ABSTRACT
Toxoplasma gondii is a human obligate intracellular parasite that has infected over 20% of the world population and has a vast intermediate host range compared to those of its nearest relatives Neospora caninum and Hammondia hammondi. While these 3 species have highly syntenic genomes (80 to 99%), in this study we examined and compared species-specific structural variations, specifically at loci that have undergone local (i.e., tandem) duplication and expansion. To do so, we used genomic sequence coverage analysis to identify and curate T. gondii and N. caninum loci that have undergone duplication and expansion (expanded loci [ELs]). The 53 T. gondii ELs are significantly enriched for genes with predicted signal sequences and single-exon genes and genes that are developmentally regulated at the transcriptional level. We validated 24 T. gondii ELs using comparative genomic hybridization; these data suggested significant copy number variation at these loci. High-molecular-weight Southern blotting for 3 T. gondii ELs revealed that copy number varies across T. gondii lineages and also between members of the same clonal lineage. Using similar methods, we identified 64 N. caninum ELs which were significantly enriched genes belonging to the SAG-related surface (SRS) antigen family. Moreover, there is significantly less overlap (30%) between the expanded gene sets in T. gondii and N. caninum than would be predicted by overall genomic synteny (81%). Consistent with this finding, only 59% of queried T. gondii ELs are similarly duplicated/expanded in H. hammondi despite over 99% genomic synteny between these species.
IMPORTANCE
Gene duplication, expansion, and diversification are a basis for phenotypic differences both within and between species. This study represents the first characterization of both the extent and degree of overlap in gene duplication and locus expansion across multiple apicomplexan parasite species. The most important finding of this study is that the locus duplications/expansions are quantitatively and qualitatively distinct, despite the high degree of genetic relatedness between the species. Given that these differential expansions are prominent species-specific genetic differences, they may also contribute to some of the more striking phenotypic differences between these species. More broadly, this work is important in providing further support for the idea that postspeciation selection events may have a dramatic impact on locus structure and copy number that overshadows selection on single-copy genes.
INTRODUCTION
Toxoplasma gondii is a category B biodefense pathogen that can be lethal in utero and in immunocompromised humans. This parasite is a candidate bioterrorism agent due to the extreme environmental stability of infective oocysts that could contaminate water or food supplies (1, 2). While infections in healthy humans are often benign, the identification of distinct Toxoplasma genotypes that are lethal in healthy adults (3) has changed the view of the bioterrorism potential of this pathogen and added to the urgency for the development of new chemotherapeutics and vaccines. Toxoplasma is unique among apicomplexans in its ability to infect, be transmitted by, and cause disease in all warm-blooded animals, a trait which has certainly contributed to its worldwide distribution (4). The genetic bases for this trait are unknown but are likely to be important given the clear link between host range expansion and increased virulence in pathogens (most clearly demonstrated in influenza virus [5]).
With the advent of whole-genome tiling arrays and, most importantly, next-generation sequencing technologies, it is now possible to examine structural differences in whole genomes both within and between closely related species. In humans, locus expansion and diversification have been linked to psychiatric disorders such as autism and schizophrenia (reviewed in reference 6) and to susceptibility to a variety of other diseases (reviewed in reference 7). Locus expansion can also be beneficial. In mammals, expansion and diversification of killer-cell immunoglobulin-like receptor genes are important for recognition of diverse pathogens (8). Laboratory studies with bacteria show that adaptation to selective conditions via gene expansion occurs much more frequently than that via point mutation (9), and in the field, copy number increases drive drug resistance in Drosophila melanogaster (10). Phenotypic impact can be driven by gene dosage, but gene duplication also allows the original copy to maintain its function while duplicated copies are free to change via mutation and selection (11).
Expanded and diversified gene families play important roles in pathogen virulence. Gene family expansions have been linked to virulence in Candida spp. (12) and Rickettsia spp. (13). The var family of genes is distributed throughout the Plasmodium falciparum genome and encodes erythrocyte membrane antigens (PfEMPs) that are under strong diversifying selection (14). Expanded genes have been linked to virulence, immune evasion (14), drug resistance (15), and host range (16) in Plasmodium spp.
Our recent work demonstrates a role for gene duplication and subsequent diversification in Toxoplasma host-pathogen interactions. The T. gondii ROP5 locus contains up to 10 copies depending on the strain, and this locus is essential for parasite virulence (17). Importantly, distinct isoforms from the ROP5 locus can have synergistic effects on parasite lethality, indicating that individual copies of the ROP5 gene have evolved subtly distinct functions. The ROP5 locus also exhibits isolate-specific copy number variation (CNV) (17). The surface antigen-1-related (SRS) and rhoptry protein 2 (ROP2) superfamilies have duplicated multiple times, and tandemly duplicated clusters of genes belonging to this family can be found throughout the genome (18). The SRS family has been implicated in immune evasion (19), and the single-copy ROP2 superfamily member ROP18 is a potent virulence factor in mice (20, 21). In T. gondii, expanded loci have impacts in other infection contexts, including the ROP4/7 locus, which has no dramatic impact on growth in vitro but affects cyst formation in vivo (23).
Less is known about CNV between species, although as early as 2007 it was postulated that differentially duplicated genes and genomic structural variations could contribute to phenotypic differences between chimps and humans and possibly have played a role in their speciation (24). Some support for this hypothesis is found in plants, where species-specific CNV is known to contribute in certain cases to reproductive isolation (25).
In this study, we used a genome-wide approach to compare the extents of locus expansion across the genomes of T. gondii, Hammondia hammondi, and Neospora caninum. These three species belong to the subfamily Toxoplasmatinae (26), and their genomes have been sequenced, revealing a high level of synteny (27, 28). T. gondii and N. caninum have distinct intermediate host ranges and different definitive hosts (felines and canines, respectively), while T. gondii and H. hammondi share the same definitive host (29). While T. gondii has a vast host range that includes birds and is virulent in mice (30), H. hammondi and N. caninum cannot infect birds and are avirulent in mice (31, 32). For three T. gondii strains (GT1, ME49B7, and VEG) and one N. caninum strain (NCLIV [27]), we used a manual pipeline to identify and curate all potentially expanded loci and to compare the degrees of overlap between them. This was facilitated by the fact that these genomes have been annotated. The H. hammondi reference genome assembly (GenBank accession no. AVCM00000000) has not yet been fully assembled into chromosomes or annotated (28). However, for a subset of expanded loci we were able to determine if they were similarly expanded in H. hammondi. Overall, we find that in contrast to the high level of synteny across these 3 species, there was a significant lack of overlap in their expanded loci. This suggests an important role for gene expansion in the evolution of these species since their divergence.
RESULTS
Fifty-three loci have increased sequence coverage in T. gondii.
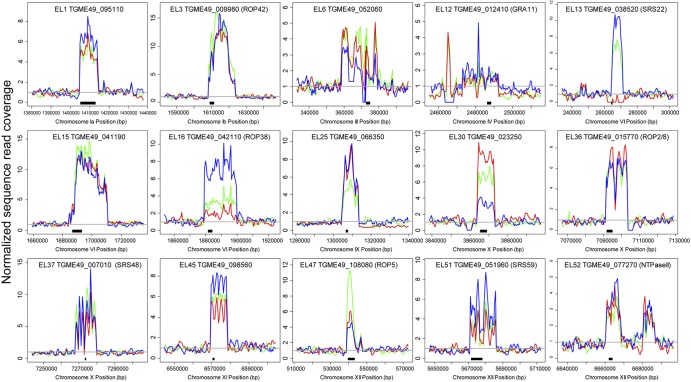
We used a manual identification and curation pipeline to identify putatively expanded loci using sequence read coverage in T. gondii (Fig. 1) and identified 53 loci of high sequence complexity in the nuclear genome of T. gondii. Average sequence coverage across the entirety of the 3 currently available Sanger-sequenced genomes differed slightly (median values, 15×, 19×, and 14× for GT1, ME49, and VEG strains, respectively) due to different numbers of raw sequenced reads (see Table S1 in the supplemental material), with 95 to 98% of the raw reads mapping to the ME49B7 genomic assembly using BLAT (33). Normalized sequence coverage across entire chromosomes from all three queried T. gondii strains was typically homogeneous, with sporadic patches of increased coverage at certain locations and telomeric sites (Fig. 2A), indicating that gene duplication/expansion was relatively infrequent. We examined gene expansion at all loci across GT1, ME49, and VEG to estimate copy number across the three strains (as in Fig. 2B). Of the 53 expanded loci, only 1 (expanded locus 13, EL13; Fig. 3 and Table 1) appeared to be entirely missing in one of the three T. gondii strains, in this case the type I strain (GT1). Otherwise, the remaining 52 loci were conserved in their expanded state in all 3 queried T. gondii strains (see Table S2). However, based on sequence coverage analysis, 22 loci exhibited CNVs of ≥3 copies across the 3 queried T. gondii strains. This list included the ROP5 locus (EL47, Fig. 3 and Table 1), which, based on sequence coverage, has ~11 copies in ME49B7 and 6 and 4 copies in GT1 and VEG, respectively. This is similar to previously published analyses using high-molecular-weight Southern blotting of the ROP5 locus in T. gondii strains RH (type I), ME49 (type II), and CTG (type III) (17).
