and mock-infected Calu-3 cells in quadruplicate was performed as previously described (41). Probe labeling and microarray slide hybridization for each biological replicate were performed using a SurePrint G3 Human Gene Expression 8×60K v2 microarray (Agilent Technologies) according to the manufacturer’s instructions. Slides were scanned on an Agilent DNA microarray scanner (model G2505B) using the XDR setting, and raw images were analyzed using the Agilent Feature Extraction software program (version 9.5.3.1). Extracted raw data were background corrected using the norm-exp method with an offset of 1 and quantile normalized using the limma software package (42) in the R environment. Replicated probes were mean summarized. All probes were required to pass Agilent quality control (QC) flags (“gIsFound,” “gIsWellAboveBG,” “gIsSaturated,” “gIsFeatNonUnifOL,” and “gIsFeatPopnOL”) for all replicates of at least one infected time point (29,382 probes passed QC filtering). For each sample, a log2FC value was calculated as the difference between log2-normalized data for this sample and the average of log2-normalized data for time-matched mocks. Microarray annotation was retrieved from the GEO data bank (GEO accession no. GPL17077).
Statistical analysis.
Differential expression was determined by comparing IAV-infected replicates to time-matched mocks, based on a linear model fit for each probe using the R software package “limma” (42). Criteria for differential expression were an absolute log 2FC of 1.5 and a q value of <0.01 calculated using a moderated t test with subsequent Benjamini-Hochberg correction. This controls the false discovery rate (FDR) associated with our results to 1%. For each viral condition, genes were classified as up- or downregulated throughout infection if they were found significantly up- or downregulated at at least one time point. Genes that were found to be both up- and downregulated at different time points were classified as genes with transient expression. A binary DE matrix with 3 columns per viral treatment (gene up- or downregulated or transient genes) and values equal to 1 for DE and 0 for not DE was used to cluster DE genes in 25 clusters by Euclidian distance and constant height cut of a hierarchical clustering dendrogram (43).
Transcriptomic distance analysis.
To visualize transcriptional similarity between samples in a 2-dimensional space, Euclidian distances were calculated with transcriptomic normalized data, and nonmetric multidimensional scaling (MDS) was performed using the MASS software package in the R Bioconductor program (44) to map the distances into the 2-dimensional space with minimal loss of information (evaluated by Kruskal’s stress). Biological replicates from the same condition were linked in convex hulls (i.e., polygons) using the function “chull” in R. Pearson correlation coefficients, Spearman correlation coefficients, total Euclidean distances, or total Manhattan distances were used to estimate similarities in the gene expression profiles between Anhui01-infected and other IAV- or mock-infected samples. The average distance between conditions was calculated as the arithmetic mean of distances between all biological replicates.
Functional enrichment and upstream regulator analysis.
Functional analysis of statistically significant gene expression changes was performed using the Ingenuity Pathways Knowledge Base (IPA; Ingenuity Systems). For all gene set enrichment analyses, a right-tailed Fisher exact test was used to calculate a P value determining the probability that each biological function assigned to that data set was due to chance alone. All enrichment scores were calculated in IPA using the probes that passed our QC filter as the background data set.
Upstream regulator analysis, which was used to predict regulators and infer their activation state, is based on prior knowledge of expected effects between regulators and their known target genes according to the IPA database. A z score is calculated and determines whether gene expression changes for known targets of each regulator are consistent with what is expected from the literature (z score > 2) or if the changes are anticorrelated with the literature (z score < −2). To predict potential antivirals, we queried the IPA database with the list of DE genes at each time point for each IAV, and we selected the molecules annotated “chemical drug,” “biologic drug,” “chemical—kinase inhibitor,” or “chemical—protease inhibitor” having a z score of <−2 on at least 2 time points and no positive z score at other time points.
Connectivity map.