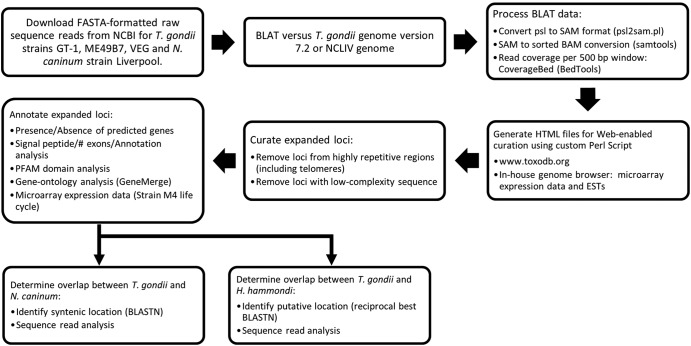
FIG 1 .
Flow chart depicting the pipeline used to identify, curate, and annotate expanded loci in Toxoplasma gondii, Neospora caninum, and Hammondia hammondi. ESTs, expressed sequence tags.
FIG 2 .
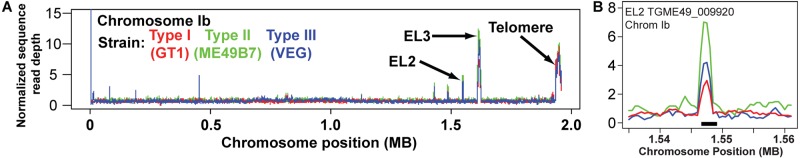
(A) Normalized sequence coverage plot of chromosome Ib for three T. gondii strains (GT1, ME49B7, and VEG). Coverage data for each 500-bp window were normalized to the average coverage for the entire chromosome for each strain. The locations of EL2 and EL3 (the expanded loci identified on chromosome Ib) and the predicted right arm telomere are indicated. (B) Detailed view of normalized sequence read coverage for EL2 for each of the three T. gondii strains, showing potential variation in copy number indicated by the read plot. Strains are color coded as in panel A.
FIG 3 .
Sequence coverage plot for 15 expanded loci in three strain types of T. gondii. Data are normalized to the read coverage in the leftmost 20 kb flanking the expanded locus, and the gray line indicates normalized sequence coverage of 1. Black bars beneath each plot indicate the location of one of the predicted T. gondii isoforms. Information for each locus can be found in Table 1.
TABLE 1 .
Properties of 18 expanded loci in T. gondii, N. caninum, and H. hammondie
| Locus | Chr. | Pos. (MB) |
Gene | Annotation |
T. gondii copy no. by straina |
No. of predicted T. gondii orthologs by strainb |
N. caninumc |
H. hammondi copy no.d |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | I | II | III | Chr. | Pos. (MB) |
Copy no. |
||||||
| EL1 | Ia | 1.41 | 095110 | ROP4/7 | 5 | 5 | 7 | 1 | 1 | 2 | Ia | 1.79 | 3 | 7–8 |
| EL3 | Ib | 1.61 | 009980 | ROP42 | 13 | 13 | 13 | 2 | 1 | 2 | Ib | 1.54 | 1 | ND |
| EL6 | III | 0.36 | 052060 | KRUF family | 4 | 4 | 4 | 2 | 1 | 1 | NA | NA | 0 | 1 |
| EL12 | IV | 2.48 | 012410 | GRA11 | 1 | 1 | 2 | 0 | 2 | 1 | IV | 2.05 | 1 | 1 |
| EL13 | VI | 0.26 | 038520 | SRS22G | 0 | 6 | 11 | 4 | 1 | 1 | VI | 0.25 | 1 | ND |
| EL15 | VI | 1.68 | 041190 | Hypothetical | 11 | 11 | 11 | 5 | 3 | 3 | VI | 1.47 | 2 | 1 |
| EL16 | VI | 1.88 | 042110 | ROP38 | 2 | 3 | 8 | 3 | 2 | 1 | VI | 1.65 | 18 | 2 |
| EL22 | VIIb | 2.79 | 059410 | SRS26A | 1 | 2 | 1 | 3 | 1 | 3 | VIIb | 2.67 | 1 | ND |
| EL23 | VIII | 6.74 | 000240 | MIC17 | 3 | 2 | 3 | 2 | 1 | 1 | VIII | 6.49 | 2 | 7–8 |
| EL25 | IX | 1.3 | 066350 | Hypothetical | 6 | 4 | 6 | 1 | 1 | 1 | NA | NA | 0 | ND |
| EL30 | X | 3.86 | 023250 | Hypothetical | 9 | 6 | 4 | 0 | 1 | 1 | X | 3.69 | 1 | ND |
| EL36 | X | 7.09 | 015770 | ROP2/8 | 6 | 6 | 6 | 1 | 1 | 2 | NA | NA | 0 | ND |
| EL37 | X | 7.27 | 007010 | SRS48 | 5 | 5 | 5 | 0 | 1 | 1 | X | 6.75 | 2 | ND |
| EL45 | XI | 6.57 | 098560 | Hypothetical | 4 | 6 | 7 | 2 | 1 | 3 | XI | 6.08 | 8 | 6–7 |
| EL47 | XII | 0.54 | 108080 | ROP5 | 3 | 14 | 4 | 1 | 1 | 1 | XII | 0.35 | 2 | 8–9 |
| EL51 | XII | 5.67 | 051960 | SRS59K | 3 | 3 | 7 | 1 | 1 | 1 | XII | 5.32 | 1 | 2–3 |
| EL52 | XII | 6.66 | 077270 | NTPase II | 3 | 3 | 4 | 0 | 1 | 1 | XII | 6.24 | 1 | 2–3 |
| EL53 | XII | 6.68 | 077240 | NTPase I | 2 | 2 | 2 | 1 | 1 | 1 | XII | 6.29 | 1 | 4–5 |
Determined using raw sequence read coverage. Type I, GT1; type II, ME49; type III, VEG.
Gene identifications are based on Toxoplasma gondii strain ME49 sequence, v7.0 (http://www.toxodb.org).
Determined using raw sequence read coverage for N. caninum Liverpool strain.
Determined using raw sequence read coverage for H. hammondi CatGER041 strain.
Abbreviations: Chr., chromosome; Pos., position; NA, not available; ND, not determined.
Expanded T. gondii loci are enriched for secretory proteins with few exons.
Of the 53 expanded loci, 42 were predicted to contain protein-coding genes (http://www.toxodb.org). In addition, one locus (EL40) that did not have a predicted gene model within it does have evidence for being a coding sequence due to the presence of expressed sequence tags that map to this locus (see Table S2 in the supplemental material). We anticipate that some of the expanded loci without an associated gene prediction will be transcribed to produce either coding or noncoding RNAs. It should be noted that, with few exceptions, the number of paralogs predicted in each of these three genome sequences greatly underestimated copy number (Table 1; see also Table S2). This was most certainly due to collapsing of the assembly in regions containing tandemly duplicated clusters of genes that are similar in sequence, as has been observed in other genomes (e.g., Homo sapiens [34] and Trichomonas vaginalis [35]).
We used existing annotations of the T. gondii genome to characterize the nature of the 42 T. gondii loci containing predicted genes. We found a significant enrichment for genes predicted to contain N-terminal signal sequences (29 out of 42 compared to the entire predicted proteome; hypergeometric distribution [HGD], P = 5.1 × 10−11 [Table 2]). In addition, these 42 genes have fewer exons (mean, 2.1; median, 1) than the rest of the predicted genes in the genome (mean, 5.2; median, 4). Kolmogorov-Smirnov (KS) analysis revealed a significant (P = 1.4 × 10−7) difference in the exon distribution between these two gene sets reflected by their cumulative distributions (see Fig. S1 in the supplemental material). In fact, 26 of the 42 expanded gene-containing loci in T. gondii have only 1 exon (a significant enrichment compared to the genome as a whole; P = 8.9 × 10−7; Table 2).
TABLE 2 .
Bioinformatic properties of duplicated/expanded loci in Toxoplasma gondii and Neospora caninum
| Property |
T. gondii |
N. caninum |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of genes |
HGD P valuea |
No. of genes |
HGD P valuea |
|||||||
| Duplicated |
All |
Duplicated |
All |
|||||||
| With property |
Total | With property |
Total | With property |
Total | With property |
Total | |||
| Signal peptideb | 29 | 42 | 1,756 | 8,103 | 5.10E−11 | 34 | 45 | 1,596 | 7,227 | 2.60E−14 |
| Single-exon genesb | 26 | 42 | 2,121 | 8,103 | 8.90E−07 | 31 | 45 | 1,514 | 7,227 | 4.70E−12 |
| Rhoptry proteinsb | 5 | 29 | 47 | 1,756 | 7.30E−04 | 2 | 34 | 30 | 1,596 | 0.11 |
| SAG-related surface antigensb | 5 | 29 | 87 | 1,756 | 0.010 | 22 | 34 | 218 | 1,596 | 4.10E−12 |
| Developmentally regulated: tachyzoite/bradyzoitec |
13 | 42 | 1,093 | 8,059 | 0.0020 | |||||
| Developmentally regulated: oocyst/sporozoitec |
10 | 42 | 1,337 | 8,059 | 0.07 | |||||
HGD, hypergeometric distribution. Values in bold are significant.
Based on version 8.2 annotation, http://toxodb.org.
Based on analysis of complete life cycle transcriptional profile of T. gondii strain M4 (38).
Expanded T. gondii loci are dominated by genes of unknown function or localization but also include predicted rhoptry proteins and surface antigens.
We examined the degree of annotation of the 42 putatively protein-coding expanded loci using ToxoDB and our own protein family searches. Of these, 29 were annotated only as hypothetical or conserved hypothetical proteins (see Table S2 in the supplemental material) and showed little similarity to previously characterized proteins from T. gondii or other eukaryotic species based on domain and BLAST analyses available on ToxoDB. We screened all 42 loci for PFAM domains (both A and B) and found 27 with a PFAM-A hit with an expect value of ≤1 (see Table S3). There were also multiple loci in this group that encode proteins with unannotated PFAM-B matches. Of those with PFAM-B hits, there were 6 PFAM-B domains that matched to multiple expanded loci, suggesting that genes with similar protein architectures have expanded into multilocus, multigene families (including EL17 and EL25 [PFAM-B 2112] and EL6 and EL50 [PFAM-B 3349]; see Table S3). Regardless, the majority of expanded T. gondii genes have yet to be characterized in terms of their function or subcellular localization and contain protein domains that have yet to be annotated and/or characterized.
Of those that were annotated, 5 were predicted rhoptry proteins, some of which have been previously characterized. These are EL1, -3, -16, -36, and -47 (ROP4/7, ROP42, ROP38, ROP2/8, and ROP5, respectively [18]; Table 1). Based on the hypergeometric distribution (HGD) (36), this was a significant enrichment in predicted rhoptry proteins over the current annotation of the genome (Table 2; P = 7.3 × 10−4), further implicating the rhoptry proteome as a target for locus expansion (18). Four expanded loci were annotated as members of the SAG1-related family of surface antigens. These are EL13, -22, -37, and -51 (SRS22, SRS26, SRS48, and SRS59, respectively [37]). One was annotated as a dense granule protein (EL12; GRA11), another as a microneme protein (EL23; MIC17), and 2 were previously characterized bradyzoite-specific nucleoside triphosphatase (NTPases) that had been determined to be found in tandem in the genome (EL52 and -53; NTPases II and I).
Expanded T. gondii loci are enriched for developmentally related genes.
We used previously published microarray expression data for T. gondii strain M4 (38) to quantitatively assess gene expression across multiple T. gondii genes during parasite development, focusing on the oocyst-to-sporocyst and tachyzoite-to-bradyzoite transitions. We found that 13 of the 42 expanded loci contained genes that were significantly up- or downregulated at the transcriptional level during the tachyzoite-to-bradyzoite transition in vitro and/or in vivo (Fig. 4). Upregulated genes included those for the rhoptry proteins ROP42 and ROP2/8, and downregulated genes included those for NTPase I and a paralog belonging to the SRS22 family. This enrichment was significant (HGD, P = 0.0019; Table 2) compared to the entire predicted transcriptome assayed by the microarray. Ten of the 42 genes were developmentally regulated during the oocyst-sporozoite transition, but this difference was not significant (HGD, P = 0.07; Table 2).
FIG 4 .
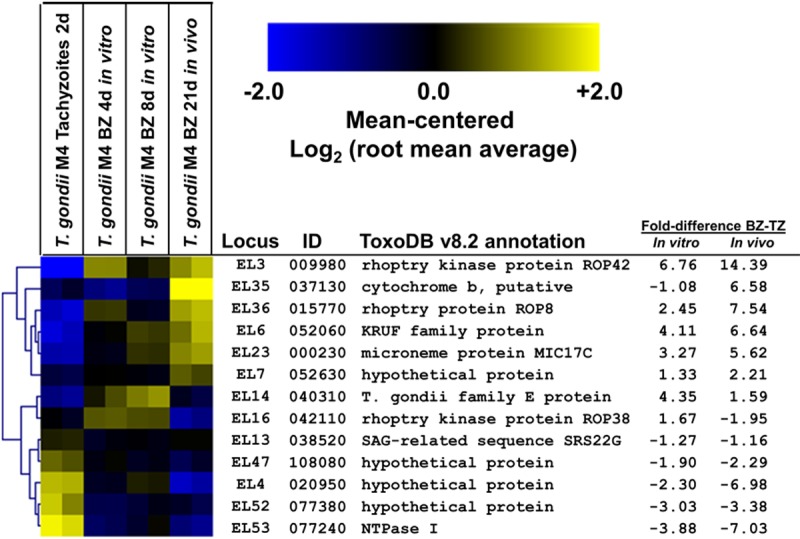
Expression profile for 13 expanded loci in tachyzoites and bradyzoites of T. gondii strain M4 (GEO database accession number GSE32427), showing an increase or decrease of transcript abundance during the tachyzoite-to-bradyzoite transition in vitro and/or in vivo.
Multiple T. gondii expanded loci exhibit within-lineage copy number variation.
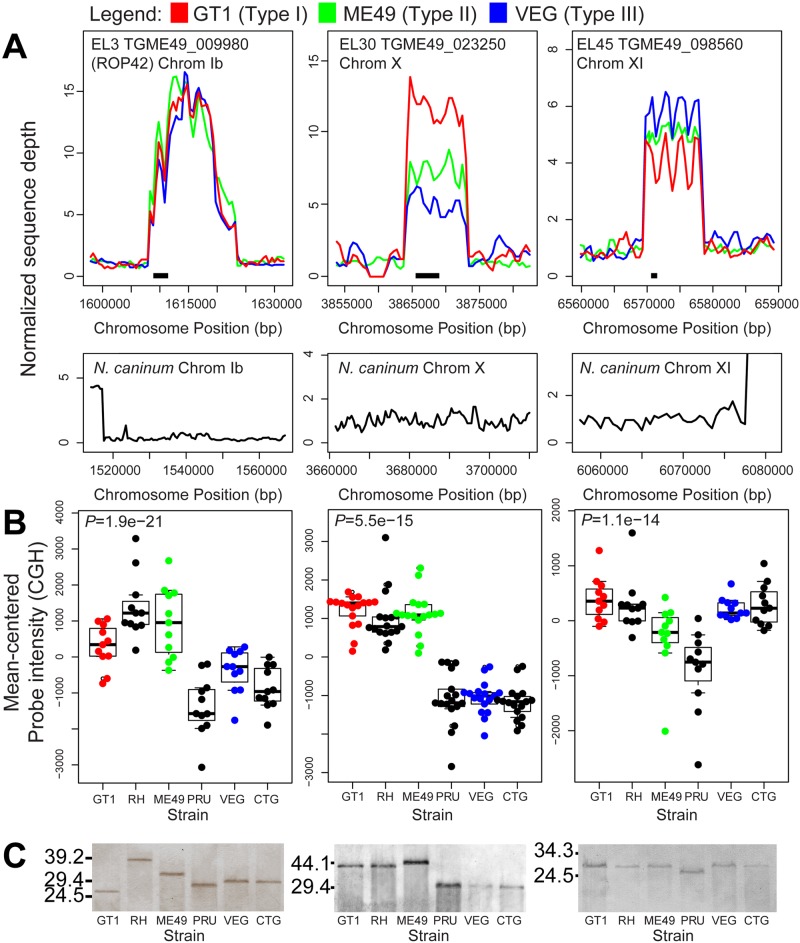
Twenty-two of the 53 expanded loci in T. gondii showed differences in sampling frequencies between the 3 representative genome strains, suggestive of copy number variation (as seen previously for T. gondii ROP5 [17]). Consistent with this, using CNV-seq to statistically assess copy number variation at all 53 loci (see Fig. S4 in the supplemental material), we identified 23 that significantly varied in copy number between GT1/VEG and ME49 (6 loci), GT1 and ME49 (12 loci), and VEG and ME49 (5 loci). This list included the ROP5 locus, for which copy number has been determined previously in multiple T. gondii strains using high-molecular-weight Southern blotting (17). We also conducted whole-genome comparative genomic hybridization (CGH) for two distinct members of each of the 3 major lineages of T. gondii. Of the 41 protein-coding expanded loci that could be surveyed by microarray, 24 had significantly higher hybridization intensity across the T. gondii strains queried (see Materials and Methods for statistical analyses). In addition to this, we observed a general correlation between copy number as estimated by sequence coverage analysis and our CGH data. For example, EL5 is predicted to have 4 to 5 copies in the type I strain GT1 and only 1 to 2 copies in type II strain ME49 and type III strain VEG, and the CGH (see Fig. S2A) and CNV-seq (see Fig. S4) analyses reflect this difference. A similar correlation can be found for EL16 (see Fig. S2A and S4). These data provide a secondary validation of locus expansion for 24 of the T. gondii loci as well as the variation in copy number observed in the sequence coverage plots. Of the remaining loci queried by the microarray, 18 had sufficient CGH data but did not show a significant increase in hybridization intensity (see Fig. S2B). We could not query 12 of the loci since they were not represented on the microarray (see Fig. S2C).