To confirm whether drugs identified in IPA induce signatures opposite to those induced by IAVs, we used the publicly available Connectivity Map (Cmap) database (build 02) (45). The Cmap is a collection of genome-wide transcriptional data from cultured human cells treated with 1,309 different compounds. We used the lists of DE genes up- and downregulated at each time point under each viral condition, limiting to the top 250 most up- and downregulated genes for lists containing a higher number of genes. Agilent probes were mapped to Affymetrix U133A probe sets using the BioMart ID converter tool (46) in order to query the Cmap database. Ten drugs out of the 26 potential antivirals identified in IPA were found in the Cmap database. We analyzed permuted results for drugs having consistent mean Cmap scores and enrichment scores (i.e., both scores were positive or both were negative). Relationships between drugs and viral signatures were depicted in a network using Cytoscape software and a circular layout (47).
Antiviral in vitro assay.
Minocycline hydrochloride (Sigma) and ribavirin (Sigma) were dissolved in dimethyl sulfoxide (DMSO). To assay minocycline cytotoxicity, Calu-3 cells were grown in 96-well plates treated with increasing concentrations of minocycline for 24 h before incubation with the WST-1 reagent (Roche) at 37°C for 1 h. A microplate spectrophotometer was used to measure the absorbance of the samples at 450 nm with 600 nm as the reference wavelength. For the antiviral assay, 1-week-old polarized Calu-3 cells were treated with drugs for 3 h and infected with Anhui01 at an MOI of 0.01 for 1 h. The inoculant was washed away, and cells were incubated with drugs for the duration of the experiment. At 24 hpi, the apical supernatants from infected Calu-3 cells were collected and titers were determined. Experiments were performed in triplicate.
Microarray data accession number.
Raw microarray data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE49840. In addition, normalized matrixes and expression values utilized in this study can be found at https://viromics.washington.edu.
SUPPLEMENTAL MATERIAL
Top 5 pathways enriched in each cluster. Download
Top 5 GO biological processes (BPs) enriched in each cluster. Download
Top 5 regulators enriched in each cluster. Download
Temporal gene expression changes for genes belonging to the antigen presentation pathway (A), cytokines (B), or eicosanoid signaling (C). (A) Twelve genes belonging to the antigen presentation pathway specifically downregulated after H7N9 infection (cluster 14) or upregulated after infection with H3N2 only (genes from cluster 8). (B) Cytokines from cluster 1 or 2. (C) Genes from cluster 7 belonging to the eicosanoid pathway. Download
Heat map of gene expression levels for genes belonging to the chromatin modification biological process (GO:0016568). Download
Calu-3 cell viability following incubation with increasing concentrations of minocycline. Download
Literature mining for in vitro and/or in vivo assays of the 26 predicted drugs for IAV infection
ACKNOWLEDGMENTS
We thank J. J. Treanor and D. J. Topham at the University of Rochester for scientific discussions about the data and S. E. Belisle and M. J. Korth for valuable feedback on the manuscript.
This project has been funded in whole or in part with federal funds from the NIH, NIAID network of Centers of Excellence in Influenza Research and Surveillance (CEIRS), under contract HHSN266200700008C.
The findings and conclusions in this report are ours and do not necessarily reflect the views of the funding agency.
Footnotes
Citation Josset L, Zeng H, Kelly SM, Tumpey TM, Katze MG. 2014. Transcriptomic characterization of the novel avian-origin influenza A (H7N9) virus: specific host response and responses intermediate between avian (H5N1 and H7N7) and human (H3N2) viruses and implications for treatment options. mBio 5(1):e01102-13. doi:10.1128/mBio.01102-13.