We also identified some loci with CGH intensity profiles suggesting a difference in copy number between members of the same clonal lineage, most notably EL30, for which CGH intensity values were distinct between T. gondii strains ME49 and PRU (Fig. 5B). To address this further, we performed high-molecular-weight Southern blotting for EL30 as well as EL3 and EL45 across 6 T. gondii strains using restriction enzymes predicted to cut outside the entire expanded locus. For all 3 loci, we observed differences in estimated copy number both between and within lineages. We observed intralineage variation in locus size for EL3 (ROP42) between GT1 and RH as well as between ME49 and PRU, and we estimated that GT1 and RH have 6 and 9 copies, respectively, and that ME49 and PRU have 8 and 6 copies, respectively. We estimated that VEG and CTG have 7 copies (Fig. 5C). We detected intralineage variation for EL30 and EL45, where ME49 and PRU had different-size loci (Fig. 5C). Southern blot data for these three loci are generally consistent with the CGH intensity values, although there are some exceptions. For example, for EL3 strain GT1 has a higher CGH intensity than would be predicted based on the Southern blot. This could be due to as-yet-unidentified cryptic restriction sites in the locus that are specific to strain GT1. In comparison, for the single-copy locus AMA1 we did not detect inter- or intralineage variation in sequence read coverage, CGH probe intensity, or locus size as estimated by Southern blotting (see Fig. S3).
FIG 5 .
Sequence read, comparative genomic hybridization, and Southern blot analysis for EL3, EL30, and EL45. (A) Sequence read analysis for three strains of T. gondii (top) and N. caninum Liverpool (bottom). Data for each strain and locus are normalized to the read coverage in the 20 kb flanking the expanded locus to the left. (B) Comparative genomic hybridization (CGH) across 6 T. gondii strains, 2 from each of the 3 canonical lineages. Only microarray probes with perfect matches in GT1, ME49, and VEG were used in the calculations. Boxes span the first and third quartiles and contain the median value for all useful probes. P values for significant differences in hybridization intensity compared to the single-copy gene AMA1 (gray horizontal line) are at the top of the graph, and individual values are shown in the bee swarm plots. (C) Southern blots for 6 T. gondii strains using DNA digested with restriction enzymes predicted to cut outside the repeat locus. Restriction enzymes were BspEI, BglI, and NotI, respectively. A similar blot for the single-copy gene AMA1 can be found in the supplemental material. Numbers at left are molecular masses in kilobases.
Three T. gondii expanded loci are not essential for in vitro growth.
We successfully knocked out 3 expanded loci (EL3 [ROP42], EL6, and EL23) in a virulent type I background (RHΔku80Δhxgprt [39]). Parasite lines with deletions at each of these loci (see Fig. S5 in the supplemental material) exhibited no obvious defects in in vitro growth, and neither RHΔku80:Δel3 or -Δel23 parasite clones showed any defects in acute virulence as measured by survival time in mouse infections with 100 tachyzoites. We did not test RHΔku80:Δel6 in mouse virulence assays.
N. caninum has a markedly different set of expanded genes that is enriched for members of the SAG1-related surface antigen family.
In order to compare gene expansion between T. gondii and its close relative N. caninum, we first examined gene expansion in N. caninum using approaches identical to those for T. gondii. We identified 65 expanded loci in N. caninum (Liverpool strain; http://www.toxodb.org), 45 of which contained predicted protein-coding genes. These loci are listed in Table S4 in the supplemental material. The set of N. caninum expanded genes was also enriched for genes encoding proteins with signal peptides (34/45) and single-exon genes (31/45) compared to the genome as a whole (HGD, P = 2.6 × 10−14 and 4.7 × 10−12, respectively; Table 2). Remarkably, nearly half (22/45; 49%) were found to contain a SAG PFAM domain, suggesting that they belong to the SRS family, and this is a significant enrichment over the annotated genome (P = 4.1 × 10−12; Table 2). Of the remaining 23 protein-coding expanded N. caninum loci, 3 were previously annotated (ROP4/7 and NTPases I and II), 17 had at least one recognized PFAM domain, and 4 were completely unannotated and had no recognizable PFAM domains. While the increased number of SRS family genes in N. caninum has been reported previously (27), our data indicate that these loci have also been subject to multiple rounds of tandem (i.e., local) duplication.
Distinct sets of genes are expanded in T. gondii and N. caninum.
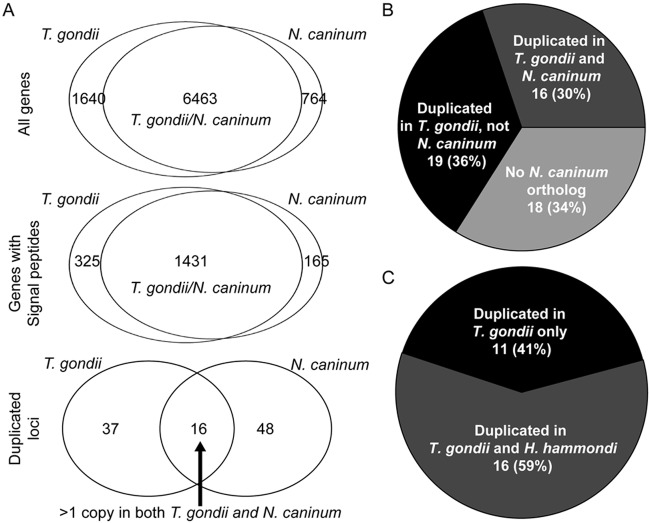
To determine the degree of overlap between genes that are expanded in T. gondii and N. caninum, we used BLASTN to identify the syntenic location for each of the 53 T. gondii loci described above and then examined that region of the genome for signatures of gene expansion in N. caninum. We found that only 16 of the 53 T. gondii loci also had evidence of expansion in N. caninum (≥2-fold-higher sequence coverage than background; Fig. 6A). This lack of overlap between T. gondii and N. caninum at these loci is in contrast to the overall gene-by-gene synteny between the T. gondii and N. caninum genomes, and this lack of overlap is significant (HGD, 16/53 versus 6,463/8,103 for T. gondii: P = 7 × 10−15; HGD, 16/64 versus 6,463/7,227 for N. caninum; Fig. 6A). One of the shared expanded loci (locus EL15; annotated in T. gondii as ROP38) had higher sequence coverage in N. caninum, while the remaining 15 had higher sequence coverage in T. gondii. Of the 37 loci that were uniquely expanded in T. gondii, 19 had syntenic orthologs in N. caninum, but these loci showed no evidence of expansion (sequence coverage was ~1×, e.g., EL3 and EL30; Fig. 5A). The remaining 18 loci did not have a syntenic ortholog based on the current T. gondii annotation based on the clusters of orthologous groups database implemented in ToxoDB (40).
FIG 6 .
(A) Overall genomic synteny between T. gondii and N. caninum predicted genes. (Top) All predicted genes; (middle) all predicted genes with predicted signal peptides; (bottom) expanded loci (53 for T. gondii and 64 for N. caninum). The top 2 Venn diagrams are based on gene-by-gene synteny, while the bottom panel for expanded loci is based on whether the locus is expanded in both N. caninum and T. gondii (16 loci) or whether it is absent or present as a single copy in one species (37 and 48 for T. gondii and N. caninum, respectively). (B) Presence and expanded state of all 53 T. gondii expanded loci compared to N. caninum. Eighteen of the 53 T. gondii loci do not have a syntenic ortholog in N. caninum, a significant enrichment compared to the entire genome. (C) Comparison of the expanded state of T. gondii loci for which a clear ortholog was identified in H. hammondi HhCatGer041. Of the 27 loci, 11 are uniquely expanded in T. gondii compared to H. hammondi, while we estimated that H. hammondi has at least 2 predicted copies for the remaining 16 loci.
T. gondii and H. hammondi share 16 of 27 expanded loci.
The H. hammondi genome has not been annotated or assembled into chromosomes, preventing a de novo analysis of gene duplication and expansion. However, we did use the recently published H. hammondi genome sequence and raw sequence reads (28) to determine which T. gondii loci were similarly expanded in H. hammondi. Of the 42 protein-coding loci, 27 had a perfect reciprocal-best-BLAST hit between T. gondii and H. hammondi. Of these 27 putative orthologous sequences, we estimated that 16 of these (59%) had more than 1 copy in H. hammondi, while data for the remaining 11 loci suggested that they were single-copy genes in H. hammondi (Fig. 6C).