REFERENCES
- 1. WHO 2013. Number of confirmed human cases of avian influenza A(H7N9) reported to WHO, global alert and response (GAR). WHO, Geneva, Switzerland [Google Scholar]
- 2. Gao HN, Lu HZ, Cao B, Du B, Shang H, Gan JH, Lu SH, Yang YD, Fang Q, Shen YZ, Xi XM, Gu Q, Zhou XM, Qu HP, Yan Z, Li FM, Zhao W, Gao ZC, Wang GF, Ruan LX, Wang WH, Ye J, Cao HF, Li XW, Zhang WH, Fang XC, He J, Liang WF, Xie J, Zeng M, Wu XZ, Li J, Xia Q, Jin ZC, Chen Q, Tang C, Zhang ZY, Hou BM, Feng ZX, Sheng JF, Zhong NS, Li LJ. 2013. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N. Engl. J. Med. 368:2277–2285. 10.1056/NEJMoa1305584 [DOI] [PubMed] [Google Scholar]
- 3. Chen Y, Liang W, Yang S, Wu N, Gao H, Sheng J, Yao H, Wo J, Fang Q, Cui D, Li Y, Yao X, Zhang Y, Wu H, Zheng S, Diao H, Xia S, Zhang Y, Chan KH, Tsoi HW, Teng JL, Song W, Wang P, Lau SY, Zheng M, Chan JF, To KK, Chen H, Li L. 2013. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet 381:1916–1925. 10.1016/S0140-6736(13)60903-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Belser JA, Bridges CB, Katz JM, Tumpey TM. 2009. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg. Infect. Dis. 15:859–865. 10.3201/eid1506.090072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yu H, Cowling BJ, Feng L, Lau EH, Liao Q, Tsang TK, Peng Z, Wu P, Liu F, Fang VJ, Zhang H, Li M, Zeng L, Xu Z, Li Z, Luo H, Li Q, Feng Z, Cao B, Yang W, Wu JT, Wang Y, Leung GM. 2013. Human infection with avian influenza A H7N9 virus: an assessment of clinical severity. Lancet 382:138–145. 10.1016/S0140-6736(13)61207-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. WHO 2011. Update on human cases of highly pathogenic avian influenza A(H5N1) virus infection, 2010. Wkly. Epidemiol. Rec. 86:161–166 http://www.who.int/wer/2011/wer8617.pdf [PubMed] [Google Scholar]
- 7. Cowling BJ, Jin L, Lau EH, Liao Q, Wu P, Jiang H, Tsang TK, Zheng J, Fang VJ, Chang Z, Ni MY, Zhang Q, Ip DK, Yu J, Li Y, Wang L, Tu W, Meng L, Wu JT, Luo H, Li Q, Shu Y, Li Z, Feng Z, Yang W, Wang Y, Leung GM, Yu H. 2013. Comparative epidemiology of human infections with avian influenza A H7N9 and H5N1 viruses in China: a population-based study of laboratory-confirmed cases. Lancet 382:129–137. 10.1016/S0140-6736(13)61171-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Evans ME, Hall KL, Berry SE. 1997. Influenza control in acute care hospitals. Am. J. Infect. Control 25:357–362. 10.1016/S0196-6553(97)90029-8 [DOI] [PubMed] [Google Scholar]
- 9. ECDC 2013. Seasonal influenza 2012/2013 in Europe. Risk assessment. ECDC, Stockholm, Sweden [Google Scholar]
- 10. Ip DK, Liao Q, Wu P, Gao Z, Cao B, Feng L, Xu X, Jiang H, Li M, Bao J, Zheng J, Zhang Q, Chang Z, Li Y, Yu J, Liu F, Ni MY, Wu JT, Cowling BJ, Yang W, Leung GM, Yu H. 2013. Detection of mild to moderate influenza A/H7N9 infection by China’s national sentinel surveillance system for influenza-like illness: case series. BMJ 346:f3693. 10.1136/bmj.f3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M. 2013. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 18:20453 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20453 [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Q, Lu L, Sun Z, Chen GW, Wen Y, Jiang S. 2013. Genomic signature and protein sequence analysis of a novel influenza A (H7N9) virus that causes an outbreak in humans in China. Microbes Infect. 15:432–439. 10.1016/j.micinf.2013.04.004 [DOI] [PubMed] [Google Scholar]
- 13. Jonges M, Meijer A, Fouchier RA, Koch G, Li J, Pan JC, Chen H, Shu YL, Koopmans MP. 2013. Guiding outbreak management by the use of influenza A(H7Nx) virus sequence analysis. Euro Surveill. 18:20460 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20460 [PubMed] [Google Scholar]
- 14. Belser JA, Gustin KM, Pearce MB, Maines TR, Zeng H, Pappas C, Sun X, Carney PJ, Villanueva JM, Stevens J, Katz JM, Tumpey TM. 2013. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature 501:556–559. 10.1038/nature12391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhu H, Wang D, Kelvin DJ, Li L, Zheng Z, Yoon SW, Wong SS, Farooqui A, Wang J, Banner D, Chen R, Zheng R, Zhou J, Zhang Y, Hong W, Dong W, Cai Q, Roehrl MH, Huang SS, Kelvin AA, Yao T, Zhou B, Chen X, Leung GM, Poon LL, Webster RG, Webby RJ, Peiris JS, Guan Y, Shu Y. 2013. Infectivity, transmission, and pathology of human-isolated H7N9 influenza virus in ferrets and pigs. Science 341:183–186. 10.1126/science.1239844 [DOI] [PubMed] [Google Scholar]
- 16. Xiong X, Martin SR, Haire LF, Wharton SA, Daniels RS, Bennett MS, McCauley JW, Collins PJ, Walker PA, Skehel JJ, Gamblin SJ. 2013. Receptor binding by an H7N9 influenza virus from humans. Nature 499:496–499. 10.1038/nature12372 [DOI] [PubMed] [Google Scholar]
- 17. Tharakaraman K, Jayaraman A, Raman R, Viswanathan K, Stebbins NW, Johnson D, Shriver Z, Sasisekharan V, Sasisekharan R. 2013. Glycan receptor binding of the influenza A virus H7N9 hemagglutinin. Cell 153:1486–1493. 10.1016/j.cell.2013.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu Y, Lu S, Song Z, Wang W, Hao P, Li J, Zhang X, Yen HL, Shi B, Li T, Guan W, Xu L, Liu Y, Wang S, Tian D, Zhu Z, He J, Huang K, Chen H, Zheng L, Li X, Ping J, Kang B, Xi X, Zha L, Li Y, Zhang Z, Peiris M, Yuan Z. 2013. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet 381:2273–2279. 10.1016/S0140-6736(13)61125-3 [DOI] [PubMed] [Google Scholar]
- 19. Zeng H, Goldsmith C, Thawatsupha P, Chittaganpitch M, Waicharoen S, Zaki S, Tumpey TM, Katz JM. 2007. Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type I interferon response in polarized human bronchial epithelial cells. J. Virol. 81:12439–12449. 10.1128/JVI.01134-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun X, Whittaker GR. 2007. Role of the actin cytoskeleton during influenza virus internalization into polarized epithelial cells. Cell. Microbiol. 9:1672–1682. 10.1111/j.1462-5822.2007.00900.x [DOI] [PubMed] [Google Scholar]
- 21. Belser JA, Zeng H, Katz JM, Tumpey TM. 2011. Infection with highly pathogenic H7 influenza viruses results in an attenuated proinflammatory cytokine and chemokine response early after infection. J. Infect. Dis. 203:40–48. 10.1093/infdis/jiq018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tseng CT, Tseng J, Perrone L, Worthy M, Popov V, Peters CJ. 2005. Apical entry and release of severe acute respiratory syndrome-associated coronavirus in polarized Calu-3 lung epithelial cells. J. Virol. 79:9470–9479. 10.1128/JVI.79.15.9470-9479.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harcourt JL, Caidi H, Anderson LJ, Haynes LM. 2011. Evaluation of the Calu-3 cell line as a model of in vitro respiratory syncytial virus infection. J. Virol. Methods 174:144–149. 10.1016/j.jviromet.2011.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Josset L, Menachery VD, Gralinski LE, Agnihothram S, Sova P, Carter VS, Yount BL, Graham RL, Baric RS, Katze MG. 2013. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio 4(3):e00165-13. 