DISCUSSION
Our previous work has demonstrated a clear and important role for gene duplication in the pathogenesis of Toxoplasma gondii (17); we showed that the ROP5 locus was tandemly expanded in multiple T. gondii strains and that this expansion led to diversification of individual copies within the locus. We were therefore interested in identifying other T. gondii loci that were tandemly expanded to determine (i) what features were shared among expanded genes and (ii) whether these loci were differentially expanded both within the T. gondii species and in comparison with its nearest sequenced relatives, Neospora caninum (27) and H. hammondi (28). Stretches of increased copy number were relatively rare in these genomes, and based on the currently released genome assemblies, all of the 42 protein-coding loci harbored multiple tandem duplications of the same gene.
An important finding of this work was that expanded loci exhibit copy number variation even between members of the same clonal lineage. In Europe and North America, T. gondii isolates are dominated by members of 3 main lineages (types I, II, and III), and isolates from within the same lineage appear to be clonal. However, based on Southern blot analysis we show that 3 loci exhibit CNV between members of the same clonal lineage, and other candidate loci with similar within-lineage variation can be identified from our CGH data. We do not assert that members of the same lineage are genetically identical; however, based on whole-genome comparisons, “true” members of the same clonal lineage are more genetically similar to one another than to other strains. For example, RH and GT1 strains have only 1,394 single-nucleotide polymorphisms (SNPs) that distinguish them, representing a polymorphism rate of ~0.002% (41), compared to a polymorphism rate between lineages that ranges from 1 to 5% (42). Therefore, we find that differences in copy number at these loci are in contrast to the overall genetic identity of the clonal strains, suggesting that these loci are changing more rapidly than the rest of the genome. However, we do not discount the impact of single-nucleotide polymorphisms in determining differences between members of the same strain type, which have been identified in RH and GT1 (41).
We have shown previously that the raw number of copies does not necessarily track with its impact on a particular phenotype (17). ROP5 alleles from type I and III T. gondii strains have a higher contribution to virulence than the type II alleles, yet the type II parental strain (ME49) has ~10 copies while types I (RH) and III (CTG) are estimated to harbor 6 and 4, respectively (17). In this case, the sequence of an individual copy is more important, and therefore, the presence or absence of even a single copy at an expanded locus could have a phenotypic impact. While we cannot yet demonstrate conclusively that these changes are driven by selection, the fact that individual structural changes in the genome are much more rare than individual mutations (6) certainly points to the possibility that this may be a selection-driven process. Data emerging from the Toxoplasma gondii GSCID project (https://sites.google.com/site/Toxoplasmagondiigscidproject/) will allow us to rapidly identify other expanded loci that differ between members of the same clonal lineage, since a number of these will be sequenced. We also do not know if within-lineage gene expansion occurs during asexual reproduction (which occurs in all intermediate hosts infected by T. gondii), sexual reproduction (which occurs only in felines), or both. In the wild, T. gondii is capable of self-mating (43), and these expansions could occur during genetic recombination.
We also successfully knocked out three expanded loci (EL3, EL6, and EL23) in a highly virulent T. gondii strain (RHΔKu80 [39]) and found no defects in their ability to replicate in vitro or, for 2 of these loci (EL3 and EL23), no defects in parasite virulence. To date, a number of expanded loci encoding secretory proteins such as those encoded in these loci have been deleted, including ROP2/8 (22) and ROP5 (17, 44), without any consequences for in vitro tachyzoite growth. The fact that these parasite lines show no defects in vitro or in vivo is not surprising given that all three of these loci are upregulated during the tachyzoite-to-bradyzoite transition (Fig. 4). Our data, however, show that these loci can indeed be deleted, facilitating future studies on their role during the chronic phase of infection where they are most highly expressed.
There was a statistically significant lack of overlap between tandemly duplicated loci in T. gondii and N. caninum. While it is tempting to hypothesize that these differentially expanded gene clusters may be responsible for the phenotypic differences between these species (such as differences in virulence in mouse and the different definitive hosts), our data cannot directly validate this claim. However, there is support for this hypothesis from other pathogenic species. In fungi, comparisons between species with different levels of pathogenesis in humans identified gene duplication and expansion as an important mechanism in the evolution of pathogenicity (reviewed in reference 12). For example, genomic comparisons between Candida albicans and Candida dubliniensis found that these species were most highly distinguished by the presence of two highly expanded gene families in pathogenic C. albicans compared to nonpathogenic C. dubliniensis (45), and these loci are under investigation as virulence factors. Findings from these studies also suggested that gene duplication and subsequent expansion may play a more important role than other mechanisms such as horizontal gene transfer in the evolution of novel traits in eukaryotic pathogens (12).
In both T. gondii and N. caninum, the expanded gene sets were statistically significantly enriched for genes predicted to encode secreted proteins with fewer exons compared to the genome as a whole. While secreted proteins make up the vast majority of T. gondii effectors, the significance of the increased propensity of genes with few exons to duplicate and expand is unknown. One possible explanation could be the fact that introns are free to mutate more freely and that while a single locus could be duplicated, subsequent distinct mutations in the introns of both copies may prevent further expansion of the locus during recombination or genome replication. The other, and not mutually exclusive, possibility is that genes with fewer exons are subjected to stronger selection, since all of the previously characterized Toxoplasma secretory proteins are known to play roles in pathogenesis, including the single-exon effector genes ROP18 (20, 21), ROP5 (17, 44), ROP16 (46), and GRA15 (47).
The minimal overlap between the loci that have expanded in T. gondii and N. caninum, and a similar lack of overlap between genes expanded in T. gondii compared to H. hammondi, is consistent with what has been reported for other closely related species with highly syntenic genomes (35). To our knowledge, this is the first comparative analysis of gene expansions across multiple apicomplexan species. In T. gondii, the majority of the uniquely expanded loci are of no known function based on primary sequence, while in N. caninum, the vast majority of the expanded loci are predicted to encode surface antigens belonging to the SRS superfamily. This was reported previously (27), although the phenotypic impact of this expansion is unknown.
MATERIALS AND METHODS
Sequence read alignments.
Raw sequence reads for T. gondii strains GT1, ME49, and VEG were downloaded from the NCBI trace archive in fasta format. T. gondii and N. caninum reads were aligned to reference genomes using BLAT with the following settings: -fastMap -minIdentity = 95, -minScore = 90 (33). Following conversion of the BLAT output file (psl format) to SAM format using the psl2sam.pl script within the BLAT distribution, the SAM file was converted to a sorted BAM file using Samtools (48). Sequence coverage was determined in each 500-bp window using coverageBed, distributed with BEDtools (49). Raw H. hammondi sequence reads were aligned to the assembled contigs for H. hammondi strain HhCatGer041 using bowtie2 (50), and sequence coverage was determined in 500-bp windows across each contig using Samtools mpileup (48) and a custom Perl script.
Output was uploaded in R statistical software as well as a locally run genome browser to generate whole-chromosome and locus-specific plots and to facilitate manual curation of the expanded loci. For locus-specific plots, data for each strain were normalized to the local sequence coverage for that strain of the 20 kb upstream of the putatively expanded locus. Reads for all three strains were aligned to the ME49 genomic assembly (version 7.0; ToxoDB).
Coverage analysis and curation.
Genome coverage plots were generated using the data above to construct chromosome-specific files that linked directly to ToxoDB or our own in-house genome browser. For visual inspection, peaks of coverage that were at least 3-fold above background were curated as follows. We removed loci containing highly repetitive sequence (such as di- and trinucleotide repeats). To begin determining whether the locus was tandemly expanded or if the increased sequence coverage was due to the presence of an identical (or nearly identical) gene somewhere else in the genome, we parsed the BLAT output to identify sequence reads that mapped to different chromosomes or to a location on the same chromosome that was at least 25 kb away from the putatively expanded locus. For the remaining loci, the chromosomal region was examined for the presence or absence of predicted genes using ToxoDB. For those regions containing predicted genes in T. gondii or N. caninum, we examined the current annotation of the locus in ToxoDB and our own genome browser and collected evidence for gene duplication based on the occurrence of multiple predicted genes in the same locus with high identity. For the curated expanded gene sets, we performed enrichment studies for a variety of features, including the number of predicted exons and the presence/absence of a predicted signal peptide. PFAM domain analyses on predicted proteins from each locus were run locally.
Comparative genomic hybridization.