10.1128/mBio.00165-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Josset L, Textoris J, Loriod B, Ferraris O, Moules V, Lina B, N’Guyen C, Diaz JJ, Rosa-Calatrava M. 2010. Gene expression signature-based screening identifies new broadly effective influenza A antivirals. PLoS One 5:e13169. 10.1371/journal.pone.0013169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tam VC, Quehenberger O, Oshansky CM, Suen R, Armando AM, Treuting PM, Thomas PG, Dennis EA, Aderem A. 2013. Lipidomic profiling of influenza infection identifies mediators that induce and resolve inflammation. Cell 154:213–227. 10.1016/j.cell.2013.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morita M, Kuba K, Ichikawa A, Nakayama M, Katahira J, Iwamoto R, Watanebe T, Sakabe S, Daidoji T, Nakamura S, Kadowaki A, Ohto T, Nakanishi H, Taguchi R, Nakaya T, Murakami M, Yoneda Y, Arai H, Kawaoka Y, Penninger JM, Arita M, Imai Y. 2013. The lipid mediator protectin D1 inhibits influenza virus replication and improves severe influenza. Cell 153:112–125. 10.1016/j.cell.2013.02.027 [DOI] [PubMed] [Google Scholar]
- 28. Watanabe T, Kiso M, Fukuyama S, Nakajima N, Imai M, Yamada S, Murakami S, Yamayoshi S, Iwatsuki-Horimoto K, Sakoda Y, Takashita E, McBride R, Noda T, Hatta M, Imai H, Zhao D, Kishida N, Shirakura M, de Vries RP, Shichinohe S, Okamatsu M, Tamura T, Tomita Y, Fujimoto N, Goto K, Katsura H, Kawakami E, Ishikawa I, Watanabe S, Ito M, Sakai-Tagawa Y, Sugita Y, Uraki R, Yamaji R, Eisfeld AJ, Zhong G, Fan S, Ping J, Maher EA, Hanson A, Uchida Y, Saito T, Ozawa M, Neumann G, Kida H, Odagiri T, Paulson JC, Hasegawa H, Tashiro M, Kawaoka Y. 2013. Characterization of H7N9 influenza A viruses isolated from humans. Nature 501:551–555. 10.1038/nature12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu X, Li T, Zheng Y, Wong K-W, Lu S, Lu H. 2013. Poor responses to oseltamivir treatment in a patient with influenza A (H7N9) virus infection. Emerg. Microbes Infect. 2:e27. 10.1038/emi.2013.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hai R, Schmolke M, Leyva-Grado VH, Thangavel RR, Margine I, Jaffe EL, Krammer F, Solórzano A, García-Sastre A, Palese P, Bouvier NM. 2013. Influenza A(H7N9) virus gains neuraminidase inhibitor resistance without loss of in vivo virulence or transmissibility. Nat. Commun 4:2854. 10.1038/ncomms3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chan-Tack KM, Murray JS, Birnkrant DB. 2009. Use of ribavirin to treat influenza. N. Engl. J. Med. 361:1713–1714. 10.1056/NEJMc0905290 [DOI] [PubMed] [Google Scholar]
- 32. Zink MC, Uhrlaub J, DeWitt J, Voelker T, Bullock B, Mankowski J, Tarwater P, Clements J, Barber S. 2005. Neuroprotective and anti-human immunodeficiency virus activity of minocycline. JAMA 293:2003–2011. 10.1001/jama.293.16.2003 [DOI] [PubMed] [Google Scholar]
- 33. Michaelis M, Kleinschmidt MC, Doerr HW, Cinatl J., Jr. 2007. Minocycline inhibits West Nile virus replication and apoptosis in human neuronal cells. J. Antimicrob. Chemother. 60:981–986. 10.1093/jac/dkm307 [DOI] [PubMed] [Google Scholar]
- 34. Mishra MK, Basu A. 2008. Minocycline neuroprotects, reduces microglial activation, inhibits caspase 3 induction, and viral replication following Japanese encephalitis. J. Neurochem. 105:1582–1595. 10.1111/j.1471-4159.2008.05238.x [DOI] [PubMed] [Google Scholar]
- 35. Brun-Buisson C, Richard JC, Mercat A, Thiébaut AC, Brochard L, REVA SRLF, A/H1N1v 2009 Registry Group 2011. Early corticosteroids in severe influenza A/H1N1 pneumonia and acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 183:1200–1206. 10.1164/rccm.201101-0135OC [DOI] [PubMed] [Google Scholar]
- 36. Yan Y, Zou Z, Sun Y, Li X, Xu KF, Wei Y, Jin N, Jiang C. 2013. Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model. Cell Res. 23:300–302. 10.1038/cr.2012.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chan RW, Yuen KM, Yu WC, Ho CC, Nicholls JM, Peiris JS, Chan MC. 2010. Influenza H5N1 and H1N1 virus replication and innate immune responses in bronchial epithelial cells are influenced by the state of differentiation. PLoS One 5:e8713. 10.1371/journal.pone.0008713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Josset L, Tisoncik-Go J, Katze MG. 2013. Moving H5N1 studies into the era of systems biology. Virus Res. 178:151–167. 10.1016/j.virusres.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McDermott JE, Shankaran H, Eisfeld AJ, Belisle SE, Neuman G, Li C, McWeeney S, Sabourin C, Kawaoka Y, Katze MG, Waters KM. 2011. Conserved host response to highly pathogenic avian influenza virus infection in human cell culture, mouse and macaque model systems. BMC Syst. Biol. 5:190. 10.1186/1752-0509-5-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stewart CE, Torr EE, Mohd Jamili NH, Bosquillon C, Sayers I. 2012. Evaluation of differentiated human bronchial epithelial cell culture systems for asthma research. J. Allergy (Cairo) 2012:943982. 10.1155/2012/943982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li C, Bankhead A, III, Eisfeld AJ, Hatta Y, Jeng S, Chang JH, Aicher LD, Proll S, Ellis AL, Law GL, Waters KM, Neumann G, Katze MG, McWeeney S, Kawaoka Y. 2011. Host regulatory network response to infection with highly pathogenic H5N1 avian influenza virus. J. Virol. 85:10955–10967. 10.1128/JVI.05792-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smyth GK. 2005. Limma: linear models for microarray data, p 397-320 In Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S. (ed), Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer, New York, NY. [Google Scholar]
- 43. Langfelder P, Zhang B, Horvath S. 2008. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24:719–720. 10.1093/bioinformatics/btm563 [DOI] [PubMed] [Google Scholar]
- 44. Venables WN, Ripley BD. 2002. Modern applied statistics with S. Springer, New York, NY. [Google Scholar]
- 45. Lamb J. 2007. The Connectivity Map: a new tool for biomedical research. Nat. Rev. Cancer 7:54–60. 10.1038/nrc2044 [DOI] [PubMed] [Google Scholar]
- 46. Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. 2009. BioMart—biological queries made easy. BMC Genomics 10:22. 10.1186/1471-2164-10-S3-S22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. 2012. A travel guide to cytoscape plugins. Nat. Methods 9:1069–1076. 10.1038/nmeth.2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Top 5 pathways enriched in each cluster. Download
Top 5 GO biological processes (BPs) enriched in each cluster. Download
Top 5 regulators enriched in each cluster. Download
Temporal gene expression changes for genes belonging to the antigen presentation pathway (A), cytokines (B), or eicosanoid signaling (C). (A) Twelve genes belonging to the antigen presentation pathway specifically downregulated after H7N9 infection (cluster 14) or upregulated after infection with H3N2 only (genes from cluster 8). (B) Cytokines from cluster 1 or 2. (C) Genes from cluster 7 belonging to the eicosanoid pathway. Download
Heat map of gene expression levels for genes belonging to the chromatin modification biological process (GO:0016568). Download
Calu-3 cell viability following incubation with increasing concentrations of minocycline. Download
Literature mining for in vitro and/or in vivo assays of the 26 predicted drugs for IAV infection