For each of the 6 strains tested, parasites were grown in human foreskin fibroblasts (HFFs), released from host cells by needle passage, washed once by centrifugation at 800 × g, and filtered through 5.0-µm polyvinylidene difluoride (PVDF) syringe filters (Millipore). Genomic DNA (gDNA) was harvested from purified parasites using DNAzol (Invitrogen) according to the manufacturer’s protocol and treated with RNase A to remove RNA contamination. After confirmation of purity by gel electrophoresis, gDNA was sheared as follows: 1 µg of gDNA was added to 750 µl of shearing buffer (Tris-EDTA [TE], pH 8.0, and 10% glycerol) and 1 µl of 20-µg/µl glycogen in a prechilled nebulizer (Invitrogen) on ice. Nebulization was performed at 40 lb/in2 for 3 min using pressurized nitrogen. The size range of the resultant gDNA fragments was confirmed to be 200 to 600 bp by gel electrophoresis. DNA fragments were precipitated using 100% isopropanol. Biotin labeling of DNA fragments was performed using the BioPrime Array CGH genomic labeling system (Invitrogen) according to the manufacturer’s protocol. The 10× dCTP nucleotide mix was used with biotin-dCTP. Purification of labeled fragments was performed with the BioPrime purification module. For each strain, 2 µg of labeled DNA was hybridized to the Affymetrix ToxoGeneChip.
Comparative genomic hybridization data analysis.
Raw Affymetrix data files for each strain were analyzed in R statistical software using the “affy” module (http://cran.us.r-project.org). Individual probe-level data were normalized using the “constant” method, and raw intensity values were exported using the expression chip definition file for the ToxoGeneChip (51). For each probe sequence from the T. gondii Affymetrix GeneChip, the number of occurrences of that 24-bp sequence in the raw genomic sequence reads for T. gondii GT1, ME49B7, and VEG was calculated, and probes not present in a particular strain were not used in subsequent calculations. This resulted in some strains having no useful probes for a given expanded locus (e.g., EL48 [see Fig. S2B in the supplemental material]). To determine whether a locus showed significantly higher hybridization intensity, which is indicative of locus duplication/expansion, data from all 6 strains were pooled and compared to data from the single-copy gene AMA1 using a Bonferroni-corrected Student t test (52). For display purposes and visual inspection of between- and within-lineage variation in CGH hybridization intensity, values for each probe were mean centered.
Identification of loci with strain-specific copy number variation using CNV-seq.
We used CNV-seq (53) to compare sequence coverage at the 53 curated loci for all 3 T. gondii strains. In addition to default settings, we used the following parameters: –genome-size = 65,000,000; minimum-windows-required = 1. CNV-seq empirically calculated the minimum window size based on sequence coverage for each strain, which was 6,158 bp for GT1 versus ME49B7 and 6,607 bp for VEG versus ME49B7. Copy number variation was determined at a significance threshold of P < 0.0001.
High-molecular-weight Southern blotting.
Genomic DNA was isolated from 6 T. gondii strains (2 representative strains from each of the 3 clonal lineages: types I, II, and III). For each strain, 20 µg of genomic DNA was digested overnight (EL3, BspEI; EL30, BglI; EL45, NotI; and AMA1, SacI) and resolved by pulsed-field gel electrophoresis using the CHEF-DR III system (Bio-Rad; run parameters, 6.0 kV, 120°, 1.3 s for 8 h and 2.7 s for 7 h). Resolved fragments were transferred onto a nylon membrane (Bio-Rad) and probed with digoxigenin (DIG)-labeled (Roche), locus-specific probes made from PCR-generated DNA fragments per the manufacturer’s protocol. Primers used for generating locus-specific probes are listed in Table S5 in the supplemental material.
Genetic knockouts and mouse infections.
To knock out loci EL3, EL6, and EL23 in the RHΔhxgprtΔku80 strain (39), 1- to 2-kb flanking regions were amplified using primers shown in Table S5 in the supplemental material. For EL3, these were cloned sequentially into plasmid pTKO-mCherry (a gift from John Boothroyd) flanking a hypoxanthine phosphoribosyltransferase minigene (HXGPRT [54]) and transfected into RHΔhxgprtΔku80. Knockout constructs for EL6 and EL23 were created using splicing by overlap extension PCR (SOE PCR [55]) as follows: the HXGPRT minigene was amplified from pTKO-mCherry with 20- to 25-bp 5′ extensions that overlapped flanking regions from the locus. Regions flanking the expanded locus were amplified from RHΔhxgprtΔku80 genomic DNA using primers with 5′ extensions identical to those of the HXGPRT primers, except that they were reverse complemented. For each locus, individual PCRs were performed, and products were gel purified and then used in equal amounts in a second PCR. Controls consisted of reaction mixtures lacking at least one of the 3 fragments. For EL23, multiple SOE reactions were performed and ethanol was precipitated and transfected directly into RHΔhxgprtΔku80. For EL6, the SOE PCR fragment was cloned into pCR2.1-Topo and then transfected into RHΔhxgprtΔku80. Strains were selected using mycophenolic acid and xanthine medium (MPA/X) as described previously (54), and individual clones were isolated by limiting dilution. Knockouts were validated using PCR for (i) integration of the plasmid into the correct location in the genome and (ii) absence of amplification from a gene within the locus (see Fig. S5). For each knockout clone, 3 to 4 mice were infected with 100 tachyzoites (along with an empty-vector control strain) to determine the effect of the knockout on parasite lethality. Mice were monitored daily for signs of morbidity according to approved IACUC protocol 12010130.
Animal experiments.
All animal experiments and procedures met the standards of the American Veterinary Association and were approved locally under IACUC protocol 12010130.
SUPPLEMENTAL MATERIAL
Cumulative distribution plots of all genes (black line) for T. gondii (top) and N. caninum (bottom) and expanded genes (gray line) showing the enrichment for genes with fewer exons in the expanded gene sets for both species. Download
Sequence read plots for all 53 expanded loci. Plots are shown for three strains of T. gondii and one strain (NCLIV) of Neospora caninum (middle panel). All read plot data were normalized for each strain/species to the average read coverage in the 20 kb flanking the expanded locus to the left. Strains are color coded as red, green, and blue (GT1, ME49, and VEG) or black in the read and CGH plots. Black bars in the N. caninum plots indicate the location of the syntenic region as identified by BLASTN. (A) Sequence read plots for loci containing predicted genes that were queried on the T. gondii microarray that had significantly higher (P ≤ 0.05) CGH intensity than did the single-copy gene AMA1. In the CGH plots, box plots indicate the first and third quartiles and contain the median, and data for all useful probes for that gene are shown in the bee swarm overlay. (B) Sequence read plots for 18 loci containing predicted genes that were queried on the T. gondii microarray that did not have significantly higher (P ≤ 0.05) CGH intensity than did the single-copy gene AMA1. (C) Sequence read plots for 12 loci without CGH data due to the lack of a predicted gene product in the genome annotation encompassed by the T. gondii Affymetrix microarray. Download
Sequence coverage, CGH, and Southern blot analysis of the single-copy AMA1 locus in T. gondii and N. caninum. (A and B) Sequence coverage analysis in T. gondii (A) and N. caninum (B) for the AMA1 locus. (C and D) Comparative genomic hybridization data for the AMA1 locus. Both raw (C) and mean-centered (D) hybridization intensity data are shown. (E) Southern blot of genomic DNA from 6 T. gondii strains digested with ScaI, separated using pulsed-field gel electrophoresis, and probed with AMA1-specific probes. Download
CNV in T. gondii GT1 and VEG compared to ME49B7 as determined by CNV-seq. Loci with statistically significant evidence for CNV based on sequence coverage analysis are shown. Arrows after each locus name indicate whether the locus had increased or decreased copy number compared to ME49B7. For the overlapping genes, the arrows indicate increased/decreased copy number for GT1 and VEG compared to ME49B7, respectively. Download
(A) Schematic of primer locations to validate homologous recombination on both the left (a + a′) and right (b + b′) flank of the locus and to validate the deletion of the gene (c + c′). (B to D) Validation of knockouts in EL3 (B), EL6 (C), and EL23 (D). Download
Mapping statistics of raw reads for T. gondii and N. caninum. Data are shown for 3 T. gondii strains and N. caninum (Liverpool strain).
Detailed information on 53 T. gondii expanded loci. The table includes current gene annotations, the number of copies predicted based on sequence coverage plot by strain, and the orthologs and their gene names that are predicted for each locus by strain, as well as the number of predicted copies in N. caninum and H. hammondi. ND, not determined; no hits, no syntenic hit for the T. gondii locus in the N. caninum genome.
PFAM analysis of expanded loci in T. gondii
Annotation and PFAM domain characterization of identified expanded loci in N. caninum
PCR primers used in the present study.
ACKNOWLEDGMENTS
We acknowledge Vern Carruthers for providing the RHΔhxgprtΔku80 strain.
This work was supported by NIH scholar development award K22 AI080977 and a Pew Scholarship in the Biomedical Sciences to J.P.B. and University of Pittsburgh Biological Sciences summer and academic year fellowships funded by the Howard Hughes Medical Institute to H.E.W. and A.L.B.
Footnotes
Citation Adomako-Ankomah Y, Wier GM, Borges AL, Wand HE, Boyle JP. 2014. Differential locus expansion distinguishes Toxoplasmatinae species and closely related strains of Toxoplasma gondii. mBio 5(1):e01003-13. doi:10.1128/mBio.01003-13.
REFERENCES
- 1. Lélu M, Villena I, Dardé ML, Aubert D, Geers R, Dupuis E, Marnef F, Poulle ML, Gotteland C, Dumètre A, Gilot-Fromont E. 2012. Quantitative estimation of the viability of Toxoplasma gondii oocysts in soil. Appl. Environ. Microbiol. 78:5127–5132. 10.1128/AEM.00246-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Moura L, Bahia-Oliveira LM, Wada MY, Jones JL, Tuboi SH, Carmo EH, Ramalho WM, Camargo NJ, Trevisan R, Graça RM, da Silva AJ, Moura I, Dubey JP, Garrett DO. 2006. Waterborne toxoplasmosis, Brazil, from field to gene. Emerg. Infect. Dis. 12:326–329. 10.3201/eid1202.041115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dardé ML. 2008. Toxoplasma gondii, “new” genotypes and virulence. Parasite 15:366–371. 10.1051/parasite/2008153366 [DOI] [PubMed] [Google Scholar]
- 4. Dubey JP, Graham DH, Dahl E, Hilali M, El-Ghaysh A, Sreekumar C, Kwok OC, Shen SK, Lehmann T. 2003. Isolation and molecular characterization of Toxoplasma gondii from chickens and ducks from Egypt. Vet. Parasitol. 114:89–95. 10.1016/S0304-4017(03)00133-X [DOI] [PubMed] [Google Scholar]
- 5. Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. 2009. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325:484–487. 10.1126/science.1177238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Malhotra D, Sebat J. 2012. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell 148:1223–1241. 10.1016/j.cell.2012.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Almal SH, Padh H. 2012. Implications of gene copy-number variation in health and diseases. J. Hum. Genet. 57:6–13. 10.1038/jhg.2011.108 [DOI] [PubMed] [Google Scholar]
- 8. Parham P, Norman PJ, Abi-Rached L, Guethlein LA. 2011. Variable NK cell receptors exemplified by human KIR3DL1/S1. J. Immunol. 187:11–19. 10.4049/jimmunol.0902332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kugelberg E, Kofoid E, Andersson DI, Lu Y, Mellor J, Roth FP, Roth JR. 2010. The tandem inversion duplication in Salmonella enterica: selection drives unstable precursors to final mutation types. Genetics 185:65–80. 10.1534/genetics.110.114074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmidt JM, Good RT, Appleton B, Sherrard J, Raymant GC, Bogwitz MR, Martin J, Daborn PJ, Goddard ME, Batterham P, Robin C. 2010. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 6:e1000998. 10.1371/journal.pgen.1000998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bergthorsson U, Andersson DI, Roth JR. 2007. Ohno’s dilemma: evolution of new genes under continuous selection. Proc. Natl. Acad. Sci. U. S. A. 104:17004–17009. 10.1073/pnas.0707158104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moran GP, Coleman DC, Sullivan DJ. 2011. Comparative genomics and the evolution of pathogenicity in human pathogenic fungi. Eukaryot. Cell 10:34–42. 10.1128/EC.00242-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ogata H, Renesto P, Audic S, Robert C, Blanc G, Fournier PE, Parinello H, Claverie JM, Raoult D. 2005. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 3:e248. 10.1371/journal.pbio.0030248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pasternak ND, Dzikowski R. 2009. PfEMP1: an antigen that plays a key role in the pathogenicity and immune evasion of the malaria parasite Plasmodium falciparum. Int. J. Biochem. Cell Biol. 41:1463–1466. 10.1016/j.biocel.2008.12.012 [DOI] [PubMed] [Google Scholar]
- 15. Rottmann M, McNamara C, Yeung BK, Lee MC, Zou B, Russell B, Seitz P, Plouffe DM, Dharia NV, Tan J, Cohen SB, Spencer KR, González-Páez GE, Lakshminarayana SB, Goh A, Suwanarusk R, Jegla T, Schmitt EK, Beck HP, Brun R, Nosten F, Renia L, Dartois V, Keller TH, Fidock DA, Winzeler EA, Diagana TT. 2010. Spiroindolones, a potent compound class for the treatment of malaria. Science 329:1175–1180. 10.1126/science.1193225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hayton K, Gaur D, Liu A, Takahashi J, Henschen B, Singh S, Lambert L, Furuya T, Bouttenot R, Doll M, Nawaz F, Mu J, Jiang L, Miller LH, Wellems TE. 2008. Erythrocyte binding protein PfRH5 polymorphisms determine species-specific pathways of Plasmodium falciparum invasion. Cell Host Microbe 4:40–51. 10.1016/j.chom.2008.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reese ML, Zeiner GM, Saeij JP, Boothroyd JC, Boyle JP. 2011. Polymorphic family of injected pseudokinases is paramount in Toxoplasma virulence. Proc. Natl. Acad. Sci. U. S. A. 108:9625–9630. 10.1073/pnas.1015980108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boothroyd JC, Dubremetz JF. 2008. Kiss and spit: the dual roles of Toxoplasma rhoptries. Nat. Rev. Microbiol. 6:79–88. 10.1038/nrmicro1800 [DOI] [PubMed] [Google Scholar]
- 19. Kim SK, Karasov A, Boothroyd JC. 2007. Bradyzoite-specific surface antigen SRS9 plays a role in maintaining Toxoplasma persistence in the brain and in host control of parasite replication in the intestine. Infect. Immun. 75:1626–1634. 10.1128/IAI.01862-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saeij JP, Boyle JP, Coller S, Taylor S, Sibley LD, Brooke-Powell ET, Ajioka JW, Boothroyd JC. 2006. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science 314:1780–1783. 10.1126/science.1133690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Taylor S, Barragan A, Su C, Fux B, Fentress SJ, Tang K, Beatty WL, Hajj HE, Jerome M, Behnke MS, White M, Wootton JC, Sibley LD. 2006. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science 314:1776–1780. 10.1126/science.1133643 [DOI] [PubMed] [Google Scholar]
- 22. Pernas L, Boothroyd JC. 2010. Association of host mitochondria with the parasitophorous vacuole during Toxoplasma infection is not dependent on rhoptry proteins ROP2/8. Int. J. Parasitol. 40:1367–1371. 10.1016/j.ijpara.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fox BA, Falla A, Rommereim LM, Tomita T, Gigley JP, Mercier C, Cesbron-Delauw MF, Weiss LM, Bzik DJ. 2011. Type II Toxoplasma gondii KU80 knockout strains enable functional analysis of genes required for cyst development and latent infection. Eukaryot. Cell 10:1193–1206. 10.1128/EC.00297-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kehrer-Sawatzki H, Cooper DN. 2007. Structural divergence between the human and chimpanzee genomes. Hum. Genet. 120:759–778. 10.1007/s00439-006-0270-6 [DOI] [PubMed] [Google Scholar]
- 25. Rieseberg LH, Blackman BK. 2010. Speciation genes in plants. Ann. Bot. 106:439–455. 10.1093/aob/mcq126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mugridge NB, Morrison DA, Jäkel T, Heckeroth AR, Tenter AM, Johnson AM. 2000. Effects of sequence alignment and structural domains of ribosomal DNA on phylogeny reconstruction for the protozoan family Sarcocystidae. Mol. Biol. Evol. 17:1842–1853. 10.1093/oxfordjournals.molbev.a026285 [DOI] [PubMed] [Google Scholar]
- 27. Reid AJ, Vermont SJ, Cotton JA, Harris D, Hill-Cawthorne GA, Könen-Waisman S, Latham SM, Mourier T, Norton R, Quail MA, Sanders M, Shanmugam D, Sohal A, Wasmuth JD, Brunk B, Grigg ME, Howard JC, Parkinson J, Roos DS, Trees AJ, Berriman M, Pain A, Wastling JM. 2012. Comparative genomics of the apicomplexan parasites Toxoplasma gondii and Neospora caninum: coccidia differing in host range and transmission strategy. PLoS Pathog. 8:e1002567. 10.1371/journal.ppat.1002567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walzer KA, Adomako-Ankomah Y, Dam RA, Herrmann DC, Schares G, Dubey JP, Boyle JP. 2013. Hammondia hammondi, an avirulent relative of Toxoplasma gondii, has functional orthologs of known T. gondii virulence genes. Proc. Natl. Acad. Sci. U. S. A. 110:7446–7451. 10.1073/pnas.1304322110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frenkel JK, Dubey JP. 1975. Hammondia hammondi: a new coccidium of cats producing cysts in muscle of other mammals. Science 189:222–224. 10.1126/science.806116 [DOI] [PubMed] [Google Scholar]
- 30. Saeij JP, Boyle JP, Boothroyd JC. 2005. Differences among the three major strains of Toxoplasma gondii and their specific interactions with the infected host. Trends Parasitol. 21:476–481. 10.1016/j.pt.2005.08.001 [DOI] [PubMed] [Google Scholar]
- 31. Collantes-Fernandez E, Arrighi RB, Alvarez-García G, Weidner JM, Regidor-Cerrillo J, Boothroyd JC, Ortega-Mora LM, Barragan A. 2012. Infected dendritic cells facilitate systemic dissemination and transplacental passage of the obligate intracellular parasite Neospora caninum in mice. PLoS One 7:e32123. 10.1371/journal.pone.0032123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dubey JP, Sreekumar C. 2003. Redescription of Hammondia hammondi and its differentiation from Toxoplasma gondii. Int. J. Parasitol. 33:1437–1453. 10.1016/S0020-7519(03)00141-3 [DOI] [PubMed] [Google Scholar]
- 33. Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–664. 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, 1000 Genomes Project. Eichler EE. 2010. Diversity of human copy number variation and multicopy genes. Science 330:641–646. 10.1126/science.1197005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, Sicheritz-Ponten T, Noel CJ, Dacks JB, Foster PG, Simillion C, Van de Peer Y, Miranda-Saavedra D, Barton GJ, Westrop GD, Müller S, Dessi D, Fiori PL, Ren Q, Paulsen I, Zhang H, Bastida-Corcuera FD, Simoes-Barbosa A, Brown MT, Hayes RD, Mukherjee M, Okumura CY, Schneider R, Smith AJ, Vanacova S, Villalvazo M, Haas BJ, Pertea M, Feldblyum TV, Utterback TR, Shu CL, Osoegawa K, de Jong PJ, Hrdy I, Horvathova L, Zubacova Z, Dolezal P, Malik SB, Logsdon JM, Henze K, Gupta A, Wang CC, Dunne RL, Upcroft JA, Upcroft P, White O, Salzberg SL, Tang P, Chiu CH, Lee YS, Embley TM, Coombs GH, Mottram JC, Tachezy J, Fraser-Liggett CM, Johnson PJ. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315:207–212. 10.1126/science.1132894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sokal RR, Rohlf FJ. 2012. Biometry: the principles and practice of statistics in biological research, 4th ed. WH Freeman & Co, New York, NY. [Google Scholar]
- 37. Wasmuth JD, Pszenny V, Haile S, Jansen EM, Gast AT, Sher A, Boyle JP, Boulanger MJ, Parkinson J, Grigg ME. 2012. Integrated bioinformatic and targeted deletion analyses of the SRS gene superfamily identify SRS29C as a negative regulator of Toxoplasma virulence. mBio 3(6):e00321-12. 10.1128/mBio.00321-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fritz HM, Buchholz KR, Chen X, Durbin-Johnson B, Rocke DM, Conrad PA, Boothroyd JC. 2012. Transcriptomic analysis of Toxoplasma development reveals many novel functions and structures specific to sporozoites and oocysts. PLoS One 7:e29998. 10.1371/journal.pone.0029998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huynh MH, Carruthers VB. 2009. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot. Cell 8:530–539. 10.1128/EC.00358-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. 10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang N, Farrell A, Niedelman W, Melo M, Lu D, Julien L, Marth GT, Gubbels MJ, Saeij JP. 2013. Genetic basis for phenotypic differences between different Toxoplasma gondii type I strains. BMC Genomics 14:467. 10.1186/1471-2164-14-467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boyle JP, Rajasekar B, Saeij JP, Ajioka JW, Berriman M, Paulsen I, Roos DS, Sibley LD, White MW, Boothroyd JC. 2006. Just one cross appears capable of dramatically altering the population biology of a eukaryotic pathogen like Toxoplasma gondii. Proc. Natl. Acad. Sci. U. S. A. 103:10514–10519. 10.1073/pnas.0510319103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Frenkel JK, Dubey JP, Miller NL. 1970. Toxoplasma gondii in cats: fecal stages identified as coccidian oocysts. Science 167:893–896. 10.1126/science.167.3919.893 [DOI] [PubMed] [Google Scholar]
- 44. Behnke MS, Khan A, Wootton JC, Dubey JP, Tang K, Sibley LD. 2011. Virulence differences in Toxoplasma mediated by amplification of a family of polymorphic pseudokinases. Proc. Natl. Acad. Sci. U. S. A. 108:9631–9636. 10.1073/pnas.1015338108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jackson AP, Gamble JA, Yeomans T, Moran GP, Saunders D, Harris D, Aslett M, Barrell JF, Butler G, Citiulo F, Coleman DC, de Groot PW, Goodwin TJ, Quail MA, McQuillan J, Munro CA, Pain A, Poulter RT, Rajandream MA, Renauld H, Spiering MJ, Tivey A, Gow NA, Barrell B, Sullivan DJ, Berriman M. 2009. Comparative genomics of the fungal pathogens Candida dubliniensis and Candida albicans. Genome Res. 19:2231–2244. 10.1101/gr.097501.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saeij JP, Coller S, Boyle JP, Jerome ME, White MW, Boothroyd JC. 2007. Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature 445:324–327. 10.1038/nature05395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rosowski EE, Lu D, Julien L, Rodda L, Gaiser RA, Jensen KD, Saeij JP. 2011. Strain-specific activation of the NF-kappaB pathway by GRA15, a novel Toxoplasma gondii dense granule protein. J. Exp. Med. 208:195–212. 10.1084/jem.20100717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup 2009. The Sequence Alignment/map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bahl A, Davis PH, Behnke M, Dzierszinski F, Jagalur M, Chen F, Shanmugam D, White MW, Kulp D, Roos DS. 2010. A novel multifunctional oligonucleotide microarray for Toxoplasma gondii. BMC Genomics 11:603. 10.1186/1471-2164-11-603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dunn OJ. 1961. Multiple comparisons among means. J. Am. Stat. Assoc. 56:52–64. 10.1080/01621459.1961.10482090 [DOI] [Google Scholar]
- 53. Xie C, Tammi MT. 2009. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics 10:80. 10.1186/1471-2105-10-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Donald RG, Carter D, Ullman B, Roos DS. 1996. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J. Biol. Chem. 271:14010–14019. 10.1074/jbc.271.24.14010 [DOI] [PubMed] [Google Scholar]
- 55. Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68. 10.1016/0378-1119(89)90359-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cumulative distribution plots of all genes (black line) for T. gondii (top) and N. caninum (bottom) and expanded genes (gray line) showing the enrichment for genes with fewer exons in the expanded gene sets for both species. Download
Sequence read plots for all 53 expanded loci. Plots are shown for three strains of T. gondii and one strain (NCLIV) of Neospora caninum (middle panel). All read plot data were normalized for each strain/species to the average read coverage in the 20 kb flanking the expanded locus to the left. Strains are color coded as red, green, and blue (GT1, ME49, and VEG) or black in the read and CGH plots. Black bars in the N. caninum plots indicate the location of the syntenic region as identified by BLASTN. (A) Sequence read plots for loci containing predicted genes that were queried on the T. gondii microarray that had significantly higher (P ≤ 0.05) CGH intensity than did the single-copy gene AMA1. In the CGH plots, box plots indicate the first and third quartiles and contain the median, and data for all useful probes for that gene are shown in the bee swarm overlay. (B) Sequence read plots for 18 loci containing predicted genes that were queried on the T. gondii microarray that did not have significantly higher (P ≤ 0.05) CGH intensity than did the single-copy gene AMA1. (C) Sequence read plots for 12 loci without CGH data due to the lack of a predicted gene product in the genome annotation encompassed by the T. gondii Affymetrix microarray. Download
Sequence coverage, CGH, and Southern blot analysis of the single-copy AMA1 locus in T. gondii and N. caninum. (A and B) Sequence coverage analysis in T. gondii (A) and N. caninum (B) for the AMA1 locus. (C and D) Comparative genomic hybridization data for the AMA1 locus. Both raw (C) and mean-centered (D) hybridization intensity data are shown. (E) Southern blot of genomic DNA from 6 T. gondii strains digested with ScaI, separated using pulsed-field gel electrophoresis, and probed with AMA1-specific probes. Download
CNV in T. gondii GT1 and VEG compared to ME49B7 as determined by CNV-seq. Loci with statistically significant evidence for CNV based on sequence coverage analysis are shown. Arrows after each locus name indicate whether the locus had increased or decreased copy number compared to ME49B7. For the overlapping genes, the arrows indicate increased/decreased copy number for GT1 and VEG compared to ME49B7, respectively. Download
(A) Schematic of primer locations to validate homologous recombination on both the left (a + a′) and right (b + b′) flank of the locus and to validate the deletion of the gene (c + c′). (B to D) Validation of knockouts in EL3 (B), EL6 (C), and EL23 (D). Download
Mapping statistics of raw reads for T. gondii and N. caninum. Data are shown for 3 T. gondii strains and N. caninum (Liverpool strain).
Detailed information on 53 T. gondii expanded loci. The table includes current gene annotations, the number of copies predicted based on sequence coverage plot by strain, and the orthologs and their gene names that are predicted for each locus by strain, as well as the number of predicted copies in N. caninum and H. hammondi. ND, not determined; no hits, no syntenic hit for the T. gondii locus in the N. caninum genome.
PFAM analysis of expanded loci in T. gondii
Annotation and PFAM domain characterization of identified expanded loci in N. caninum
PCR primers used in the present study.